Abstract
We hypothesized that evolution of salivary gland secretory proteome has been important in adaptation to insectivory, the most common dietary strategy among Chiroptera. A submandibular salivary gland (SMG) transcriptome was sequenced for the little brown bat, Myotis lucifugus. The likely secretory proteome of 23 genes included seven (RETNLB, PSAP, CLU, APOE, LCN2, C3, CEL) related to M. lucifugus insectivorous diet and metabolism. Six of the secretory proteins probably are endocrine, whereas one (CEL) most likely is exocrine. The encoded proteins are associated with lipid hydrolysis, regulation of lipid metabolism, lipid transport, and insulin resistance. They are capable of processing exogenous lipids for flight metabolism while foraging. Salivary carboxyl ester lipase (CEL) is thought to hydrolyze insect lipophorins, which probably are absorbed across the gastric mucosa during feeding. The other six proteins are predicted either to maintain these lipids at high blood concentrations or to facilitate transport and uptake by flight muscles. Expression of these seven genes and coordinated secretion from a single organ is novel to this insectivorous bat, and apparently has evolved through instances of gene duplication, gene recruitment, and nucleotide selection. Four of the recruited genes are single-copy in the Myotis genome, whereas three have undergone duplication(s) with two of these genes exhibiting evolutionary ‘bursts’ of duplication resulting in multiple paralogs. Evidence for episodic directional selection was found for six of seven genes, reinforcing the conclusion that the recruited genes have important roles in adaptation to insectivory and the metabolic demands of flight. Intragenic frequencies of mobile- element-like sequences differed from frequencies in the whole M. lucifugus genome. Differences among recruited genes imply separate evolutionary trajectories and that adaptation was not a single, coordinated event.
Introduction
Among mammals, bats exhibit the greatest intraordinal diversification in diet. In general terms, diets range from insectivory and carnivory to frugivory and nectarivory and sanguivory [1]–[3]. Thus, pathways of bat adaptive radiation are defined by lineage differences in diet-associated enzymes, morphology (especially of the rostrum), dentition, salivary glands, and digestive tracts [4]–[10]. Flight is the centerpiece of bat biology. The constraints imposed by the morphological and physiological requirements of flight are interrelated to the diverse chiropteran dietary adaptations. Flight is metabolically the most expensive form of locomotion [11], [12]. Moreover, recent physiological studies have shown that insectivorous bats use exogenous lipids to fuel their flight within minutes of onset of feeding [13]. How this feat is achieved, is unknown although it is obvious that either the digestive process is accelerated or that alternative metabolic pathways are involved.
Salivary glands evolve rapidly, are involved in dietary adaptations, and release both exocrine and endocrine products [4], [7], [14]–[16]. We hypothesized that the submandibular salivary glands (also known in literature as ‘submaxillary’ glands) have had a direct role in bat adaptation to insectivory and possibly an indirect role in flight, given the rapid processing of exogenous lipids required for refueling flight muscles while feeding. The evolution of this capability could involve recruitment and expression of genes encoding secretory proteins with lipid-processing functions. If salivary glands had an important role in adaptive radiation in mammals, as has been proposed, it probably is due to the fact that additions and deletions to a salivary gland secretory proteome in a particular species could occur quickly and regularly [15]. Salivary glands have been described as a test bed for new, adaptive, roles for secretory proteins [15].
In addition to evidence that the secretory proteome is not conserved, there also are generic differences in salivary gland phenotypic characters (cell ultrastructure, histology, and histochemistry) that correlate with phylogenetic topologies and can be diet-associated [4]. For example, lysozyme-c, which is hypothesized to function both as a pH-dependent chitinase and as an antibacterial enzyme, is produced by different cells and in different salivary glands in various species of bats. Patterns such as these are compatible with the hypothesis that salivary glands offer multiple opportunities for gene recruitment and expression [14]. In terms of expression sites in particular salivary gland secretory cells, orthologous genes have had independent evolutionary trajectories in different bat lineages [14]. Thus, in some insectivorous species lysozyme-c-like immunoreactivity is associated only with acinar cells, whereas in other species in different bat families expression is associated with intercalated duct cells [14].
We think that insectivory was the original microbat diet and that the evolution of flight, echolocation, digestive tract, excretory system, and metabolic physiology all are related to exploiting lipid-rich insects as the primary source of energy. To test our hypothesis about the role of the submandibular salivary gland, we sequenced the transcriptome from the principal submandibular salivary gland (SMG) of the little brown bat, Myotis lucifugus. We then used this transcriptome data set to identify a putative secretory proteome for this gland in this species. The large, paired, principal submandibular salivary glands in Myotis lucifugus are positioned medial to the angular process of the mandible. The gland has a conserved histological structure and the secretory endpieces, intercalated ducts, and striated ducts all are involved in regulated secretion [17]. Although histological structure is preserved, the secretory endpieces in the Myotis lucifugus SMG are unusual in comparison to bats in other families because they consist of mucous tubules capped by seromucous demilunes (Fig. 1A). Transmission electron microscopy reveals a variety of secretory granules in the cell cytoplasm and differences in granule size and morphology among cell types (Fig. 1B). Ultrastructural diversity in secretory granule contents within a particular SMG is a consequence of physiochemical differences among secretory products [4], [18].
Figure 1. Optical micrograph of a semithin section of a secretory endpiece in a Myotis lucifugus principal submandibular gland, (A).
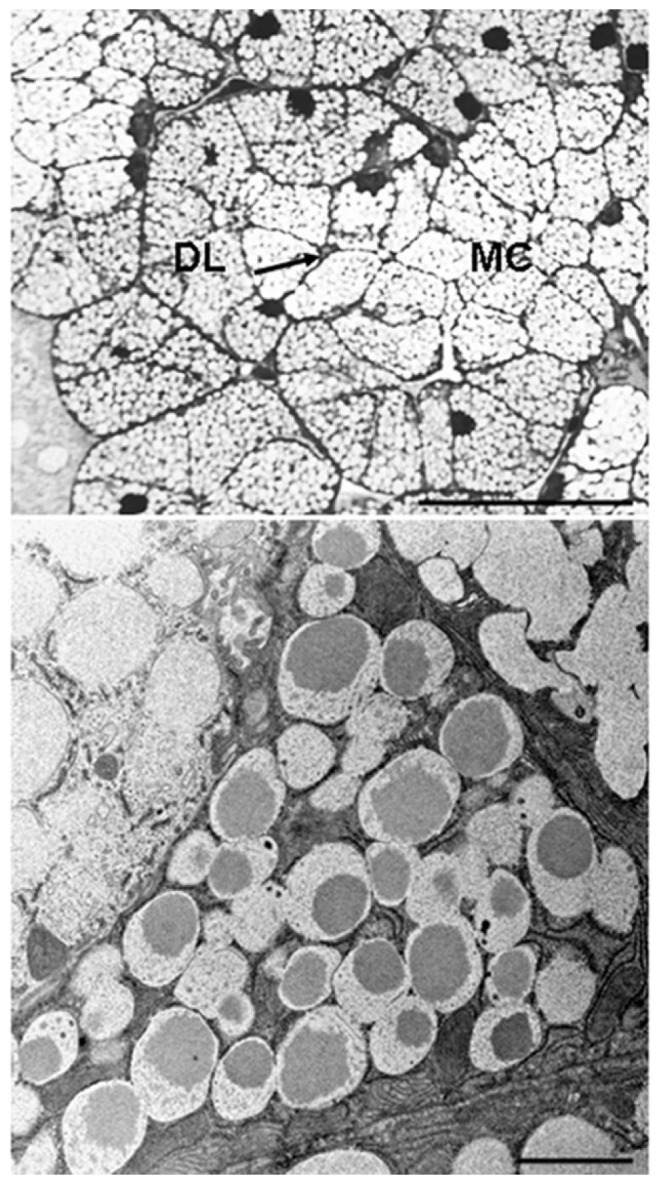
The more central mucous cells (MC) are surrounded by slightly darker demilunar seromucous cells (DL). The endpiece lumen is indicated by the arrow. Toluidine blue. Scale bar = 40 µm. B. Transmission electron micrograph of a demilunar seromucous cell flanked by mucous cells. Note the difference in the structure of the secretory granules in the respective cell types. Scale bar = 2 µm.
Myotis lucifugus is an insectivorous bat. It feeds on soft-bodied insects, especially moths, captured in flight or by gleaning from vegetation [19]. Nutritionally, moths are a rich source of lipids. A specialized fat body accumulates and stores lipids in the pupa. The stored lipids are the energy source for metamorphosis and the fat body in the adult provides energy for flight and reproduction [20]–[22]. The genetic and physiological adaptations that have enabled insectivorous bats to use this insect resource for fueling flight and basic metabolism are currently unknown. Specifically, we selected M. lucifugus because: 1) the histology and mucous histochemistry of its SMG have been analyzed [17]; 2) there are many unresolved metabolic and energetic questions; and because 3) the M. lucifugus genome is available (www.ensembl.org) as a reference point for genetic analyses.
Transcriptomes of selected organs or tissues are an effective way of comparing gene expression, discovering interspecies differences, and identifying putative intracellular and secretory proteins. This is particularly the case in glands that have both exocrine and endocrine secretory functions. Gene expression in such glands is dynamic and interspecific differences are commonplace [23]. In our study, we surveyed the SMG transcriptome for genes that encode secretory proteins known to be associated in mammals with lipid hydrolysis, transport, hyperlipidemia, and energy metabolism. Expression of a cluster of such genes in the SMG would support our hypothesis and would illuminate specific adaptations to a lipid-rich insectivorous diet. Potentially, such results would also help explain the remarkably rapid use of exogenous lipids during foraging flights.
Results and Discussion
Seven Key Genes in the Secretory Proteome
The intact SMG consists of a duct system with four epithelial cell types known in Myotis lucifugus to exhibit regulated secretion (demilune, acinar, intercalated, and striated duct cells). In addition to data for these cell types, the SMG transcriptome sequences included products of genes expressed in myoepithelial and endothelial cells, fibroblasts, cells in blood plasma, and a substantial number and variety (in terms of types) of neurons.
To construct a putative secretory proteome for the salivary gland itself, we focused on expressed genes encoding products known to be associated with a) regulated secretory cells (rather than constitutive secretory cells) such as acinar or pancreatic beta cells or intestinal epithelium or b) extracellular (post-secretion) lipid metabolism. A set of 23 genes met criterion ‘a’ and thus constitute our putative secretory proteome; seven of these 23 genes in the M. lucifugus SMG transcriptome fit both ‘a’ and ‘b’ criteria (Table 1). Co-expression of these seven genes in the secretory proteome of M. lucifugus SMG is known only from Myotis and thus is unique among mammals for which we have comparative SMG data either from literature or Expressed Sequence Tags (EST). We propose that these seven genes have had adaptive roles in terms of salivary gland evolution, lipid metabolism, and, in general terms, the insectivorous diet and metabolism of M. lucifugus. Expression levels of these seven genes varied from a FPKM value (fragments per kilobase of exon model per million fragments mapped) of 355.6 to 30.2 as calculated using Cufflinks [24].
Table 1. Summary for seven genes recruited to the secretory proteome of the submandibular salivary gland in the little brown bat, Myotis lucifugus.
| Gene | Protein | FPKM value (mRNA) | Pathway | Typical Expression Sites | Lipid-associated Function(s) |
| RETNLB | Resistin-like β | 355.6 | Endocrine | Intestinal epithelium (restricted) | Lipidemia and insulin resistance |
| LCN2 | Lipocalin 2 | 353.4 | Endocrine | Liver and adipose cells | Lipid transport |
| C3 | Complement 3 | 191.4 | Endocrine | Liver and adipose cells | Lipidemia, lipid transport |
| PSAP | Prosaposin | 139.9 | Endocrine | Kidney tubules, mammary glands | Lipid transport |
| CLU | Clusterin | 60.9 | Endocrine | Intestine, pancreas, liver | Lipid transport |
| APOE | Apolipoprotein E | 35.1 | Endocrine | Liver, adipose cells, macrophages | Lipid transport |
| CEL | Carboxyl ester lipase | 30.2 | Exocrine | Pancreas, lactating mammary gland | Lipid hydrolysis |
The predicted secretory pathway (exocrine or endocrine) is based on protein function. Expression sites selected as typical are based on a combination of literature and EST profile.
Four of the seven secretory protein genes listed in Table 1 are single-copy genes in the Myotis genome. The other three genes (LCN2, C3, CEL) have undergone duplications. In each case only one of the duplicated paralogous genes is expressed in the SMG. Expression sites for the other LCN2, C3, and CEL paralogs in Myotis are presently unknown. All seven of the genes vary substantially in terms of their ‘typical’ tissue expression sites across mammals. We used UniGene (www.ncbi.nlm.nih.gov/UniGene), Expressed Sequence Tag (EST) profiles, and literature mining to determine approximate expression patterns for the seven genes of interest. EST profiles are available for ‘salivary glands’ (presumably the submandibular or parotid gland, or both) of humans, mice, cow, and swine. From the profiles we know that the seven genes of interest are not expressed as a set in salivary glands of these four species.
Single-copy Genes
Resistin-like B protein (RETNLB; ENSMLUG00000001459)
Mammals typically have a single RETNLB gene. In Myotis lucifugus the gene spans 2.2 kb, is structured in three exons separated by two introns, and encodes a 115-amino acid cysteine-rich hormone. Analysis of intron 1 in the M. lucifugus RETNLB gene revealed a SINE-like sequence, whereas intron 2 contains a DNA/hAT-like sequence. Under the REV model, codon 7 in the N-terminal of the predicted M. lucifugus RETNLB protein is under positive selection. The EST profile for the RETNLB gene in mammals indicates typical expression only in intestinal epithelium (especially colon). There has been debate about the multiple functions attributed to RETNLB protein. The subject is complicated because although the RETNLB gene typically is expressed in intestinal epithelium, the RETN gene (which encodes resistin) is expressed in adipose tissue [25], [26]. Expression of the RETNLB gene in M. lucifugus SMG is unique and further complicates understanding of functional similarities and differences and roles of resistin-like B and resistin. It is relevant to our assessment of adaptation in insectivorous bats that circulating resistin-like B hormone has been shown to increase insulin resistance and to be associated with hyperlipidemia [26]. It also has been shown to regulate lipid energy metabolism and homeostasis [27]. This hormone probably is an endocrine product of M. lucifugus SMG (Table 1).
Prosaposin (PSAP; ENSMLUG00000015746)
Mammals typically have a single PSAP gene. In M. lucifugus, the gene spans 15.4 kb, is structured in 14 exons separated by 13 introns, and encodes a 523 amino acid precursor protein. Mobile element-like sequences occur in introns 1–3 and 5–8, but are absent from intron 4 and introns 9–13. The REV model did not detect any statistical evidence of prosaposin codons under episodic directional selection in M. lucifugus. The predicted prosaposin protein in M. lucifugus is 87% identical to the ortholog in the fruit bat, Pteropus alecto. Data are not currently available for comparison with other bat species. The EST profile indicates nearly ubiquitous PSAP gene expression in humans, laboratory mice, and swine. The gene is expressed at very low levels in human and mouse salivary glands, but not in swine salivary glands. The PSAP protein is thought to be secreted before its final processing and the secreted form serves as a lipid transporter that delivers bound sphingolipids to cell plasma membranes and into an endocytotic pathway [28]. The intracellular prosaposin peptides —referred to as saposins (termed A, B, C, and D)—enhance lysosomal hydrolytic activity [29].
Clusterin (CLU; ENSMLUG00000006721)
Mammals typically have a single CLU gene. The M. lucifugus CLU gene spans 12.6 kb, is structured in eight exons separated by seven introns, and encodes a 449 amino acid protein. Mobile element-like sequences occur in introns 1–7. The predicted CLU protein is 95% identical to the ortholog in congeneric M. davidii and 78% identical to the Pteropus alecto ortholog. The REV model identified one codon (238) as under episodic directional selection in the Myotis lucifugus CLU gene. The EST profile for CLU estimates that it is expressed principally in brain, joints, lymph, and pituitary glands, but also appears in numerous other tissues including liver. The clusterin protein has both intra- and extracellular functions [30], [31]. Numerous functions have been ascribed to clusterin, including service as a chaperone protein [31]. In blood, the CLU protein binds with and transports high-density lipoprotein (HDL) particles to the megalin cell surface receptor [32].
Apolipoprotein E (APOE; ENSMLUG00000006546)
Mammals typically have a single APOE gene. In M. lucifugus, the gene spans 3.3 kb, is structured in three exons separated by two introns, and encodes a 310 amino acid protein. No mobile element-like sequences were found in the intragenic introns of the M. lucifugus APOE gene. The REV model identified episodic directional selection in the ancestral gene tree branch leading to both Myotis and a fruit bat Pteropus vampyrum for which a genome database is available. The predicted APOE protein in M. lucifugus shares 87% identity with the orthologous protein in congeneric M. davidii, and only 70% with the orthologous protein in the vampire bat, Desmodus rotundus. The EST profile documents APOE expression principally in liver and adipose cells, but the gene is widely expressed at lower levels. The secreted form of APOE consists of two domains—the NH2-terminal domain that binds to the low-density lipoprotein (LDL) cell surface receptor and the COOH-terminal that binds to LDLs [33]. In humans, three APOE alleles encode proteins that are associated with differing lipoprotein plasma concentrations [34]. The APOE protein probably is secreted as an endocrine product from M. lucifugus SMG (Table 1).
Multiple-copy (duplicated) Genes
Lipocalin 2 (LCN2; ENSMLUG00000016210)
Duplications of the LCN2 gene have occurred independently in several mammalian lineages (particularly rodents and carnivores). In M. lucifugus, a single duplication has produced two LCN2 genes (ENSMLUG00000016210 expressed in SMG and ENSMLUG00000029693 with unknown expression site(s)). The LCN2 gene expressed in the SMG spans 3.4 kb, is structured in six exons separated by five introns, and encodes a 199 amino acid protein. No mobile element-like sequences were identified in the introns. Gene 1 (ENSMLUG00000016210) is the most conserved paralog of the two LCN2 genes in M. lucifugus. Gene 1 shares 85% identity with the orthologous gene in M. davidii and 80% identity with the Pteropus alecto ortholog. The REV model estimates that the gene tree branch separating the duplicated LCN2 genes in M. lucifugus from the single copy LCN2 gene in Pteropus vampyrum has been under episodic directional selection. The EST profile for LCN2 indicates that the gene is expressed, albeit at low levels, in a variety of tissues and cells. In humans, the highest expression level is in bone marrow but it also is expressed in liver and adipose tissue.
Lipocalin 2 is a cytokine with a role in regulating lipid metabolism and increasing insulin resistance [35], [36]. The protein has the β-barrel motif similar to other lipocalins, including an array of secretory and intracellular lipid-binding proteins [37], [38]. In the extracellular milieu, the LCN2 protein can bind to fatty acids or iron and has been investigated from a health perspective in connection with obesity, insulin resistance, and inflammation [39], [40]. It also modulates the peroxisome proliferator-activated receptor-γ (PPAR- γ), which in turn regulates energy expenditure and lipid homeostasis in adipocytes [41]. The Myotis lucifugus SMG protein isoform most likely is secreted as an endocrine product (Table 1).
Complement component 3 (C3; ENSMLUG00000011254)
In most (35/40 genera) mammals for which we have genome databases, C3 is a single copy gene. Myotis lucifugus is one of 5 exceptions to the rule; in the genome of this bat, six gene duplications account for seven paralogous C3 genes (Fig. 2). The fruit bat, Pteropus vampyrum, has only a single copy of the C3 gene in its genome. The C3 gene expressed in the M. lucifugus SMG (ENSMLUG00000011254, Gene 1) spans 32.2 kb, is structured in 42 exons separated by 41 introns, and encodes a large, 1664 amino acid glycoprotein. The size and structure of Gene 1 and primary structure of the encoded protein in Myotis lucifugus are conserved relative to orthologous C3 genes in other mammals. The predicted M. lucifugus Gene 1 protein is 78% identical to the single ortholog in Pteropus alecto (data unavailable for M. davidii). Mobile element-like sequences and simple repeat motifs occur in 14 of 41 introns (34%) in Myotis C3 Gene 1. The REV model identified episodic directional selection in Gene 1. In mammals with a single copy of the C3 gene, EST profiles indicate expression level is highest in liver, but that C3 also can be expressed at low levels in a variety of other tissues. Low level expression is seen in mouse salivary gland, but not in human or swine submandibular glands.
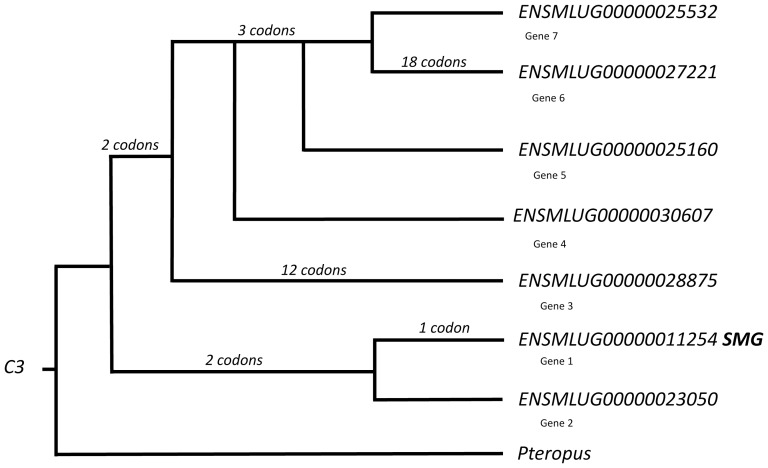
Figure 2. Gene tree for the C3 genes in Myotis lucifugus.
Numbers of codons under episodic positive selection are shown for each lineage where they occur. Gene ENSMLUG00000011254 is expressed in the principal submandibular gland (labeled SMG). Genes are numbered G1-G7 to correspond to Figure 5 and text.
The mammalian C3 gene encodes a complex precursor molecule [42]. Cell-specific processing of the large precursor protein results in one of two potential peptides that are cleaved from the precursor protein [43]–[45]. In hepatocytes, enzymatic cleavage within the N-terminal of the nascent C3 protein produces a 77 amino acid anaphylatoxin peptide (C3a) with immunological functions [43], [46]. The C3 protein that remains after cleavage is termed C3b. Consequently, hepatocytes secrete two proteins—the anaphylatoxin peptide (C3a) and a large C3b protein—both of which are processed from a single precursor. An alternative processing occurs in adipose cells. In these cells the C-terminal arginine is cleaved from the anaphylatoxin peptide. This modification of the anaphylatoxin peptide creates a functionally different peptide known as acylation-stimulating protein (ASP; C3adesArg) [47]–[49]. Removal of the terminal arginine eliminates any of the immunological properties associated with anaphylatoxin peptide. Instead, ASP is associated with triacylglycerol clearance from the blood [50]. In summary, in humans, laboratory mice, and rats, adipose cells secrete ASP and C3b and hepatocytes secrete anaphylatoxin peptide (C3a) and the C3b (complement component 3) protein.
The cell-specific ASP pathway in adipose cells requires co-expression of two other genes, complement factor B (CFB) and complement factor D (CFD), that encode cytoplasmic (non-secretory) proteins involved in the processing of the C3 gene secretory product [48]. With regard to expression of a C3 gene in M. lucifugus SMG, we asked whether the salivary gland uses the adipose cell ASP pathway or the alternate anaphylatoxin (C3a–C3b) processing pathway typically seen in hepatocytes. Based on the transcriptome data, the CFB gene (ENSMLUG00000000609) is expressed in the Myotis SMG, whereas the CFD gene is not expressed in the M. lucifugus submandibular gland (although it is present in the genome). Thus, we hypothesize that the M. lucifugus SMG secretes the anaphylatoxin peptide, C3a, as in liver rather than ASP as in adipose cells. In terms of known functions, anaphylatoxin peptide is immunological, the alternative protein, ASP, is associated with triacylglycerol clearance, and the C3b protein is associated with insulin resistance and FFA (free fatty acid) trapping. Over-production of the C3b protein has been linked to hyperlipidemia in humans [51]. If the same is true for Myotis, then hyperlipidemia resulting from the abundant secretion of the C3b protein would be advantageous in processing and using insect lipids while foraging. The C3 protein probably is an endocrine secretory product from M. lucifugus SMG (Table 1).
Carboxyl ester lipase (CEL; ENSMLUG00000006710)
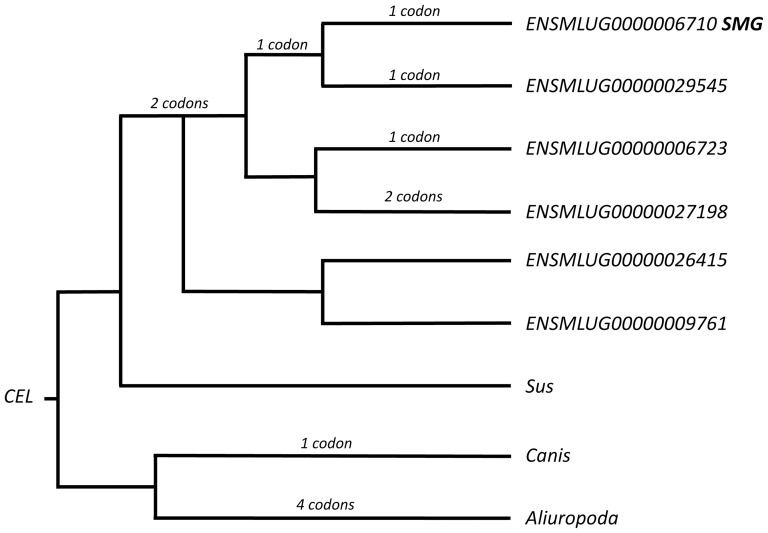
Mammals typically have a single CEL gene. Myotis lucifugus is 1 of 2 species (out of 33 for which genome data are available) that exhibits CEL gene duplications. The fruit bat, Pteropus vampyrum, has a single copy of the CEL gene. In the M. lucifugus genome, five hypothesized gene duplications have produced a total of six paralogous CEL genes, one of which (ENSMLUG00000006710, labeled Gene 1; Fig. 3) is expressed in the SMG. Gene 1 spans 18.4 kb and is structured in 11 exons separated by 10 introns. Expression in the M. lucifugus SMG is predicted to produce two transcripts (ENSMLUT00000006710 and ENSMLUT00000022459) that result in protein isoforms of 573 and 560 amino acids, respectively. In comparison to M. davidii, which also has 6 paralogous CEL genes [52], Gene 1 in M. lucifugus shares 82–91% identity. Gene 1 is only 76% identical to the CEL gene in Pteropus alecto.
Figure 3. Gene tree for the CEL genes in Myotis lucifugus.
Numbers of codons under episodic positive selection are shown for each lineage where they occur. Gene ENSMLUG00000006710 is expressed in the principal submandibular gland (labeled SMG).
Multiple mobile element-like sequences occur in introns in Gene 1. The REV model identified episodic directional selection in this gene (Fig. 3). The EST profiles show that in mammals a CEL gene typically is expressed in pancreatic acinar cells. Expression also has been reported in lactating mammary glands of humans and laboratory rodents and in rat SMG striated ducts [53]. Striated duct cells frequently are secretory in the SMG and diet-associated patterns do occur [7]. Data from rats and mice and broad interspecific comparisons illustrate some of the difficulty in comparing possible expression across species in absence of transcriptome data [7]. Functionally, CEL encodes a bile-salt activated pancreatic acinar cell enzyme that hydrolyzes lipids and is associated with their intestinal absorption [54], [55]. It also has a role in neonate lipid metabolism and gastric absorption [56]. Presumably, the isoforms of carboxyl ester lipase secreted by the Myotis SMG are exocrine and thus are components of saliva in this bat.
Evolutionary Mechanisms
Seven genes with interrelated lipid-associated functions are expressed in the M. lucifugus submandibular salivary gland transcriptome. The expression of these seven genes in a salivary gland secretory proteome is unique to M. lucifugus (among mammals for which we have comparable data) and probably is the result of multiple evolutionary mechanisms. We presently have no equivalent transcriptome-based secretory proteome data for any other insectivorous bat, or bats with any other diet for that matter. Consequently, we have no basis for estimating the evolutionary history of the SMG secretory proteome found in Myotis. A testable hypothesis based on the current research is that events resulting in expression of a group of seven genes with lipid-associated secretory products occurred early in the evolution of microbats, probably predating the origin of vespertilionids and certainly predating the origin of the genus Myotis.
The term ‘recruitment’ is suitable for describing the acquisition of gene expression in a novel cellular or tissue location. We assume that the seven genes of interest were recruited by various means for SMG expression early in microbat evolution. Although recruitment of gene expression is an important mechanism of adaptation, basic questions about the process are unanswered. For example, is recruitment of gene expression stochastic? Does the opportunity for novel salivary gland expression await some event such as insertion of an ERV/LTR transposon? Amylase expression in human salivary glands is an example of transposon involvement [57], [58]. But Cabej [59] has argued that under certain circumstances gene recruitment could be directly controlled by innervation, which is consistent with the role of innervation in salivary gland gene regulation and secretion [60]. Although acquisition of seven genes in the Myotis SMG proteome was important, it is unknown whether it was a coordinated process or the consequence of multiple independent stochastic events. In our opinion, the observation that recruited genes have different histories of duplication and expression profiles across tissues supports the argument that the Myotis SMG proteome evolved through multiple complementary, if not concerted, events rather than unrelated (independent) stochastic events.
Single Copy Gene Recruitment to the SMG
Among the single copy genes, recruitment of RETNLB expression to Myotis SMG perhaps is the most interesting example. Typically, expression of the RETNLB gene is regarded as specific to intestine [25]. Expression recruitment of single-copy genes, as opposed to duplicates, likely conserves protein primary sequence. RETNLB is a peptide hormone and recruitment of the gene for SMG expression presumably conserves the peptide's interactions with receptors and its function(s) as well. But what would be the adaptive advantage of recruiting a gene whose secretory product normally appears in the circulation anyway? Endocrine secretion from the Myotis SMG would place resistin-like B hormone directly into the circulation independent of the intestinal pathway to the liver. Its normal secretion is triggered by lipids in the intestine and in fact RETNLB gene expression is up-regulated by a high fat diet [27]. Secretion from the SMG would link release into the circulation with feeding rather than being dependent on lipids reaching the intestine. Such an explanation is consistent with a rapid response to the availability of exogenous lipids.
Gene Duplications
Gene duplications are an essential mechanism of genetic adaptation. Zhang et al. [52] reported that gene duplications appear to have played unusually important roles, perhaps including speciation, in Myotis. Gene duplications are believed to provide opportunities for structural and functional diversity in proteins insofar as paralogs can follow independent evolutionary trajectories [61], [62]. The C3 and CEL genes in Myotis exhibit multiple paralogs requiring 5–6 duplication events. Their gene trees portray ‘evolutionary bursts’ because of the large number of paralogs produced. Closer examination of each of the paralogs revealed that gene truncations are common and that some of the paralogous genes have premature stop codons probably rendering them nonfunctional (Figs. 2, 3). Among the Myotis C3 genes, three of the seven paralogs (Genes 1, 3, 6; Fig. 2) are structurally-conserved full-length genes, three of the genes (Genes 4, 5, 7; Fig. 2) are truncated and have early stop-codons, and one paralog (Gene 2; Fig. 2) is truncated so that it encodes a conserved version of the C3b protein without a premature stop codon. This latter gene presumably is functional. Among the three full-length Myotis C3 paralogous genes, the codons under episodic directional selection (P>0.05; EBF>20) are asymmetrically distributed in terms of gene tree topology. Most of these codons are found in two lineages, one leading to Gene 6 (ENSMLUG00000027221) and the other leading to Gene 3 (ENSMLUG 00000028875). Our conclusion that three of the seven C3 paralogs in Myotis have diverged under positive selection is consistent with the expectation for gene duplication [62]. An evolutionary burst and divergence of three paralogous C3 genes in Myotis might be indicative of the important biological roles for the proteins encoded by this gene. It also is consistent with the conclusion that the C3 gene and its encoded secretory product are an important part of the history of dietary and metabolic adaptation in insectivorous bats.
The carboxyl ester lipase gene (CEL) in Myotis also exhibits an evolutionary burst. In this instance gene duplications have resulted in six paralogs (Fig. 3). One to two codons under episodic directional selection (P>0.05; EBF>20) were identified in four of the six gene lineages (Fig. 3). Unlike the case of the C3 gene, it does not appear that CEL paralogs are rapidly diverging under positive selection.
Novel Regulation for a Recruited Gene
The NR5A2 (previously LRH-1) gene has been shown to regulate carboxyl ester lipase (CEL) expression in pancreatic acinar cells [55]. Gene duplication and recruitment of expression to the M. lucifugus SMG did not include the pancreatic regulatory gene, NR5A2. The absence of NR5A2 from the salivary gland transcriptome indicates CEL gene expression in the SMG is regulated differently than is CEL gene expression in pancreatic acinar cells. So far, at least, it is typical that genes recruited to salivary gland secretory proteomes arrive without their regulatory systems. Rennin and amylase are just two examples from human and rodent salivary glands [63], [64]. The consequence is that recruitment is more than just novel gene expression and protein secretion from the salivary gland. The data suggest that de novo regulation of a recruited gene adds another opportunity for evolutionary diversification.
Genomic locations of SMG-expressed paralogs
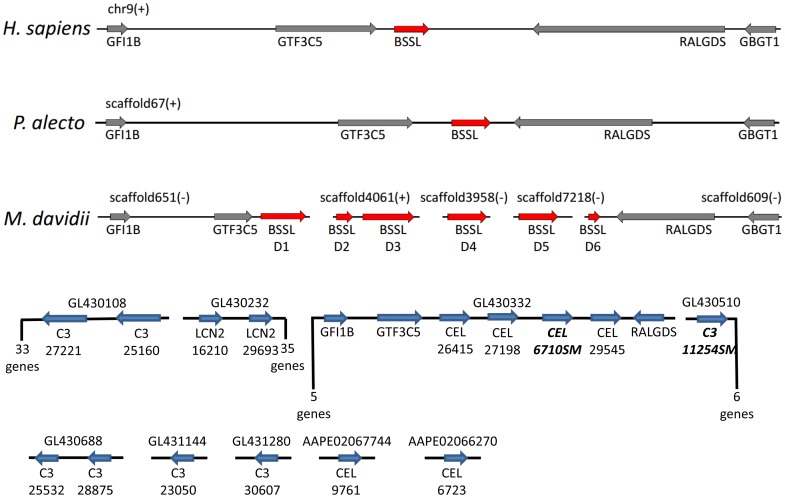
Although we do not yet know the chromosomal location(s) of genes expressed in the Myotis lucifugus SMG, we used the order of assembled contigs in the Ensembl genomic database to explore relative positions of some of the secretory protein genes expressed in M. lucifugus SMG. The analogous region in humans is on chromosome 9 (9q34-34.3), which has a cluster that includes single copies of LCN2 and CEL along with GTF3C5, RALGDS, and GBGT1. Zhang et al. [52] partially aligned their M. davidii data with the human genome and the fruit bat, Pteropus alecto, and showed the location of six copies of the CEL gene in this Myotis species (Fig. 4). Currently for M. davidii, the six CEL paralogs are assembled into five scaffolds, two of which include GTF3C5 and RALGDS, but spatial relationships among scaffolds are unknown [56]. The Ensembl scaffolding for the M. lucifugus genome places four of six CEL paralogs on a scaffold with GTF3C5 and RALGDS (Fig. 4). The other two CEL genes are on separate scaffolds. These comparisons suggest genomic rearrangements among the CEL gene cluster within the genus Myotis (Fig. 4). Additional investigations of patterns of congeneric chromosomal rearrangements are warranted because such studies will shed light on evolutionary rate and because bat chromosomal rearrangements already have been explored in substantial detail. Evidence points to rearrangements as important aspects of evolution and speciation [65], [66].
Figure 4. Comparison of relative genomic locations and gene order based on shared scaffolds in Myotis lucifugus (bottom in blue) with similar data from M. davidii, Pteropus alecto, and human beings.
The comparative data are from Zhang et al. [52], who used the symbol ‘BSSL’ for the carboxyl ester lipase (CEL) genes.
Mobile elements
Genomic databases and bioinformatic algorithms provide substantial new insights in the possible role(s) of mobile elements in shaping the mammalian genome [67]–[69]. Mobile elements (ME) had an important role in genome evolution specifically in Myotis and some categories of transposons possibly are still active in these bats [70], [71]. This particularly is the case with DNA/hAT elements, which are abundantly represented in Myotis [70]. In terms of salivary glands, for more than 20 years it has been known that a combination of gene duplications and transposable elements (especially LTR-retrotransposons) are associated with the recruitment of genes encoding secretory proteins [63], [64], [72], [73].
With the foregoing in mind, we determined the identities of ME-like sequences occurring in introns within each of five genes (PSAP, CLU, RETNLB, C3, CEL) containing such sequences. For comparison, we used the published data on the percentages of Class 1 and 2 MEs identified in the entire Myotis genome [74]. In making such a comparison it is essential to remember that our data are intragenic, whereas Pagán et al. [74] analyzed the entire genome, including intergenic sequence.
The set of C3 paralogous genes consists of three full-length and four truncated genes. Moreover, two of the full-length genes have 12–18 codons under episodic directional selection. In comparison to the overall genome-wide distribution of MEs summarized by Pagán et al. [74], the intragenic distribution of ME-like sequences in the C3 paralogs differed significantly (P>0.05) in three ways: 1) percentage-wise the C3 paralogs had more intragenic Class 1 LINE (1 and 2) elements than the genome (24.17vs 17.45±4.27%); 2) more intragenic DNA/hAT element-like sequences (19.43vs 12.55±3.95%); and 3) more intragenic DNA/Tigger-like sequences (7.11vs 0.27±2.56%)(95%CI = p±1.96(p(1-p)/N)0.5). Additionally, the frequency of helitrons in the Myotis genome is 16.25%, whereas helitron-like sequences were absent from intragenic introns in the C3 paralogs. Given the diversity and evolutionary history of the Myotis C3 paralogs (truncated vs full-length; functional vs non-functional, positive episodic selection vs no statistical evidence of selection), we also asked if there were significant differences among ME frequencies in these genes. But no statistical difference was found (Χ 2 = 47.5; d.f. = 40; P>0.2). Overall, then, the relative intragenic percentages of various Class 1 and 2 mobile elements among paralogous C3 genes is conserved.
The carboxyl ester lipase (CEL) genes provide us with a second example. In this instance the six paralogs differ remarkably from each other; one gene (Gene 1, expressed in the Myotis SMG) exhibits multiple intronic ME-like sequences, but all five of its paralogs have only 2–5 intronic MEs. MER20 (DNA-hAT/Charlie) sequences are the exception. In M. lucifugus, a MER20 sequence is conserved in all four of the functional CEL paralogs. MER20 has been associated with recruitment and creation of novel regulatory pathways [75]. In contrast, no MER20 sequences were found in the C3 paralogs. The differences in the relative frequencies of Class 1 and 2 intragenic MEs in the duplicated CEL and C3 genes in M. lucifugus is evidence of independent evolutionary histories for these genes, both of which are expressed in the SMG.
Likewise, differences among the single copy genes (PSAP, CLU, RETNLB) document that they also have had independent evolutionary histories (Table 2; Fig. 4).
Table 2. Summary of frequency (given in percentages) of mobile-element-like sequences in the Myotis lucifugus genome (from Pagán et al. ref 74) compared to intragenic frequencies (given in percentages) in the introns of four genes recruited to the submandibular salivary gland proteome.
| Genome | C3 | CEL | CLU | PSAP | |
| Class1 | |||||
| SINES | 39.86 | 41.23 | 42.86 | 31.58 | 41.6 |
| LINES | 17.49 | 24.17 | 0 | 10.53 | 16.6 |
| ERV/LTR | 9.7 | 8.06 | 23.81 | 15.79 | 8.33 |
| Class2 | |||||
| DNA/hAT | 12.55 | 19.43 | 14.29 | 42.11 | 33.3 |
| DNA/Tigger | 0.27 | 7.11 | 0 | 0 | 8.33 |
| Helitrons | 16.25 | 0 | 14.29 | 0 | 0 |
| Mariner | 3.32 | 0 | 4.76 | 0 | 0 |
| PiggyBac | 0.65 | 0 | 0 | 0 | 0 |
The data for C3 are for all 7 paralogs. The data for CEL are from Gene 1 (based on 21 mobile element sequences in this gene), which is expressed in the SMG. The 5 paralogous CEL genes have only 2–5 mobile sequences/gene. See Fig. 5 for graphical presentation.
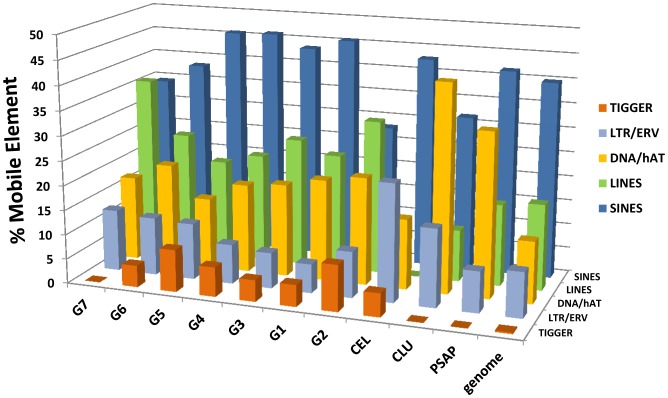
No single pattern of difference in ME distribution emerged from our comparisons among five recruited genes (including two that exhibited evolutionary bursts of duplication). At the same time, all five differed dramatically from the respective percentages of Class 1 and 2 mobile elements in whole genomic dataset. In the case of C3, the differences could be tested and were found to be statistically significant. The SINE-like sequences provided the only conserved pattern (Fig. 4). In the whole genome dataset, SINES comprise 39.86% of the elements and in all paralogs of C3 and CEL and in PSAP and CLU, SINES range from 31.58 to 42.86% of the elements (Table 2). One striking difference between our intragenic data and the genome overall is the low frequency of LINES (1 and 2) sequences in two of the genes, CEL (0) and CLU (10.53), compared to the genome overall (17.45). DNA/Tigger-like sequences were prominent in introns in these genes, both of which had undergone an evolutionary burst (C3, CEL). Helitron-like sequences made up 14.29% of the ME-like sequences in the CEL gene recruited and expressed in Myotis lucifugus SMG (Table 2; Fig. 5). One relatively recent and demonstrably active DNA/transposon (piggyBat) known from the M. lucifugus genome [71], [76], was not detected in our intragenic dataset.
Figure 5. Histogram comparing percentages of Class 1 and 2 mobile element-like sequences in intragenic introns of selected Myotis lucifugus genes.
The relative percentages of mobile element-like sequences in the total genome are shown on the right (from Pagán et al. [74]). Seven paralogs of C3 are shown, labeled G1–G7 (see Figure 2). See text for a full discussion of the data.
In summary, the occurrence of significant differences between intragenic and whole genomic distributions of certain MEs is evidence that MEs are not randomly positioned within the genome. Whether or not MEs had a role in recruitment of expression to the Myotis lucifugus SMG is unknown, although the data do support the hypothesis that all of these seven genes of interest have had independent evolutionary histories.
Function and Proposed Adaptive Role(s)
Bats depend on flight for obtaining nutrients, but flight is metabolically expensive. The fact that bats typically spend nearly all of their non-foraging time in energy-saving physiological torpor with lowered body temperature and respiration shows that they live in a delicate metabolic balance. Bat flight muscles rely on fat as the oxidative substrate, whereas the capacity for glycogen metabolism is low or essentially nonexistent [77]–[80]. The emphasis on fat in bat metabolism has been attributed to its high density of energy storage (8× more density efficient) as compared to glycogen [13]. It also is the case that stored glycogen is heavier than stored fat because of the water co-located with glycogen. This in turn raises the physiological question of water balance in bats, which might limit glycogen storage. Moreover, exogenous rather than stored lipids are used to sustain foraging flight in insectivorous bats within minutes of consuming insects [13]. In a typical mammal, exogenous nutrients of all types are processed through a slow postprandial digestive pathway. Fats, for example, are hydrolyzed in the intestine, absorbed by enterocytes, and often converted to chylomicrons in a multistep intracellular process [81]. In fatty diets, chylomicron formation also inhibits gastric emptying through a feedback loop involving cholecystokinin [82]. Chylomicrons are exported from the enterocytes and carried away from the intestine via the lymphatic system. Usually the next steps are circulatory system clearance and storage in adipose cells or liver. In contrast, stable carbon isotope ratios have been used in insectivorous bats to show the rapid assimilation of exogenous lipids [13].
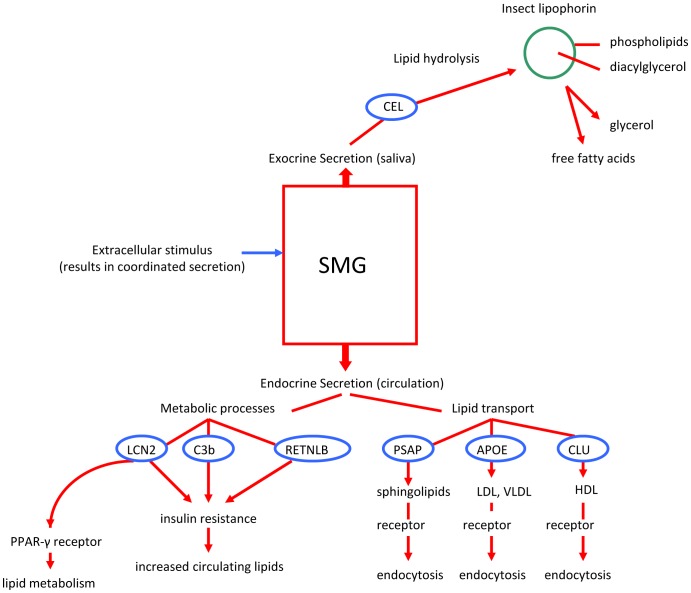
The expression in Myotis SMG of seven genes whose encoded secretory products are associated with lipid metabolism is compatible with the hypothesis that this gland has an important adaptive role in processing exogenous insect lipids. It is logical to anticipate that salivary glands have roles in dietary adaptation, but in this instance its adaptive role apparently includes flight metabolism. Among the predicted SMG secretory proteins, one (CEL) hydrolyzes lipids, three (C3b, LCN2, RETNLB) are associated with insulin resistance, regulation of lipid metabolism, and fatty acids, and the remaining three (PSAP, APOE, CLU) all are associated with lipid transport and receptor-mediated endocytosis (Fig. 6). Insulin resistance is important because insulin promotes the clearance and storage of lipids while inhibiting their release into the circulation [83]. In fact, in addition to insulin resistance in particular, overproduction of any of these secretory proteins is associated with metabolic syndrome (type 2 diabetes) in humans [84], [85]. Coordinated SMG secretion of these proteins makes them quickly available in circulation. In M. lucifugus, adaptation has produced a metabolic strategy that also favors extremely rapid movement of lipids into the circulation followed by a period of sustained hyperlipidemia. It is clear that there are fundamental, but normal, metabolic differences between insectivorous bats, such as M. lucifugus, and humans.
Figure 6. Diagram showing proposed adaptive roles of each of the genes recruited to expression in the secretory proteome of the Myotis lucifugus SMG.
We conclude that the seven genes expressed in Myotis SMG (Table 1; Fig. 6) compose an adaptive package as the encoded secretory proteins have interrelated—or possibly coordinated—functions. Orthologs to these seven genes are expressed in a variety of tissues in other mammals, but never linked together in a single organ or tissue. Co-expression of this set of genes in an organ where secretion is linked to feeding elucidates one evolutionary mechanism underlying adaptation. In effect, recruitment of genes ordinarily expressed and regulated in different tissues (ranging from pancreas and intestine to liver and adipose cells) and their subsequent expression in a single organ represents adaptation through creation of novel gene and protein networks. Dropping or adding proteins to an existing network, or creation of a novel network, has important adaptive potential [86]. Finally, it also is the case that salivary glands have both endocrine and exocrine secretory pathways and this adds significantly to their importance in adaptation [15], [87]. This conclusion about the adaptive role of salivary glands in mammals is strongly supported by elegant studies of mice that demonstrated a correlation between aggressive behavior and fighting and endocrine release of products including nerve growth factor from the SMG [88].
Among predicted secretory products, carboxyl ester lipase is a likely candidate for making exogenous lipids available quickly. In insects, most lipids are stored in the form of lipophorins, which consist primarily of diacylglycerol cores with phospholipid surfaces [21], [22]. The conventional carboxyl ester lipase (the enzyme in humans) has been shown to be especially effective with mono- and diacylglycerols and with phospholipids [56]. The human and rodent CEL isoforms retain their function when present in the stomach. If the Myotis SMG carboxyl ester lipase is released into saliva and if it also retains its functional capabilities in the stomach, it then is reasonable to hypothesize that at least the medium chain length insect lipids can be absorbed directly across the gastric mucosa, following the same pathway as some milk lipids in neonate humans and laboratory rodents [89]–[92]. In neonatal rats, medium chain lipids in milk pass through the stomach wall and can be detected in circulation within five minutes of feeding [89]. This rate approximates the rate at which insectivorous bats begin to use exogenous lipids to fuel their flight [13]. But milk and insects are obviously different physically. In order for rapid gastric absorption of lipids to occur in a flying insectivorous bat, the ingested prey must be finely comminuted before it reaches the stomach. This requirement is consistent with the conservative coronal morphology and complex occlusion patterns characteristic of molar dentition in insectivorous bats [5], [11], [93], [94]. The molar teeth have sharp, W-shaped shearing cusps that readily reduce prey to fine particles before swallowing.
Our study illustrates how an organ, the submandibular salivary gland, that at first blush has no obvious connection to the central feature of bat existence, namely, powered flight, plays a key and heretofore unexpected adaptive role in the evolution and maintenance of this unique ability among mammals.
Methods and Materials
Our research was conducted in accordance with protocol number 26592 for wild animals approved prior to our study by the Institutional Animal Care and Use Committee (IACUC) of Pennsylvania State University. The IACUC follows NIH and USDA guidelines for animal research and every effort is made to minimize discomfort or pain and to use the minimal number of individuals. A male specimen of Myotis lucifugus was collected under scientific collecting permit #00098 (issued to Michael Gannon by the Pennsylvania Game Commission) at Shaver's Creek Environmental Center, Huntingdon County, Pennsylvania. This collection locality was chosen so that the SMG transcriptome would come from a bat within the same genetic lineage of Myotis lucifugus used for the Ensembl genome project. The bat was euthanized according to the approved protocol and then the right and left intact submandibular salivary glands were dissected in the field and immediately placed in liquid nitrogen. Tissue samples from this bat are archived in the Genetics Resources Collection at the Museum of Texas Tech University. In the laboratory, one entire gland weighting approximately 60 mg was homogenized in 1 ml of Trizol Reagent and total RNAs were isolated following manufacturer protocol (Life Technologies, Carlsbad, California, USA). RNA integrity was verified using a Bioanalyzer (Santa Clara, California, USA). The fragment library preparation was developed from Poly(A) selected RNAs and sequenced for one lane of SOLEXA GAIIx 75 base-pair paired end sequencing, yielding 29.3 million pairs of reads. Sequence data are available through GenBank Short Read Archive (accession number SRP031492).
The Ensembl Myotis lucifugus assembly and annotation Myoluc2.0.67 [95] was used for whole transcriptome analysis. A total of 29.3 million RNA-seq read pairs (58.6 m read fragments) were mapped to the assembly using Tophat v1.2.0 [96] (and Bowtie v0.12.7) with default parameters, and Rna-SeQC v1.1.7 [97] was used to obtain read mapping statistics. Table 3 provides statistics describing the mapped reads. ∼13 m read pairs and ∼39 m read fragments were mapped to the genome, with ∼93% of read pairs and ∼85% of read fragments mapping uniquely. ∼25% of reads mapped to known exons, and ∼47% mapped to intragenic regions. These statistics indicate that the sequencing data is robust and the read mapping was successful, especially given that the Myoluc genome is quite incomplete at more than 11,500 scaffolds. The mapped reads were assembled using Cufflinks version 1.0.3 with the Myoluc2.0.67 gene annotation as the reference annotation and otherwise using default parameters [98].
Table 3. Read mapping statistics.
| Category | Value |
| Total Reads | ∼2.9 M. read pairs/∼5.9 M. read fragments |
| Mapped Reads | ∼1.3 M read pairs (45.8%)*/∼1.2 unique pairs (45.1% of total, 92.9% of mapped pairs)/∼3.9 total fragments (66.9%)*/∼3.3 uniquely mapped fragments (56.7% of total, 84.8% of mapped fragments) |
| Mapped Reads' Mismatch Rate | 1.4% |
| Mapping to Gene Annotation | 24.8% mapped to exons, 22.2% mapped to introns, 53% intergenic |
From the assembled transcriptome we produced a putative secretory proteome for the Myotis SMG. Although we use the term ‘secretory proteome,’ we did not independently confirm the presence of any of the secretory proteins in SMG exocrine or endocrine secretions. For gene names in all cases we followed the HUGO Gene Nomenclature Committee's symbols and names.
To examine the possibility of adaptive amino acid replacements in the genes (or gene families) of functional interest detected in the transcriptomes, the Myotis loci identified in ENSEMBL were aligned with those from other mammal species in the same or sister clades in the ENSEMBL gene tree (s). In most instances this included orthologous genes in the megabat Pteropus using the ClustalW for codons routinely available in MEGA 5.05 [99]. Minor refinements of automated alignments were done manually. Decisions on which genes are included and on the corresponding alignments are critical to results of this analysis: to facilitate assessment or reanalysis, final alignments are available in Support Information 1. Analyses of episodic directional selection on codons then were carried out using the MEME routine implemented in http://www.datamonkey.org, using the general codon time-reversible model (REV, see 98) substitution model [100]. Sites were posited to be under directional selection at selected branches if they were: 1) significant at the P>0.05 level; and if they 2) also showed an Empirical Bayes Factor>20.
RepeatMasker web service was implemented to determine the overall occurrence of repetitive elements within genes of interest in the secretory proteome (RepeatMasker website. Available; http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker, Accessed 2012 October 16). The complete nucleotide sequences for each gene were obtained from ENSEMBL and paralogs were aligned against the ‘ancestral’ sequence with ClustalW. These sequences were used as input with individualized introns and exons in the notation. The cross-match search engine and mammal DNA source option were implemented during analysis.
Acknowledgments
The bat salivary gland project was conceived by participants at a Texas Tech University Genomics Symposium funded by the Office of the Vice President for Research. We acknowledge Robert Bradley, Adam Freedman, Scott Edwards, Mohamed Noor, Fedrico Hoffmann, Anton Nekrutenko, Oliver Ryder, and John Novembre for discussions on experimental design. We thank Ronald K. Chesser and David A. Ray for assistance with mobile element analyses.
Funding Statement
Financial and curatorial support was provided by the Texas Biological Database Program and the Natural Science Research Laboratory and Genetic Resources Center at The Museum of Texas Tech University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baker RJ, Bininda-Emonds ORP, Mantilla-Meluk H, Porter CA, Van Den Bussche RA (2012) Molecular timescale of diversification of feeding strategy and morphology in New World leaf-nosed bats (Phyllostomidae): A phylogenetic perspective. In: Gunnell GF, Simmons NB, editors. Evolutionary History of Bats: Fossils, Molecules and Morphology. Cambridge Studies in Molecules and Morphology – New Evolutionary Paradigms, Cambridge Univ Press. pp. 385–409.
- 2. Baker RJ, Bradley LC, Bradley RD, Dragoo JW, Engstrom MD, et al. (2003) Revised checklist of North American mammals north of Mexico, 2003. Occ Papers Mus Texas Tech Univ 229: 1–23. [Google Scholar]
- 3. Dumont ER, Dávalos LM, Goldberg A, Santana SE, Rex K, et al. (2012) Morphological innovation, diversification and invasion of a new adaptive zone. Proc Royal Soc B 212: 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips CJ, Tandler B, Nagato T (1993) Evolutionary divergence of salivary gland acinar cells: a format for understanding molecular evolution. In: Dobrosielski-Vergona K, editor. Biology of the Salivary Glands. CRC Press. pp. 39–80. [Google Scholar]
- 5.Phillips CJ (2000) A theoretical consideration of dental morphology, ontology, and evolution in bats. In: Adams R, Pedersen S, editors. Ontogeny, Functional Ecology, and Evolution of Bats. Cambridge Univ Press. pp. 247–274. [Google Scholar]
- 6. Phillips CD, Butler B, Fondon JW, Mantilla-Meluk H, Baker RJ (2013) Contrasting evolutionary dynamics of the developmental regulator PAX9, among bats, with evidence for a novel post-translational regulatory mechanism. PLoS One e57649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tandler B, Gresik EW, Nagato T, Phillips CJ (2001) Secretion by striated ducts of mammalian major salivary glands: a review from an ultrastructural, functional, and evolutionary perspective. Anat Rec 264: 121–145. [DOI] [PubMed] [Google Scholar]
- 8. Liu Y, Xi H, Yuan X, Rossiter SJ, Zhang S (2012) Multiple adaptive losses of alanine-glyoxlate aminotransferase mitochondrial targeting in fruit eating bats. Mol Biol Evol 10.1093/molbev/mss013 [DOI] [PubMed] [Google Scholar]
- 9. Shen B, Han X, Zhang J, Rossiter SJ, Zhang S (2012) Adaptive evolution in the glucose transporter 4 gene slc2a4 in old world fruit bats (Family: Pteropodidae). PLoS One 7: e33197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santana SE, Strait S, Dumont ER (2011) The better to eat you with: functional correlations of tooth structure in bats. Functional Ecol doi: 10.1111 [Google Scholar]
- 11. Chappell MA, Roverud RC (1990) Temperature effects on metabolism, ventilation, and oxygen extraction in a Neotropical bat. Respir Physiol 81: 401–412. [DOI] [PubMed] [Google Scholar]
- 12. Winter Y, von Helversen O (1998) The energy cost of flight: do small bats fly more cheaply than birds? J Comp Physiol B 168: 105–111. [DOI] [PubMed] [Google Scholar]
- 13. Voigt CC, Sörgel K, Dechmann DKN (2010) Refueling while flying: Foraging bats combust food rapidly and directly to power flight. Ecology 91: 2908–2917. [DOI] [PubMed] [Google Scholar]
- 14. Phillips CJ, Weiss A, Tandler B (1998) Plasticity and patterns of evolution in mammalian salivary glands: comparative immunohistochemistry of Lysozyme in bats. Eur J Morph 36: 19–26. [PubMed] [Google Scholar]
- 15.Phillips CJ (1996) Cells, molecules, and adaptive radiation in mammals. In: Baker RJ, Genoways HH, editors. Contributions in Mammalogy: A Memorial Volume Honoring Dr. J. K. Jones, Jr. Mus Texas Tech Univ, Lubbock. pp. 1–24.
- 16. Tandler B, Pinkstaff CA, Phillips CJ (2006) Interlobular excretory ducts of mammalian salivary glands: structural and histochemical review. Anat Rec 288A: 498–526. [DOI] [PubMed] [Google Scholar]
- 17. Pinkstaff CA, Tandler B, Cohan RP (1982) Histology and histochemistry of the parotid and the principal and accessory submandibular glands of the little brown bat. J Morph 172: 271–285. [DOI] [PubMed] [Google Scholar]
- 18. Tandler B, Phillips CJ (1993) Structure of serous cells in salivary glands. Micros ResTech 26: 32–48. [DOI] [PubMed] [Google Scholar]
- 19. Ratcliffe JM, Dawson JW (2003) Behavioural flexibility: the little brown bat, Myotis lucifugus, and the northern long-eared bat, M. septentrionalis, both glean and hawk prey. Animal Behav 66: 847–856. [Google Scholar]
- 20. Downer RGH, Matthews RJ (1976) Patterns of lipid distribution and utilization in insects. Am Zool 16: 733–745. [Google Scholar]
- 21. Soulages JL, van Antwerpen R, Wells MA (1996) Role of diacylglycerol and apolipophorin-III in regulation of the physiochemical properties of the lipophorin surface: metabolic implications. Biochem 35: 5191–5198. [DOI] [PubMed] [Google Scholar]
- 22. Arrese EL, Canavoso LE, Jouni ZE, Pennington JE, Tsuchida K, et al. (2001) Lipid storage and mobilization in insects: current status and future directions. Insect Biochem Mol Biol 31: 7–17. [DOI] [PubMed] [Google Scholar]
- 23. Ozyildirim AM, Wistow GJ, Gao J, Wang J, Dickinson DP, et al. (2005) The lacrimal gland transcriptome is an unusually rich source of rare and poorly characterized gene transcripts. Invest Ophthalmol Vision Sci 46: 1572–1580. [DOI] [PubMed] [Google Scholar]
- 24. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification of RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotech 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steppan CM, Brown EJ, Wright CM, Bhat S, Banerjee RR, et al. (2001) A family of tissue-specific resistin-like molecules. PNAS 98: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rajala MW, Obici S, Scherer PE, Rossetti L (2003) Adipose-derived resistin and gut-derived resistin-like molecule-β selectively impair insulin action on glucose production. J Clin Invest 111: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hildebrandt MA (2009) High fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137: 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hiraiwa M, Soeda S, Kishimoto Y, O'Brien JS (1992) Binding and transport of gangliosides by prosaposin. PNAS 89: 11254–11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan W, Qi X, Tsang P, Kang S-K, Illarionov PA, et al. (2007) Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. PNAS 104: 5551–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bailey R, Griswold MD (1999) Clusterin in the male reproductive system: localization and possible function. Mol Cell Endocrinol 151: 17–23. [DOI] [PubMed] [Google Scholar]
- 31. Wilson MR, Easterbrook-Smith SB (2000) Clusterin is a secreted mammalian chaperone. Research Online http://ro.uow.edu.au/scipapers/16. [DOI] [PubMed] [Google Scholar]
- 32. Calero M, Tokuda T, Rostagno A, Kumar A, Zlokovic B, et al. (1999) Functional and structural properties of lipid-associated apolipoprotein J (clusterin). Biochem J 344: 375–383. [PMC free article] [PubMed] [Google Scholar]
- 33. Mahley RW (1998) Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 240: 622–630. [DOI] [PubMed] [Google Scholar]
- 34. Elousa R, Demissie S, Cupples LA, Meigs JB, Wilson PWF, et al. (2003) Obesity modulates the association among APOE genotypes, insulin, and glucose in men. Obesity Res 11: 1502–1508. [DOI] [PubMed] [Google Scholar]
- 35. Guo H, Jin D, Zhang Y, Wright W, Bazuine M, et al. (2010) Lipocalin-2 deficiency impairs thermogenesis and potentiates diet-induced resistance in mice. Diabetes 59: 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin D, Guo H, Bu SY, Zhang Y, Hannaford J, et al. (2012) Lipocalin 2 is a selective modulator of peroxisome proliferator-activated receptor-y activation and function in lipid homeostasis and energy expenditure. FASEB J 25: 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flower DR (1996) Review Article the lipocalin protein family: structure and function. Biochem J 318: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flower DR, North ACT, Sansom CE (2000) The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 1482: 9–24. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y, Lam KSL, Kraegen EW, Sweeney G, Zhang J, et al. (2007) Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem 53: 34–41. [DOI] [PubMed] [Google Scholar]
- 40. Zhang J, Wu Y, Zhang Y, LeRoith D, Bernlohr DA, et al. (2008) The role of lipocalin 2 in the regulation of inflammation in adipocytes and macrophages. Mol Endocrinol 22: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Spiegelman BM (1998) Perspectives in diabetes PPAR-γ: Adipogenic regulator and thiazolidinedione receptor. Diabetes 47: 507–514. [DOI] [PubMed] [Google Scholar]
- 42. Thai C-T, Ogata RT (2003) Expression and characterization of the C345C/NTR domains of complement components C3 and C5. J Immunol 171: 6565–6573. [DOI] [PubMed] [Google Scholar]
- 43. Caporale LH, Tippett PS, Erickson BW (1980) The active site of C3a anaphylatoxin. J Biol Chem 255: 10758–10763. [PubMed] [Google Scholar]
- 44. Cianflone K (1997) Acylation stimulating protein and the adipocyte. J Endocrinol 155: 203–206. [DOI] [PubMed] [Google Scholar]
- 45. Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, et al. (2005) Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437: 505–510. [DOI] [PubMed] [Google Scholar]
- 46. De Bruijn MHL, Frey GH (1985) Human complement component C3: cDNA coding sequence and derived primary structure. PNAS 82: 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choy LN, Rosen BS, Spiegelman BM (1992) Adipsin and an endogenous pathway of complement from adipose cells. J Biol Chem 267: 12736–12741. [PubMed] [Google Scholar]
- 48. Baldo A, Sniderman AD, St-Luce S, Avramoglu RK, Maslowska M, et al. (1993) The adipsin-acylation stimulating protein system and regulation of intracellular triglyceride synthesis. J Clin Invest 92: 1543–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zwiner J, Werfel T, Wilken H-C, Theile E, Götze O (1998) Anaphylatoxin C3a but not C3a(desARG) is a chemotoxin for the mouse macrophage cell line J774. Eur J Immunol 28: 1570–1577. [DOI] [PubMed] [Google Scholar]
- 50. Cianflone K, Zakarian R, Couillard C, Delplanque B, Despres J-P, et al. (2004) Fasting acylation-stimulating protein is predictive of postprandial triglyceride clearance. J Lipid Res 45: 124–131. [DOI] [PubMed] [Google Scholar]
- 51. Verseyden C, Meijssen S, van Dijk H, Jansen H, Cabezas MC (2003) Effects of atorvastin on fasting and postprandial complement component 3 response in familial combined hyperlipidemia. J Lipid Res 44: 2100–2108. [DOI] [PubMed] [Google Scholar]
- 52. Zhang G, Cowled C, Shi Z, Huang Z, Baker ML, et al. (2013) Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poorkhalkali N, Lidmer A-S, Lundberg LG, Dalrymple MA, Gibson Y, et al. (1998) Bile salt-stimulated lipase (BSSL) distribution in rat, mouse and transgenic mouse expressing human BSSL. Histochem Cell Biol 110: 367–376. [DOI] [PubMed] [Google Scholar]
- 54. Shamir R, Johnson WJ, Mortlock-Fitzpatrick K, Zolfaghari R, Li L, et al. (1996) Pancreatic carboxyl ester lipase: a circulating enzyme that modifies normal and oxidized lipoproteins in vitro. J Clin Invest 97: 1696–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fayard E, Schoonjans K, Annicotte J-S, Auwerx J (2003) Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J Biol Chem 278: 35725–35731. [DOI] [PubMed] [Google Scholar]
- 56. Hui DY, Howles PN (2002) Carboxyl ester lipase: structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res 43: 2017–2030. [DOI] [PubMed] [Google Scholar]
- 57. Samuelson LC, Wiebauer K, Gumucio DL, Meisler MH (1988) Expression of the human amylase genes: recent origin of a salivary amylase promoter from an actin pseudogene. Nucleic Acids Res 16: 8261–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Samuelson LC, Wiebauer K, Snow CM, Meisler MH (1990) Retroviral and pseudogene insertion site reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol Cell Biol 10: 2413–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cabej NR (2010) Neural control of gene recruitment in metazoans. Devel Dynamics 240: 1–8. [DOI] [PubMed] [Google Scholar]
- 60.Proctor GB (1998) Secretory protein synthesis and constitutive (vesicular) secretion by salivary glands. In: Garrett JR, Ekström J, Anderson LC editors. Glandular Mechanisms of Salivary Secretion. Frontiers Oral Biology, Karger Press, Basel. pp. 73–88. [Google Scholar]
- 61. Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc Royal Soc London B 256: 119–124. [DOI] [PubMed] [Google Scholar]
- 62. Zhang J (2003) Evolution by gene duplication: an update. TRENDS Ecol Evol 18: 292–298. [Google Scholar]
- 63. Abel KJ, Howles PN, Gross KW (1992) DNA insertions distinguish the duplicated rennin genes of the DBA/2 and M. hortulanus mice. Mamm Genet 2: 32–40. [DOI] [PubMed] [Google Scholar]
- 64. Meisler MH, Ting C-N (1993) The remarkable evolutionary history of the human amylase genes. Crit Rev Oral Biol Med 4: 503–509. [DOI] [PubMed] [Google Scholar]
- 65.Baker RJ (2006) Order Chiroptera. In: O'Brien SJ, Menninger JC, Nasg WG editors. The Atlas of Mammalian Chromosomes. John Wiley & Sons, Inc., Hoboken, New Jersey. pp. 378–380. [Google Scholar]
- 66. Baker RJ, Bickham JW (1986) Speciation by monobrachial centric fusions. PNAS 83: 8245–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deninger PL, Moran JV, Batzer MA, Kazazian HH Jr (2003) Mobile elements and mammalian genome evolution. Curr Opinion Genet Devel 13: 651–658. [DOI] [PubMed] [Google Scholar]
- 68. Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Ann Rev Genet 41: 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Muotri AR, Marchetto MCN, Coufai NG, Gage FH (2007) The necessary junk: new functions for transposable elements. Human Mol Genet 16: R159–R167. [DOI] [PubMed] [Google Scholar]
- 70. Ray DA, Pagán HJT, Thompson ML, Stevens RD (2007) Bats with hATs: Evidence for recent DNA transposon activity in genus Myotis . Mol Biol Evol 24: 632–639. [DOI] [PubMed] [Google Scholar]
- 71. Ray DA, Feschotte C, Pagan HJT, Smith JD, Pritham EJ, et al. (2008) Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus . Genet Res 18: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dickinson DP, Mirels L, Tabak LA, Meisler KW (1988) Expression of the human amylase genes: recent origin of a salivary amylase promoter from an actin pseudogene. Nucleic Acids Res 16: 8261–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lin HH, Ann DK (1991) Molecular characterization of rat multigene family encoding proline-rich proteins. Genetics 10: 102–113. [DOI] [PubMed] [Google Scholar]
- 74. Pagán HJT, Macas J, Novák P, McCulloch ES, Stevens RD, et al. (2012) Survey sequencing reveals elevated DNA transposon activity, novel elements, and variation in repetitive landscapes among vesper bats. Genet Biol Evol 4: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lynch VJ, Leclerc RD, May G, Wagner GP (2011) Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nature Genet 43: 1154–1159. [DOI] [PubMed] [Google Scholar]
- 76. Mitra R, Li X, Kopusta A, Mayhew D, Mitra RD, et al. (2013) Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. PNAS 110: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yacoe ME, Cummings JW, Myers P, Creighton GK (1982) Muscle enzyme profile, diet, and flight in South American bats. Am J Physiol 242: R189–194. [DOI] [PubMed] [Google Scholar]
- 78. Foehring RC, Hermanson JW (1984) Morphology and histochemistry of flight muscles in free-tailed bats, Tadarida brasiliensis . J Mamm 65: 388–394. [Google Scholar]
- 79.Altenback JS, Hermanson JW (1987) Bat flight muscle function and the scapulo-humeral lock. In: Fenton MB, Racy P, Rayner JMV editors. Recent Advances in the Study of Bats. Cambridge Univ Press NY. pp. 100–118. [Google Scholar]
- 80. Powers LV, Kandarian SC, Kunz TH (1991) Ontogeny of flight in the little brown bat, Myotis lucifugus: behavior, morphology, and muscle histochemistry. J Comp Physiol A 168: 675–685. [Google Scholar]
- 81. Karmen A, Whyte M, Goodman DS (1963) Fatty acid esterification and chylomicron formation during fat absorption: 1. Triglycerides and cholesterol esters. J Lipid Res 4: 312–321. [PubMed] [Google Scholar]
- 82. Raybould HE, Meyer JH, Tabrizi Y, Liddle RA, Tso P (1998) Inhibition of gastric emptying in response to intestinal lipid is dependent on chylomicron formation. Am J Physiol 274: R1834–R1838. [DOI] [PubMed] [Google Scholar]
- 83. Saltiel AR, Kahn CR (2001) Insulin signaling and the regulation of glucose and lipid metabolism. Nature 414: 799–806. [DOI] [PubMed] [Google Scholar]
- 84. Chapman MJ (2007) Metabolic syndrome and type 2 diabetes: lipid and physiological consequences. Dyslipidaemia 4: S5–S8. [DOI] [PubMed] [Google Scholar]
- 85. Van Oostrom AJHM, Alipour A, Plokker TWM, Sinderman AD, Cabezas MC (2007) The metabolic syndrome in relation to complement component 3 and postprandial lipemia in patients from an outpatient lipid clinic and healthy volunteers. Atherosclerosis 190: 167–173. [DOI] [PubMed] [Google Scholar]
- 86. Yu H, Kim PM, Sprecher E, Trifonov V, Gerstei M (2007) The importance of bottlenecks in protein networks:correlation with gene essentiality and expression dynamics. PLOS Comp Biol 3: e59 10.1371/journal.pcbi.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Isenman L, Liebow C, Rothman S (1999) The endocrine secretion of mammalian digestive enzymes by exocrine glands. Am J Physiol 39: E223–E232. [DOI] [PubMed] [Google Scholar]
- 88. Aloe L, Alleva E, Böhm A, Levi-Montalcini R (1986) Aggressive behavior induces release of nerve growth factor from mouse salivary gland into the bloodstream. PNAS 83: 6184–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aw TY, Grigor MR (1980) Digestion and absorption of milk triacylglycerols in 14-day-old suckling rats. J Nutrit 110: 2133–2140. [DOI] [PubMed] [Google Scholar]
- 90. Carey MC, Small DM, Bliss CM (1983) Lipid digestion and absorption. Annu Rev Physiol 45: 651–677. [DOI] [PubMed] [Google Scholar]
- 91. Hamosh M, Bitman J, Liao TH, Mehta NR, Buczek RJ, et al. (1989) Gastric lipolysis and fat absorption in preterm infants: effect of medium-chain triglyceride or long-chain triglyceride-containing formula. Pediatrics 83: 86–92. [PubMed] [Google Scholar]
- 92. Freeman PW (1979) Specialized insectivory: beetle-eating and moth-eating molossid bats. J Mamm 60: 467–479. [Google Scholar]
- 93. Freeman PW (1981) Correspondence of food habits and morphology in insectivorous bats. J Mamm 62: 166–173. [Google Scholar]
- 94. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, et al. (2013) Ensembl 2013. Nucl Acids Res 10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics doi:10.1093 [DOI] [PubMed] [Google Scholar]
- 97. Deluca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, et al. (2012) RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28: 1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Delport W, Scheffler K, Botha G, Gravenor MB, Muse SV, et al. (2010) CodonTest: Modeling Amino Acid Substitution Preferences in Coding Sequences. PLoS Comput Biol 6 (8) e1000885 10.1371/journal.pcbi.1000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Murrell B, Wertheim JD, Noola S, Weighill T, Scheffler K, et al. (2012) Detecting individual sites subject to episodic diversifying selection. PLoS Genetics 8: e1002764 10.1371/journal.pgen.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]