Abstract
Bladder cancer is the fourth most common cancer in men in the United States, and its recurrence rate is highest among all malignancies. The unmet need for improved strategies for early detection, treatment, and monitoring of the progression of this disease continues to translate into high mortality and morbidity. The quest for advanced diagnostic, therapeutic, and prognostic approaches for bladder cancer is a high priority, which can be achieved by understanding the molecular mechanisms of the initiation and progression of this malignancy. Aberrant DNA methylation in single or multiple cancer-related genes/loci has been found in human bladder tumors and cancer cell lines, and urine sediments, and correlated with many clinicopathological features of this disease, including tumor relapse, muscle-invasiveness, and survival. The present review summarizes the published research on aberrant DNA methylation in connection with human bladder cancer. Representative studies are highlighted to set forth the current state of knowledge, gaps in the knowledgebase, and future directions in this prime epigenetic field of research. Identifying the potentially reversible and ‘drugable’ aberrant DNA methylation events that initiate and promote bladder cancer development can highlight biological markers for early diagnosis, effective therapy and accurate prognosis of this malignancy.
Keywords: aberrant DNA methylation, animal models, epigenetics, transitional cell carcinoma, tumorigenesis
Introduction
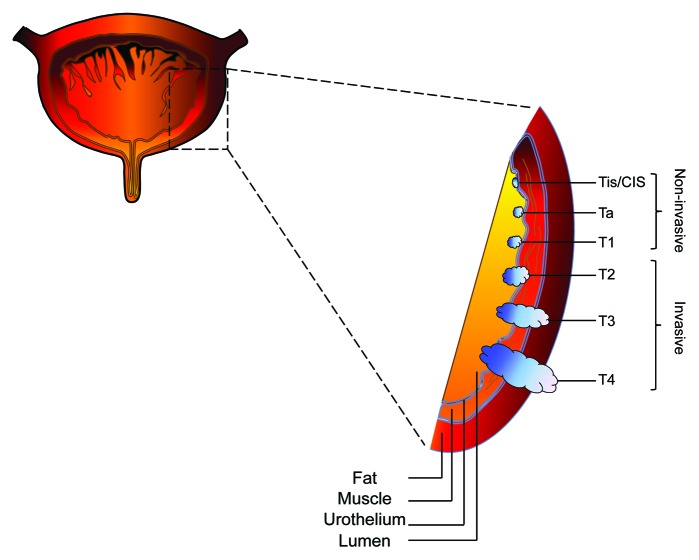
Bladder cancer is a major public health problem, causing significant mortality and morbidity worldwide.1-3 In the US, bladder cancer is the fourth most common cancer in men, with an estimated 72,570 new cases and 15,210 deaths in 2013.2 The vast majority (90%) of bladder cancer cases are transitional cell carcinomas (TCC), of which 75–85% present as non-muscle invasive tumors at the time of first diagnosis (Tis/CIS, Ta, and T1).4-6 The prognosis of these non-invasive tumors is favorable although up to 80% of cases will recur after complete transurethral resection, and up to 45% of cases will progress to invasive cancer (T2-T4) within 5 y (Fig. 1).5,7-10 The gold standard method for bladder cancer diagnosis is cystoscopy followed by biopsy of suspicious lesions. However, this approach is highly invasive and costly, and can miss up to 30% of malignant cases.10-15 The non-invasive tests for bladder cancer diagnosis include voided urine cytology,16 cytogenetic analysis by fluorescence in situ hybridization, and detection of genetic mutations in urine.17-22 However, these tests have reported sensitivities of 54–86%, and specificities of 61–90%.17-19,21 Thus, there is a need to improve the non-invasive tests of bladder cancer detection and progression.23 The quest for sensitive, specific, and non-invasive diagnostic, therapeutic, and prognostic tools for bladder cancer is a high priority research area, and can be achieved by understanding the molecular mechanisms underlying the initiation and progression of this malignancy.24
Figure 1. Diagram of progressive stages of transitional cell carcinoma of the bladder. Non-muscle invasive transitional cell carcinoma is staged at Tis/CIS, Ta, and T1, whereas muscle invasive transitional cell carcinoma is staged at T2, T3, and T4. Tis/CIS, carcinoma in situ.
Epigenetics is a fast growing field in cancer biology, and has enormous potential for clinical and translational research.25-27 Cancer epigenetics refers to inheritable, yet reversible, changes associated with gene dysregulation, which manifest in a (pre) malignant phenotype where sequence of the genome is not altered28-30. Aberrant DNA methylation, dynamic changes in chromatin structure through post-translational modification of histones, nucleosome positioning, and microRNA-mediated modulation of gene expression are known epigenetic changes associated with carcinogenesis.25,28,31-33 The occurrence of epigenetic changes prior to malignant transformation, which frequently manifests in target and non-invasively obtainable surrogate organs, and the reversibility of these changes through pharmacologic and genetic interventions30,33-40 provide a unique opportunity for cancer research, ultimately leading to the discovery of non-intrusive diagnostic, therapeutic, and prognostic approaches for human malignancies. Characterizing the epigenetic changes that initiate and promote bladder cancer development can help identify biological markers that can be used for early detection, treatment, and monitoring of the progression of this malignancy. The continuous shedding of bladder lining cells into the urine is highly advantageous for non-invasive surveillance of the epigenetic changes occurring during bladder carcinogenesis.39-41 From a therapeutic standpoint, the anatomical confinement of the bladder makes this organ amenable to targeted epigenetic therapies whose complications and side effects should be fewer and less severe than those of systemic treatments.29,33,37,42,43
Aberrant DNA methylation is the most extensively studied epigenetic change associated with all types of human cancer, including bladder cancer.25,32,33,44 Aberrant DNA methylation was initially found in single genes/loci of relevance to carcinogenesis in human bladder tumors and cancer cell lines. Subsequent advances in genome-scale technologies for DNA methylation analysis and bioinformatics approaches have enabled a comprehensive characterization of DNA methylation patterns in human bladder cancer.39-41,44-51 The present review summarizes the published research on aberrant DNA methylation in connection with human bladder cancer. Representative studies are highlighted to set forth the current state of knowledge, gaps in the knowledgebase, and future directions in this prime epigenetic field of research. The utility of the in vivo animal models of induced-bladder tumorigenesis, and the need for population-based studies in ‘at risk’ individuals for bladder cancer development are also discussed. A synopsis of research on microRNA (miRNA) methylation and human bladder cancer, and an overview of the promises and pitfalls of epigenetic therapy for solid tumors are also provided.
Aberrant DNA Methylation and Carcinogenesis
In mammalian genomes, DNA methylation occurs almost exclusively in the context of 5′-CpG dinucleotides (CpGs).31,52 A family of DNA methyl transferases (DNMTs) comprised of DNMT1, DNMT3A, and DNMT3B catalyzes this reaction by transferring a methyl group from the donor S-adenosyl methionine to the fifth carbon position of the cytosine pyrimidine ring.27,53-55 In humans, the vast majority (80–90%) of CpGs in the genome are normally methylated.52,53,56 The remaining methylation-free CpGs are found in sequence stretches, termed CpG islands, that are > 500 base pairs long, and have a GC content of > 55% and an observed/expected CpG ratio of ≥ 0.65.57
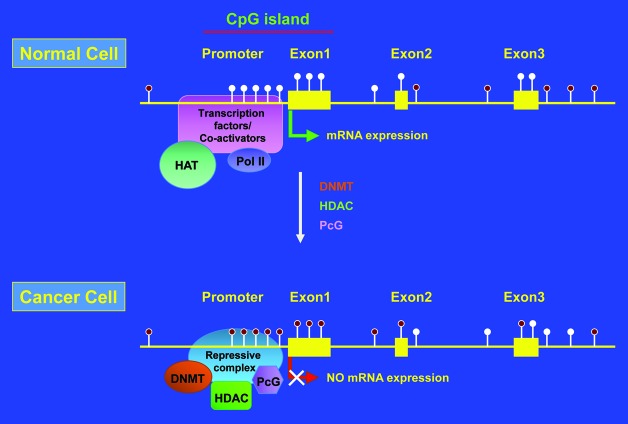
Aberrant DNA methylation has been found in almost all types of human cancer.25,31-33 Aberrant DNA methylation is characterized by global loss of DNA methylation (hypomethylation) and locus-specific gain/loss of DNA methylation (hyper/hypomethylation).25,31,58-61 Whereas DNA hypomethylation is thought to contribute to oncogenesis by reactivation of latent retrotransposons, induction of genomic instability, and activation of proto-oncogenes,62,63 DNA hypermethylation is believed to elicit tumorigenesis by dysregulation of gene expression, e.g., through transcriptional silencing of tumor suppressor genes.26,54,56,64 Hypermethylation of CpG islands, clustered at the promoter, untranslated 5′- region and exon 1 of known genes (promoter CpG islands) or localized within gene bodies (intragenic CpG islands) is a common event in human carcinogenesis (Fig. 2).25,32,33,56,65,66 Global DNA hypomethylation at repetitive DNA elements, such as long- and short interspersed nuclear elements (LINE and SINE, respectively), and long-terminal repeat retrotransposons (LTR) is also a frequent occurrence in human cancers.63,67,68
Figure 2. Schematic representation of promoter CpG island hypermethylation in cancer. Filled and unfilled lollypops represent methylated and unmethylated CpGs, respectively. HAT, histone acetyltransferase; Pol II, DNA polymerase II; DNMT, DNA methyl transferase; HDAC, histone deacetylase; PcG, polycomb group proteins.
Overview of Research on DNA Methylation and Human Bladder Cancer
The evolution of classic single-gene DNA methylation detection assays to genome-wide microarray based technologies, and most recently, next-generation sequencing platforms coupled with cutting-edge bioinformatics approaches has provided an unprecedented opportunity to investigate the role of aberrant DNA methylation in the genesis and progression of human cancers.32,65,69,70 Thus far, single- and multi-locus aberrant DNA methylation has been demonstrated in human bladder tumors and cancer cell lines, and urine sediments.39-41,44-51 In the following, we provide a synopsis of representative research on aberrant DNA methylation in connection with human bladder carcinogenesis.
Markl et al.45 used a semi-quantitative methylation-sensitive arbitrarily primed polymerase chain reaction (MS.AP.PCR) technique to investigate the methylation status of GC-rich regions in the genome of metachronous tumors and their derived cell lines from two patients with TCC of the bladder as compared with normal urothelium from five disease-free individuals. The authors detected 17% changes in the methylation status of the 214 evaluable sequences between tumors and normal urothelia from disease-free subjects. These methylation changes were cancer-specific (3%: common in all tumors from both patients), patient-specific (13%: present in all tumors from one patient only), and tumor-specific (1%: present in one tumor from one patient only). When compared with the normal urothelia and tumors concurrently, there were also 3% cell-line specific methylation changes.45
Maruyama et al.46 used the methylation-specific PCR (MSP) assay to investigate the promoter methylation profile of 10 different cancer-related genes, including CDH1, RASSF1A, APC, CDH13, FHIT, RARβ, GSTP1, p16INK4a, DAPK, and MGMT, in 98 bladder tumors in connection to clinicopathological features of the aggressiveness of the disease. Except for one squamous cell carcinoma, all the tumors were TCC of the bladder of varying grades and stages. High methylation frequency, expressed as median methylation index (MI), was detected for four genes, including RASSF1A (35%), APC (35%), and two members of the cadherin family, CDH1 (36%), and CDH13 (29%), which significantly correlated with various parameters of poor prognosis, such as tumor grade, growth pattern, muscle invasion, tumor stage, and ploidy status. In Kaplan-Meier analysis, the methylation of CDH1 and FHIT significantly associated with shortened survival. In a multivariate analysis adjusted for tumor grade, papillary or non-papillary histology, muscle invasion, and MI index of other genes, CDH1 methylation was independently associated with poor prognosis.46
Sathyanarayana et al.47 used the MSP assay to analyze the promoter methylation of three invasion and metastasis-related genes (LAMA3, LAMB3, and LAMC2) in bladder tumors (n = 128) and exfoliated cells (bladder washes and voided urine; n = 128), and correlated the results to clinicopathological findings. All the tumors were TCC except for two squamous cell carcinomas of the bladder. The methylation frequencies of these three genes in bladder tumors and urinary exfoliated cells, respectively, were as follows: 45% and 39% for LAMA3, 21% and 19% for LAMB3, and 23% and 15% for LAMC2. There was excellent concordance in methylation between tumors and corresponding exfoliated cell samples, including 88% for LAMA3 (p = 0.0006), 92% for LAMB3 (p = 0.002), and 83% for LAMC2 (p = 0.003). The LAMA3 and LAMB3 methylation significantly correlated with several parameters of poor prognosis, whereas the LAMC2 methylation was independently associated with shortened survival (p = 0.03; 95% CI = 1.14 – 10.84). Of significance, the frequency of LAMA3 methylation was significantly higher in invasive tumors than in non-invasive tumors (80% vs 10%; p = 0.0001).47
Christoph et al.48 used quantitative real-time MSP analysis to establish promoter CpG island methylation in four pro-apoptotic TP53 target/effector genes, including the APAF-1, CASP-8, DAPK-1, and IGFBP-3, in non-invasive and invasive TCC of the bladder (n = 110) as compared with normal urothelium from patients without urological malignancy (n = 20). Hypermethylation of the promoter regions of the APAF-1, CASP-8, and DAPK-1 was detected in 100%, 74%, and 66%, respectively, of all tumors, with no or very low levels of methylation being detectable in normal urothelia. The APAF-1 methylation significantly correlated with both tumor stage (p < 0.01) and tumor grade (p = 0.04). The APAF-1 and IGFBP-3 methylation were predictors of tumor relapse by differentiating tumors at higher recurrence risk from those at low-risk of recurrence. In multivariate analysis adjusted for tumor stage, tumor grade, and MI index of other genes, APAF-1 and IGFBP-3 methylation were independent prognostic factors for recurrence in superficial bladder tumors.48
Hoque et al.49 used quantitative real-time MSP analysis to investigate hypermethylation of promoter regions of nine cancer-related genes, including APC, ARF, CDH1, GSTP1, MGMT, CDKN2A, RARβ2, RASSF1A, and TIMP3, in primary tumors and urine sediments from 15 bladder cancer patients and 25 control urine sediments from subjects with no history of genitourinary malignancy. For all paired tumors and urine samples from bladder cancer patients, there was a perfect match in promoter methylation of all the tested genes. Four genes (ARF, GSTP1, MGMT, and CDKN2A) displayed promoter hypermethylation in all the samples from bladder cancer patients and no methylation in samples from controls (100% specificity). The methylation status of this four-gene panel was further investigated in the urine sediments of an additional 160 bladder cancer patients with tumors of varying stages and grades (90% with TCC histology) as compared with 69 urine samples from age-matched control subjects. Promoter hypermethylation was detectable for CDKN2A in 45% (95% CI = 38 – 53%), GSTP1 in 43% (95% CI = 35 – 51%), MGMT in 35% (95% CI = 28 – 42%), and ARF in 28% (95% CI = 21 – 35%) of all samples from bladder cancer patients, with no detectable level of methylation in the respective gene promoters in control samples. A two-stage prediction model based on the methylation of the four-gene panel followed by logistic regression analysis of the methylation of the remaining five genes was developed, which produced an internally validated Receiver Operating Characteristic (ROC) with an overall sensitivity of 82% (95% CI = 75 – 87%) and specificity of 96% (95% CI = 90 – 99%). The ROC Curve represents the trade-off between the false negative and false positive rates for every possible cut off points of a diagnostic test.49
Chung et al.39 used bisulfite pyrosequencing to investigate promoter hypermethylation of ten cancer-related genes, including A2BP1, NPTX2, SOX11, PENK, NKX6–2, DBC1, MYO3A, HSPB9, NPY2R, and CA10, in 26 primary bladder tumors and 6 bladder cancer cell lines. This panel of ten genes was previously identified by methylated CpG island amplification and microarray (MCAM) analysis of 85 primary bladder tumors and 12 bladder cancer cell lines.71,72 Eight genes were highly methylated in bladder tumors, with very low levels of methylation detectable in normal controls (bladder and leukocyte DNA from three individuals). The methylation frequencies in these genes in declining order were 92% (PENK), 88% (NPTX2), 85% (CA10), 77% (SOX11), 69% (DBC1 and NKX6–2), 65% (MYO3A), and 62% (A2BP1). Subsequently, a quantitative real-time MSP assay was used to analyze the 8-gene set in urine sediments from 128 bladder cancer patients (86% TCC histology) and 110 age-matched control subjects with no history of bladder malignancy. Using combinatorial analysis of methylation of these 8 genes, a predictive model was developed for bladder cancer detection, which showed 81% sensitivity and 97% specificity based on a panel of four genes (MYO3A, CA10, NKX6-2, and DBC1 or SOX11), and 85% sensitivity and 95% specificity based on a panel of five genes (MYO3A, CA10, NKX6-2, DBC1, and SOX11 or PENK). Analyzing the data by cancer invasiveness, predictive models of ≥ 3-gene panel showed 81% sensitivity and 95% specificity for non-muscle invasive tumors (Ta, Tis, and T1), and 90% sensitivity and 95% specificity for muscle invasive tumors (T2, T3, and T4).39
Wolff et al.40 used the Illumina GoldenGate methylation assay to interrogate 1,370 autosomal loci (784 genes) in 49 non-invasive urothelial tumors (Ta-T1), 38 invasive tumors (T2-T4) and their matched normal-appearing urothelia, and control urothelia from 12 age-matched individuals with no history of urothelial cancer. There were distinct patterns of aberrant DNA methylation in non-invasive tumors vs controls and invasive tumors vs. controls; whereas hypermethylation was more pronounced in the invasive tumors, hypomethylation occurred more frequently in the non-invasive tumors. Relative to control samples from urothelial cancer-free patients, invasive tumors had 526 hypermethylated loci (38%), whereas non-invasive tumors had 132 hypermethylated loci (10%) of which 117 (89%) overlapped with those found in the invasive tumors (p < 0.0001). Normal-appearing urothelia taken at least 5 cm away from their corresponding invasive tumors had 169 hypermethylated loci (12%) of which 142 (89%) were common to those found in the invasive tumors (p < 0.0001), indicating an epigenetic field defect. Most of the identified hypermethylated loci were located in the context of CpG islands. Conversely, non-invasive tumors had more unique hypomethylated loci (217 = 16%) than invasive tumors (41 = 3%) as compared with control urothelial cancer-free tissues (p < 0.001), in addition to the 253 hypomethylated loci (18%) that were common to both tumor types. The majority of the identified hypomethylated loci were located outside of CpG islands.40
Investigating the Mechanistic Involvement of Aberrant DNA Methylation in Human Bladder Carcinogenesis
The salient findings of the studies highlighted in the preceding section are: (1) aberrant DNA methylation is a common event in human bladder carcinogenesis; (2) aberrant DNA methylation is likely to occur in the early stages of human bladder carcinogenesis; and (3) detection of aberrant DNA methylation in the urine sediments can be potentially used as a diagnostic tool for human bladder cancer detection, and a prognostic tool for monitoring of the progression of this malignancy. However, the above findings are limited to clinically diagnosed bladder cancer patients only. Ideally, bladder cancer detection should be achievable in the general population before clinical manifestation of the disease.10,41,73,74 In addition, prognosis of bladder cancer and prediction of the recurrence of tumors should be attainable in apparently disease-free individuals who have undergone therapeutic regimens.10,41,73,74 Thus, a major goal of future research should be to find the role of aberrant DNA methylation in the initiation and progression of bladder cancer before clinical symptoms of the disease become manifest.
To achieve this goal, three questions need to be answered:
(1) Timing: How early in the process of bladder carcinogenesis does aberrant DNA methylation occur, and when is it manifest at a detectable level?
(2) Scale: What is the genomic distribution of aberrant DNA methylation in bladder carcinogenesis (genome-wide and locus-specifically)?
(3) Origin: Is aberrant DNA methylation a cause or a consequence of bladder carcinogenesis?
The answers to these questions should be sought in proof-of-principal experiments in in vivo models of induced-bladder tumorigenesis, where different stages of carcinogenesis can be intercepted. The existing animal models of bladder tumorigenesis include rodents, rabbits, and dogs in which bladder tumors can be induced in vivo by administering known bladder carcinogens, e.g., aromatic amines75-78 and arsenic.78,79 The advent of high throughput genome-wide technologies for DNA methylation detection will be instrumental in establishing the genomic distribution of aberrant DNA methylation at various stages of bladder carcinogenesis.32,44,65,69 Such an approach will verify the mechanistic involvement of aberrant DNA methylation in the evolution of bladder cancer from an early stage non-invasive neoplasia to late and aggressive malignancy.
Obviously, the use of in vivo animal models of induced-bladder tumorigenesis can best serve the above purpose because experimental exposure of humans to known bladder carcinogens is unethical and simply out of the question.80 This said, however, one should remain mindful of the need to validate the in vivo findings in animal models in follow up studies in human populations. Decades of experimental studies in animal models of human cancer have provided invaluable information on various aspects of human carcinogenesis.81-83 Nonetheless, the “incomplete” comparability of these models to humans underscores the need to validate the in vivo data in confirmatory research in humans.80 Thus, a requisite for animal studies of induced-bladder tumorigenesis is to recapitulate the in vivo findings in disease-free human populations, e.g., in case-control studies in populations with known susceptibility to bladder cancer development, e.g., smokers.84-86 For illustrative purposes, the utility of animal models of aromatic amine-induced bladder tumorigenesis, and the need for validation studies in smokers at high risk of developing bladder cancer are discussed below.
Smoking-Related Bladder Cancer and Tobacco Smoke-Derived Aromatic Amines
Bladder cancer represents a unique model of chemical carcinogenesis in humans.87 Unlike other types of human cancer with unknown or less well-defined etiologic agent(s), bladder cancer is primarily linked to tobacco smoking.84-86 The smoking-attributable bladder cancer is widely believed to originate from exposure to aromatic amines, a family of proven bladder carcinogens present in tobacco smoke.87-90 The elevated risk of bladder cancer in smokers of black (air-cured) tobacco relative to blond (flue-cured) tobacco is ascribed to the richer content of aromatic amines in the former tobacco products.89-91 Although the bladder-specific carcinogenicity of aromatic amines is well-established, the underlying mechanisms of action of these chemicals in the genesis and progression of bladder cancer are not fully delineated.24
A genotoxic mode of action for aromatic amines has been demonstrated that involves the induction of DNA damage and mutations.92-95 However, the genotoxicity of aromatic amines is not exclusive to the target organ of tumorigenesis.24,94-98 This suggests that an alternative mode of action (epigenetics) may also exist that singly or in combination with genotoxicity can explain the bladder-specific tumorigenicity of aromatic amines.24 The concept of a chemical carcinogen exerting its biological effects through epigenetic changes is very novel, and has never been investigated comprehensively. Historically, investigations of the mode of action of chemical carcinogens have been dominated by genotoxicity studies.93,99-101 Exploring the ability of aromatic amines to induce epigenetic changes, such as aberrant DNA methylation, can emerge as a paradigm shifting research in chemical carcinogenesis. Establishing the alterations of DNA methylome in vivo in animal models of aromatic amine-induced bladder tumorigenesis followed by confirmatory research in ‘at risk’ populations, such as smokers, can help elucidate the underlying mechanisms of the initiation and progression of human bladder cancer.
Although smoking is the main source of exposure to aromatic amines in the general population,84-86 occupational exposure to these chemicals also occurs in a wide range of industries, including rubber, cable, and textile manufacturing, aluminum transformation, and gas, coal, pesticide, and cosmetics production.102-104 Furthermore, other sources of exposure to aromatic amines also exist, including dietary (e.g., pesticides in food), life style (e.g., hair dye use), and environmental (e.g., engine exhaust) sources.102-104 Thus, the widespread exposure of human populations to carcinogenic aromatic amines constitutes a major public health problem.102,103 The unmet need for biomonitoring of humans exposed to aromatic amines86 can be addressed by the use of mechanistic markers that can chronicle the initiation and progression of bladder cancer.
Earlier research on aberrant DNA methylation in smoking-attributable bladder cancer has provided initial clues on the underlying mechanisms of the initiation and progression of this malignancy. For example, Wolff et al.50 conducted an elegant study in which smoking proved to be independently associated with aberrant DNA methylation in a unique genomic locus in bladder cancer patients. The authors used quantitative real-time MSP analysis to investigate the promoter methylation of nine cancer-related genes, including RUNX3, BCL2, PTGS2 (COX2), DAPK, CDH1 (ECAD), EDNRB, RASSF1A, TERT, and TIMP3, in 342 matched bladder tumors (TCC) and corresponding normal-appearing mucosa in comparison to urolthelia from age-matched cancer-free patients undergoing prostatectomy. In matched sets of bladder tumors and control mucosa, the methylation frequencies were 65% vs. 15% for EDNRB, 60% vs. 11% for RASSF1A, 39% vs. 9% for BCL2, and 30% vs. 4% for %TERT, although for RUNX3 the respective values were 56% vs. 0%, which was of most specificity (two normal-appearing mucosa showed RUNX3 methylation patterns that were nearly identical to their matched high-grade and invasive tumors, suggesting that these bladders had epigenetic aberrancies throughout or that the invasive tumor had spread across the bladder; the two samples were excluded from the above analysis). No or very low levels of methylation were detectable in normal urothelia of control subjects for all the nine genes tested. Of significance, methylation of RUNX3 preceded methylation of the other eight genes (p < 0.001), and its prevalence increased as a function of age at diagnosis (p = 0.031; adjusted for sex, smoking history, tumor stage and tumor grade), and history of smoking (p = 0.015; adjusted for age, sex, tumor stage, and tumor grade). It was suggested that because RUNX3 methylation increases with age, is absent in normal urothelium, and occurs early in tumorigenesis, it can be used as a molecular clock to determine the age of a bladder tumor. Doing so, tumors from smokers appear to be “older” than tumors from nonsmokers (p = 0.009) due to either initiating earlier or undergoing faster cell divisions. Since RUNX3 methylation is acquired early on in tumorigenesis, its detection in biopsy or urine specimens may provide a marker to screen smokers long before any symptoms of bladder cancer become manifest.50
miRNA Methylation and Human Bladder Cancer
miRNAs are a class of short noncoding RNAs (~22 nt) that negatively regulate gene expression at the post-transcriptional level through sequence-specific targeting of mRNA.105-107 The recognition of the target mRNA is based on complementarity of the seed sequence of the miRNA (i.e., 7–8 nt at the 5′-end) to a specific sequence motif within the 3′-untranslated region (3′-UTR) of the mRNA.108,109 Whereas partial base pairing between the seed region of the miRNA and the 3′-UTR of the target mRNA results in translational inhibition, (near) perfect complementarity causes degradation of the mRNA by endonuclease cleavage.107,110 Approximately 30% of human genes are putative targets of miRNAs.111-114 A single miRNA can regulate up to 200 different target mRNAs, whereas one individual mRNA can be controlled by multiple miRNAs.111,115,116 Thus, the influence of miRNAs in crucial biological processes, such as cell cycle regulation, differentiation, proliferation, apoptosis, development, metabolism, and aging, is unassailable.117,118 Accumulating evidence suggests that dysregulation of miRNA expression contributes to a wide range of human diseases, including cancer.106,118-121 A growing body of literature supports the involvement of miRNAs in the pathogenesis of human bladder cancer.122-125 For example, aberrant expression of various miRNAs has been observed in human bladder tumors and cancer cell lines, and urine sediments, and correlated with various clinicopathological characteristics of this disease.126-130 Although the mechanisms underlying miRNA dysregulation in human bladder cancer are not fully determined, proof-of-principle experiments have established a tight link between epigenetic changes, such as aberrant DNA methylation and histone modifications, and altered miRNA expression in bladder carcinogenesis.131-133 For instance, Saito et al.134 have shown that simultaneous treatment of T24 human bladder cancer cells with 5-aza-2′-deoxycytidine (5-Aza-CdR) and 4-phenylbutyric acid, which inhibit DNA methylation and histone deacetylase, respectively, resulted in upregulation (> 3-fold) of 17 out of 313 (5.4%) human miRNAs examined. Epigenomic studies in various types of human cancer, including bladder cancer, have shown that the number of dysregulated miRNA genes with aberrant DNA methylation at their promoter CpG islands and/or the flanking CpG “shores” (regions with less CpG density) is rapidly rising.135-137 Also, an increasing body of evidence suggests that detection of aberrantly methylated miRNA genes in human bladder cancer can serve as biomarkers for disease detection, evaluation of prognosis, and prediction of response to therapy.122-125 Furthermore, emerging research supports the notion that restoration of epigenetically dysregulated miRNAs in human bladder cancer, e.g., through drugs that mitigate aberrant DNA methylation and histone modifications, can constitute an effective therapeutic strategy for this disease.138,139 For more detailed information, we refer the readers to recent comprehensive reviews.118,120,140,141
Promises and Pitfalls of Epigenetic Therapy for Solid Tumors
Tumors display widespread epigenetic aberrancies, including alterations in DNA methylation patterns, histone modifications and variants, miRNAs dysregulation, chromatin remodeling, and nucleosome positioning.25,28,31-33 These epigenetic abnormalities are potentially reversible as the enzymes maintaining the epigenetic state are regulatable through pharmacologic interventions, i.e., epigenetic therapy.28,29,142,143 To date, the best-characterized and only Food and Drug Administration (FDA) approved drugs for epigenetic therapy are DNA demethylating agents and histone deacetylase inhibitors (HDACi).33,144,145 As cytosine analogs, Decitabine (5-Aza-CdR) and Vidaza (5-Azacytidine; 5-Aza-CR) target aberrant DNA methylation by getting incorporated into DNA and sequestering DNMTs, thus, resulting in depletion of these enzymes and global hypomethylation upon cell divisions.37,42,146 Targeting histone deacetylases (HDACs) for epigenetic therapy is challenging because this group of enzymes has multiple subclasses with mechanisms of action still under contention.147-149 More than a dozen HDACi are currently undergoing preclinical and clinical investigations for the treatment of both hematological malignancies and solid tumors, including bladder cancer.150-153 The common mechanism of action of HDACi is the chelation of Zn2+ ions, which are critical to the activity of these enzymes.149,154 As single agents, two HDACi, including Vorinostat and Romidepsin have FDA approval for the treatment of cutaneous T-cell lymphoma.155 More recently, the utility of synthetic anti-miRNA oligonucleotides with tumor suppressive function as epigenetic regulators has also been investigated.156-160 For instance, locked nucleic acids (LNAs)-anti-miRNA-based therapy has been explored in nonhuman primates for inhibition of oncogenic miRNAs.161
Thus far, epigenetic therapy has shown encouraging results for the treatment of hematological malignancies; however, the promise of targeting epigenetic aberrancies in solid tumors has not been realized yet.143,162,163 Major challenges include the delivery of epigenetic drugs, maintenance of a pharmacodynamics response, and achievement of a therapeutic index.164,165 Obviously, a better understanding of the mechanisms of action of the epigenetic drugs and greater insights into tumor biology will help improve the efficacy of epigenetic therapy for solid tumors. The goal is to combine epigenetic therapy with other chemotherapeutic approaches, such as hormonal therapies, immunomodulatory therapies, and standard chemotherapy, to help sensitize tumor cells to cytotoxic effects of targeted/systemic therapies or to durably slow or reverse resistance to these therapies. For more detailed information, we refer the readers to recent comprehensive reviews.121,166-169
Concluding Remarks: Potential Challenges and Future Directions
Bladder cancer continues to be a major public health problem in the US and throughout the world.1-3 The ineffective strategies for early detection, treatment, and monitoring of the progression of this disease have translated into significant mortality and morbidity.23 A requisite for alleviating the global burden of this disease is to understand the underlying mechanisms of the initiation and progression of this disease.24 As the most extensively studied epigenetic change associated with human cancers, aberrant DNA methylation in single or multiple genes/loci of relevance to carcinogenesis has been found in human bladder tumors and cancer cell lines, and urine sediments. In addition, detection of aberrant DNA methylation in cancer-related genes/loci in the above specimens has prognosticated many clinicopathological features of this disease, including tumor relapse, muscle-invasiveness, and survival.39-41,44-51
The advent of high throughput genome-wide technologies for epigenomics and genomics studies has provided a unique opportunity to identify global changes in DNA methylation with functional impact on gene expression, which can best predict bladder cancer initiation and progression.32,44,65,69,70 Future exploratory research should focus on characterization of aberrant DNA methylation, on a genome-wide scale, in experimental animals wherein bladder tumors can be induced by administering known bladder cancer-causing agents. Confirmatory research in humans at high risk for bladder cancer development should validate the exploratory research in animal models. The ultimate goal will be to construct the whole DNA methylome of bladder cancer at various stages of bladder carcinogenesis, including the initiation and progression stages. Identifying the potentially reversible and ‘drugable’ aberrant DNA methylation events that initiate and promote bladder cancer development can serve as biological markers for early detection, effective treatment and accurate prognosis of this malignancy.
Acknowledgments
This work was supported by grants from the American Cancer Society (RSG-11-083-01-CNE) and the University of California Tobacco Related Disease Research Program (20XT-0116) to AB.
Glossary
Abbreviations:
- 3′-UTR
untranslated region
- 5-Aza-CR
5-Azacytidine
- 5-Aza-CdR
5-aza-2′-deoxycytidine
- FDA
Food and Drug Administration
- HDACs
histone deacetylases
- HDACi
histone deacetylase inhibitors
- LNAs
locked nucleic acids
- MCAM
methylated CpG island amplification and microarray
- MI
methylation index
- miRNA
microRNA
- MSP
methylation-specific polymerase chain reaction
- MS.AP.PCR
methylation-sensitive arbitrarily primed polymerase chain reaction
- ROC
Receiver Operating Characteristic
- TCC
transitional cell carcinomas
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/25927
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society (ACS). Cancer Facts & Figures 2013. ACS National Home Office, Atlanta, GA, 2013. [Google Scholar]
- 3.World Health Organization (WHO). WHO report on the global tobacco epidemic, 2008: the MPOWER package. 2008.
- 4.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J, et al. European Association of Urology (EAU) EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur Urol. 2011;59:997–1008. doi: 10.1016/j.eururo.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 5.Carradori S, Cristini C, Secci D, Gulia C, Gentile V, Di Pierro GB. Current and emerging strategies in bladder cancer. Anticancer Agents Med Chem. 2012;12:589–603. doi: 10.2174/187152012800617768. [DOI] [PubMed] [Google Scholar]
- 6.Prasad SM, Decastro GJ, Steinberg GD, Medscape Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat Rev Urol. 2011;8:631–42. doi: 10.1038/nrurol.2011.144. [DOI] [PubMed] [Google Scholar]
- 7.Kurth KH, Denis L, Bouffioux C, Sylvester R, Debruyne FM, Pavone-Macaluso M, et al. Factors affecting recurrence and progression in superficial bladder tumours. Eur J Cancer. 1995;31A:1840–6. doi: 10.1016/0959-8049(95)00287-S. [DOI] [PubMed] [Google Scholar]
- 8.Allard P, Bernard P, Fradet Y, Têtu B. The early clinical course of primary Ta and T1 bladder cancer: a proposed prognostic index. Br J Urol. 1998;81:692–8. doi: 10.1046/j.1464-410x.1998.00628.x. [DOI] [PubMed] [Google Scholar]
- 9.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–5, discussion 475-7. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Zhang S, MacLennan GT, Williamson SR, Lopez-Beltran A, Montironi R. Bladder cancer: translating molecular genetic insights into clinical practice. Hum Pathol. 2011;42:455–81. doi: 10.1016/j.humpath.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Kriegmair M, Baumgartner R, Knüchel R, Stepp H, Hofstädter F, Hofstetter A. Detection of early bladder cancer by 5-aminolevulinic acid induced porphyrin fluorescence. J Urol. 1996;155:105–9, discussion 109-10. doi: 10.1016/S0022-5347(01)66559-5. [DOI] [PubMed] [Google Scholar]
- 12.Denzinger S, Burger M, Walter B, Knuechel R, Roessler W, Wieland WF, et al. Clinically relevant reduction in risk of recurrence of superficial bladder cancer using 5-aminolevulinic acid-induced fluorescence diagnosis: 8-year results of prospective randomized study. Urology. 2007;69:675–9. doi: 10.1016/j.urology.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 13.Zaak D, Kriegmair M, Stepp H, Stepp H, Baumgartner R, Oberneder R, et al. Endoscopic detection of transitional cell carcinoma with 5-aminolevulinic acid: results of 1012 fluorescence endoscopies. Urology. 2001;57:690–4. doi: 10.1016/S0090-4295(00)01053-0. [DOI] [PubMed] [Google Scholar]
- 14.Stenzl A, Cowan NC, De Santis M, Kuczyk MA, Merseburger AS, Ribal MJ, et al. European Association of Urology (EAU) Treatment of muscle-invasive and metastatic bladder cancer: update of the EAU guidelines. Eur Urol. 2011;59:1009–18. doi: 10.1016/j.eururo.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Morgan TM, Keegan KA, Clark PE. Bladder cancer. Curr Opin Oncol. 2011;23:275–82. doi: 10.1097/CCO.0b013e3283446a11. [DOI] [PubMed] [Google Scholar]
- 16.Papanicolaou GN, Marshall VF. Urine Sediment Smears as a Diagnostic Procedure in Cancers of the Urinary Tract. Science. 1945;101:519–20. doi: 10.1126/science.101.2629.519. [DOI] [PubMed] [Google Scholar]
- 17.Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology. 2003;61:109–18, discussion 118. doi: 10.1016/S0090-4295(02)02136-2. [DOI] [PubMed] [Google Scholar]
- 18.van Rhijn BW, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol. 2005;47:736–48. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Hajdinjak T. UroVysion FISH test for detecting urothelial cancers: meta-analysis of diagnostic accuracy and comparison with urinary cytology testing. Urol Oncol. 2008;26:646–51. doi: 10.1016/j.urolonc.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Parker J, Spiess PE. Current and emerging bladder cancer urinary biomarkers. ScientificWorldJournal. 2011;11:1103–12. doi: 10.1100/tsw.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt J, Cowan N, Protheroe A, Crew J. Recent advances in urinary bladder cancer detection. Expert Rev Anticancer Ther. 2012;12:929–39. doi: 10.1586/era.12.73. [DOI] [PubMed] [Google Scholar]
- 22.Gakis G, Schwentner C, Todenhöfer T, Stenzl A. Current status of molecular markers for prognostication and outcome in invasive bladder cancer. BJU Int. 2012;110:233–7. doi: 10.1111/j.1464-410X.2011.10839.x. [DOI] [PubMed] [Google Scholar]
- 23.Netto GJ, Cheng L. Emerging critical role of molecular testing in diagnostic genitourinary pathology. Arch Pathol Lab Med. 2012;136:372–90. doi: 10.5858/arpa.2011-0471-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besaratinia A, Tommasi S. Genotoxicity of tobacco smoke-derived aromatic amines and bladder cancer: current state of knowledge and future research directions. FASEB J. 2013;27:2090–100. doi: 10.1096/fj.12-227074. [DOI] [PubMed] [Google Scholar]
- 25.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 29.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21:502–17. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grønbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115:1039–59. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 31.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 32.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 33.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 34.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 35.Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–71. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 36.Jacinto FV, Ballestar E, Esteller M. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene. 2009;28:4212–24. doi: 10.1038/onc.2009.267. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–46. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibragimova I, Ibáñez de Cáceres I, Hoffman AM, Potapova A, Dulaimi E, Al-Saleem T, et al. Global reactivation of epigenetically silenced genes in prostate cancer. Cancer Prev Res (Phila) 2010;3:1084–92. doi: 10.1158/1940-6207.CAPR-10-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung W, Bondaruk J, Jelinek J, Lotan Y, Liang S, Czerniak B, et al. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev. 2011;20:1483–91. doi: 10.1158/1055-9965.EPI-11-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–78. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Carbayo M. Hypermethylation in bladder cancer: biological pathways and translational applications. Tumour Biol. 2012;33:347–61. doi: 10.1007/s13277-011-0310-2. [DOI] [PubMed] [Google Scholar]
- 42.Momparler RL. Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–51. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 43.O’Rourke CJ, Knabben V, Bolton E, Moran D, Lynch T, Hollywood D, et al. Manipulating the epigenome for the treatment of urological malignancies. Pharmacol Ther. 2013;138:185–96. doi: 10.1016/j.pharmthera.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kandimalla R, van Tilborg AA, Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nat Rev Urol. 2013;10:327–35. doi: 10.1038/nrurol.2013.89. [DOI] [PubMed] [Google Scholar]
- 45.Markl ID, Cheng J, Liang G, Shibata D, Laird PW, Jones PA. Global and gene-specific epigenetic patterns in human bladder cancer genomes are relatively stable in vivo and in vitro over time. Cancer Res. 2001;61:5875–84. [PubMed] [Google Scholar]
- 46.Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zöchbauer-Müller S, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61:8659–63. [PubMed] [Google Scholar]
- 47.Sathyanarayana UG, Maruyama R, Padar A, Suzuki M, Bondaruk J, Sagalowsky A, et al. Molecular detection of noninvasive and invasive bladder tumor tissues and exfoliated cells by aberrant promoter methylation of laminin-5 encoding genes. Cancer Res. 2004;64:1425–30. doi: 10.1158/0008-5472.CAN-03-0701. [DOI] [PubMed] [Google Scholar]
- 48.Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Miller K, et al. Regularly methylated novel pro-apoptotic genes associated with recurrence in transitional cell carcinoma of the bladder. Int J Cancer. 2006;119:1396–402. doi: 10.1002/ijc.21971. [DOI] [PubMed] [Google Scholar]
- 49.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 50.Wolff EM, Liang G, Cortez CC, Tsai YC, Castelao JE, Cortessis VK, et al. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68:6208–14. doi: 10.1158/0008-5472.CAN-07-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishiyama N, Arai E, Chihara Y, Fujimoto H, Hosoda F, Shibata T, et al. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–40. doi: 10.1111/j.1349-7006.2009.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bird A, Macleod D. Reading the DNA methylation signal. Cold Spring Harb Symp Quant Biol. 2004;69:113–8. doi: 10.1101/sqb.2004.69.113. [DOI] [PubMed] [Google Scholar]
- 54.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 55.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 57.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–19. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 63.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 64.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 66.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–97. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 69.Nagarajan RP, Fouse SD, Bell RJ, Costello JF. Methods for cancer epigenome analysis. Adv Exp Med Biol. 2013;754:313–38. doi: 10.1007/978-1-4419-9967-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Estécio MR, Issa JP. Tackling the methylome: recent methodological advances in genome-wide methylation profiling. Genome Med. 2009;1:106. doi: 10.1186/gm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Estécio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–36. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dudziec E, Goepel JR, Catto JW. Global epigenetic profiling in bladder cancer. Epigenomics. 2011;3:35–45. doi: 10.2217/epi.10.71. [DOI] [PubMed] [Google Scholar]
- 74.Kim WJ, Kim YJ. Epigenetics of bladder cancer. Methods Mol Biol. 2012;863:111–8. doi: 10.1007/978-1-61779-612-8_6. [DOI] [PubMed] [Google Scholar]
- 75.Huepper WC, Wiely FH, Wolfe HD, Ranta KE, Leming MF, Blood FR. Experimental production of bladder tumors in dogs by administration of β-naphthylamine. J Ind Hyg Toxicol. 1938;•••:46–84. [Google Scholar]
- 76.Spitz S, Maguigan WH, Dobriner K. The carcinogenic action of benzidine. Cancer. 1950;3:789–804. doi: 10.1002/1097-0142(1950)3:5<789::AID-CNCR2820030505>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 77.Walpole AL, Williams MHC, Roberts DC. Tumours of the urinary bladder in dogs after ingestion of 4-aminodiphenyl. Br J Ind Med. 1954;11:105–9. doi: 10.1136/oem.11.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wanibuchi H, Salim EI, Kinoshita A, Shen J, Wei M, Morimura K, et al. Understanding arsenic carcinogenicity by the use of animal models. Toxicol Appl Pharmacol. 2004;198:366–76. doi: 10.1016/j.taap.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 79.Kitchin KT. Recent advances in arsenic carcinogenesis: modes of action, animal model systems, and methylated arsenic metabolites. Toxicol Appl Pharmacol. 2001;172:249–61. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- 80.Besaratinia A, Pfeifer GP. Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis. 2006;27:1526–37. doi: 10.1093/carcin/bgi311. [DOI] [PubMed] [Google Scholar]
- 81.Ahmad I, Sansom OJ, Leung HY. Exploring molecular genetics of bladder cancer: lessons learned from mouse models. Dis Model Mech. 2012;5:323–32. doi: 10.1242/dmm.008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Washington MK, Powell AE, Sullivan R, Sundberg JP, Wright N, Coffey RJ, et al. Pathology of rodent models of intestinal cancer: progress report and recommendations. Gastroenterology. 2013;144:705–17. doi: 10.1053/j.gastro.2013.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langdon SP. Animal modeling of cancer pathology and studying tumor response to therapy. Curr Drug Targets. 2012;13:1535–47. doi: 10.2174/138945012803530152. [DOI] [PubMed] [Google Scholar]
- 84.U.S. Department of Health and Human Services. The health consequences of smoking: a report of the Surgeon General. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Atlanta, GA, 2004. [Google Scholar]
- 85.Boffetta P. Tobacco smoking and risk of bladder cancer. Scand J Urol Nephrol Suppl. 2008;218:45–54. doi: 10.1080/03008880802283664. [DOI] [PubMed] [Google Scholar]
- 86.Kiriluk KJ, Prasad SM, Patel AR, Steinberg GD, Smith ND. Bladder cancer risk from occupational and environmental exposures. Urol Oncol. 2012;30:199–211. doi: 10.1016/j.urolonc.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 87.Vineis P, Pirastu R. Aromatic amines and cancer. Cancer Causes Control. 1997;8:346–55. doi: 10.1023/A:1018453104303. [DOI] [PubMed] [Google Scholar]
- 88.Patrianakos C, Hoffman D. Chemical studies on tobacco smoke LXIV. On the analysis of aromatic amines in cigarette smoke. J Anal Chem. 1979;3:150–4. [Google Scholar]
- 89.D’Avanzo B, Negri E, La Vecchia C, Gramenzi A, Bianchi C, Franceschi S, et al. Cigarette smoking and bladder cancer. Eur J Cancer. 1990;26:714–8. doi: 10.1016/0277-5379(90)90124-C. [DOI] [PubMed] [Google Scholar]
- 90.Vineis P, Esteve J, Hartge P, Hoover R, Silverman DT, Terracini B. Effects of timing and type of tobacco in cigarette-induced bladder cancer. Cancer Res. 1988;48:3849–52. [PubMed] [Google Scholar]
- 91.Bartsch H, Malaveille C, Friesen M, Kadlubar FF, Vineis P. Black (air-cured) and blond (flue-cured) tobacco cancer risk. IV: Molecular dosimetry studies implicate aromatic amines as bladder carcinogens. Eur J Cancer. 1993;29A:1199–207. doi: 10.1016/S0959-8049(05)80315-6. [DOI] [PubMed] [Google Scholar]
- 92.Besaratinia A, Bates SE, Pfeifer GP. Mutational signature of the proximate bladder carcinogen N-hydroxy-4-acetylaminobiphenyl: inconsistency with the p53 mutational spectrum in bladder cancer. Cancer Res. 2002;62:4331–8. [PubMed] [Google Scholar]
- 93.Kadlubar FF, Beland FA, Beranek DT, Dooley KL, Heflich RH, Evans FE. Arylamine-DNA adduct formation in relation to urinary bladder carcinogenesis and Salmonella typhimurium mutagenesis. In: Sugimura T, Kondo S, Takebe H, eds. Environmental mutagens and carcinogens. New York, NY: Alan R. Liss, 1982:385-96. [Google Scholar]
- 94.Talaska G, al-Juburi AZ, Kadlubar FF. Smoking related carcinogen-DNA adducts in biopsy samples of human urinary bladder: identification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl as a major adduct. Proc Natl Acad Sci U S A. 1991;88:5350–4. doi: 10.1073/pnas.88.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoon JI, Kim SI, Tommasi S, Besaratinia A. Organ specificity of the bladder carcinogen 4-aminobiphenyl in inducing DNA damage and mutation in mice. Cancer Prev Res (Phila) 2012;5:299–308. doi: 10.1158/1940-6207.CAPR-11-0309. [DOI] [PubMed] [Google Scholar]
- 96.Thompson PA, DeMarini DM, Kadlubar FF, McClure GY, Brooks LR, Green BL, et al. Evidence for the presence of mutagenic arylamines in human breast milk and DNA adducts in exfoliated breast ductal epithelial cells. Environ Mol Mutagen. 2002;39:134–42. doi: 10.1002/em.10067. [DOI] [PubMed] [Google Scholar]
- 97.Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper P, Yu MC, et al. Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis. 2007;28:342–9. doi: 10.1093/carcin/bgl142. [DOI] [PubMed] [Google Scholar]
- 98.Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, et al. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–25. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- 99.Besaratinia A, Pfeifer GP. DNA damage and mutagenesis induced by polycyclic aromatic hydrocarbons. In: Luch A, ed. The carcinogenic effects of polycyclic aromatic hydrocarbons. London, UK: Imperial College Press, 2005:171-210. [Google Scholar]
- 100.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–25. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 101.Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat Rev Cancer. 2004;4:630–7. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 102.International Agency for Research on Cancer (IARC). Some aromatic amines, hydrazine and related substances, N-nitroso compounds and miscellaneous alkylating agents. 2-Naphthylamine. Lyon (France), 1974:97-111. [Google Scholar]
- 103.Parkes HG, Evans AEJ. Epidemiology of aromatic amine cancers. In: Searle CE, ed. Chemical carcinogens. Washington D.C.: American Chemical Society, 1984:277-301. [Google Scholar]
- 104.Steineck G, Plato N, Norell SE, Hogstedt C. Urothelial cancer and some industry-related chemicals: an evaluation of the epidemiologic literature. Am J Ind Med. 1990;17:371–91. doi: 10.1002/ajim.4700170310. [DOI] [PubMed] [Google Scholar]
- 105.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 107.Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 108.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–92. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 111.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 113.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 114.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 116.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 117.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 119.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- 121.Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 122.Ayala de la Peña F, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile are insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma. J Biol Chem. 2011;286:20778–87. doi: 10.1074/jbc.M110.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 124.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 126.Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–81. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Neely LA, Rieger-Christ KM, Neto BS, Eroshkin A, Garver J, Patel S, et al. A microRNA expression ratio defining the invasive phenotype in bladder tumors. Urol Oncol. 2010;28:39–48. doi: 10.1016/j.urolonc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 128.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–9. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 130.Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107:123–8. doi: 10.1038/bjc.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 132.Vogt M, Munding J, Grüner M, Liffers ST, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–22. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 133.Zhu J, Jiang Z, Gao F, Hu X, Zhou L, Chen J, et al. A systematic analysis on DNA methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS One. 2011;6:e28223. doi: 10.1371/journal.pone.0028223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 135.Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, Glover M, et al. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res. 2011;17:1287–96. doi: 10.1158/1078-0432.CCR-10-2017. [DOI] [PubMed] [Google Scholar]
- 136.Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–34. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 137.Shimizu T, Suzuki H, Nojima M, Kitamura H, Yamamoto E, Maruyama R, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol. 2013;63:1091–100. doi: 10.1016/j.eururo.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 138.Yoshitomi T, Kawakami K, Enokida H, Chiyomaru T, Kagara I, Tatarano S, et al. Restoration of miR-517a expression induces cell apoptosis in bladder cancer cell lines. Oncol Rep. 2011;25:1661–8. doi: 10.3892/or.2011.1253. [DOI] [PubMed] [Google Scholar]
- 139.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–18. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liep J, Rabien A, Jung K. Feedback networks between microRNAs and epigenetic modifications in urological tumors. Epigenetics. 2012;7:315–25. doi: 10.4161/epi.19464. [DOI] [PubMed] [Google Scholar]
- 141.Schaefer A, Stephan C, Busch J, Yousef GM, Jung K. Diagnostic, prognostic and therapeutic implications of microRNAs in urologic tumors. Nat Rev Urol. 2010;7:286–97. doi: 10.1038/nrurol.2010.45. [DOI] [PubMed] [Google Scholar]
- 142.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 143.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–78. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mund C, Lyko F. Epigenetic cancer therapy: Proof of concept and remaining challenges. Bioessays. 2010;32:949–57. doi: 10.1002/bies.201000061. [DOI] [PubMed] [Google Scholar]
- 145.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–69. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 147.Gray SG, Ekström TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 148.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 149.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–8. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 150.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–68. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 151.Mercurio C, Minucci S, Pelicci PG. Histone deacetylases and epigenetic therapies of hematological malignancies. Pharmacol Res. 2010;62:18–34. doi: 10.1016/j.phrs.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 152.Sachs MD, Ramamurthy M, Poel Hv, Wickham TJ, Lamfers M, Gerritsen W, et al. Histone deacetylase inhibitors upregulate expression of the coxsackie adenovirus receptor (CAR) preferentially in bladder cancer cells. Cancer Gene Ther. 2004;11:477–86. doi: 10.1038/sj.cgt.7700726. [DOI] [PubMed] [Google Scholar]
- 153.Pong RC, Roark R, Ou JY, Fan J, Stanfield J, Frenkel E, et al. Mechanism of increased coxsackie and adenovirus receptor gene expression and adenovirus uptake by phytoestrogen and histone deacetylase inhibitor in human bladder cancer cells and the potential clinical application. Cancer Res. 2006;66:8822–8. doi: 10.1158/0008-5472.CAN-05-4672. [DOI] [PubMed] [Google Scholar]
- 154.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 155.Quintás-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25:226–35. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 156.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–85. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 157.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 159.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 160.Robert C, Rassool FV. HDAC inhibitors: roles of DNA damage and repair. Adv Cancer Res. 2012;116:87–129. doi: 10.1016/B978-0-12-394387-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 161.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 162.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 163.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–80. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 164.Cherblanc F, Chapman-Rothe N, Brown R, Fuchter MJ. Current limitations and future opportunities for epigenetic therapies. Future Med Chem. 2012;4:425–46. doi: 10.4155/fmc.12.7. [DOI] [PubMed] [Google Scholar]
- 165.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–13. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours–lessons from the past. Nature reviews. Clin Oncol. 2013;10:256–66. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 168.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 169.Han H, Wolff EM, Liang G. Epigenetic alterations in bladder cancer and their potential clinical implications. Adv Urol. 2012;2012:546917. doi: 10.1155/2012/546917. [DOI] [PMC free article] [PubMed] [Google Scholar]