Abstract
Prions are protein conformations that “self-seed” the misfolding of their non-prion iso-forms into prion, often amyloid, conformations. The most famous prion is the mammalian PrP protein that in its prion form causes transmissible spongiform encephalopathy. Curiously there can be distinct conformational differences even between prions of the same protein propagated in the same host species. These are called prion strains or variants. For example, different PrP variants are faithfully transmitted during self-seeding and are associated with distinct disease characteristics. Variant-specific PrP prion differences include the length of the incubation period before the disease appears and the deposition of prion aggregates in distinct regions of the brain.1 Other more common neurodegenerative diseases (e.g., Alzheimer disease, Parkinson disease, type 2 diabetes and ALS) are likewise caused by the misfolding of a normal protein into a self-seeding aggregate.2-4 One of the most important unanswered questions is how the first prion-like seed arises de novo, resulting in the pathological cascade.
Keywords: yeast, prion, [PIN+], [PSI+]
Our recent article uses a yeast model system to investigate the hypothesis that one prion aggregate cross-seeds the de novo aggregation of a heterologous protein.5 In support of the hypothesis we find that different prion variants of one yeast protein preferentially promote the de novo appearance of different prion variants of a heterologous yeast protein. This would be expected if the shapes of the cross-seeding prion variants preferentially converted the heterologous prion protein into variants with conformations similar to themselves.
In this article addendum we show that the preferential induction of specific prion variants depends not only of the variant of the cross-seeding prion, but also upon the temperature of incubation during the cross-seeding.
The Yeast Model System
[PSI+] is the prion form of the translational release factor, Sup35.6 Variants of [PSI+] are frequently distinguished on the basis of the level of cellular Sup35 protein in the aggregated prion vs. soluble non-prion form.7-9 In addition, other properties can further distinguish variants that have identical levels of soluble Sup35.10 Strong [PSI+] variants have less soluble Sup35 than do weak [PSI+] variants. Thus the strong variants have less functional Sup35 termination factor available and cause readthrough of premature nonsense codon mutations (i.e., nonsense suppression) more efficiently than weak [PSI+] variants. While strong [PSI+] variants have more aggregated prion protein, the protein aggregates are composed of detergent resistant oligomers that are smaller in size than the corresponding oligomers in weak [PSI+] variants.11,12 Indeed, there are on average more prion seeds in strong [PSI+] variant cells than in weak [PSI+] variant cells.13 The larger number but smaller size of the strong [PSI+] seeds,12 is because their fibers break more easily than the weak [PSI+] fibers.14 Since prions grow at their fiber ends, strong [PSI+], grows more rapidly than weak [PSI+] and thus have less soluble/functional Sup35.15,16
[PIN+], the prion form of the Rnq1 protein,17-19 was initially uncovered by its ability to dramatically increase the de novo appearance of [PSI+] when Sup35 is overexpressed. [PIN+] variants were first distinguished on the basis of the (low, medium, high, and very high) efficiency with which they promoted [PSI+] appearance.20 [PIN+] variants also differ in the amount of aggregated vs. soluble Rnq1 protein and the size and number of fluorescent dots that appear when Rnq1:GFP is overexpressed. However, these characteristics do not correlate with the efficiency with which the variants promote the induction of [PSI+] (see Table 1).
Table 1. Distinguishing properties of low, high, and very high [PIN+] variants.
| Distinguishing characteristics of [PIN+] variants | Low [PIN+] | High [PIN+] | Very high [PIN+] | |
|---|---|---|---|---|
| Effects on [PSI+] | Efficiency of [PSI+] induction | + | +++ | ++++ |
| Predominant [PSI+] variant induced in vivo at 30 °C | weak | weak | strong | |
| Predominant [PSI+] variant induced in vivo at 4 °C | Not tested | strong | Not tested | |
| Other features | Level of soluble Rnq1 | ++ | + | +++ |
| Single (s.d.) or multi (m.d.) fluorescent Rnq1:Gfp dots | s.d. | m.d. | s.d. | |
| Average [PIN+] seed (propagon) number/cell | 25 ± 5 | 96 ± 17 | 50 ± 5 | |
| Oligomers break into sub-particles at 60 °C12 | no | yes | no |
Testing the Mechanism of Variant-Specific Cross-Seeding Efficiencies
The ability of [PIN+] to promote the de novo appearance of [PSI+] could not result from inactivation of the aggregated Rnq1 protein because a deletion of RNQ1 does not promote the efficient induction of [PSI+].21 We thus proposed that in addition to self-seeding, [PIN+] can also cross-seed the de novo conversion of the heterologous Sup35 prion protein into the [PSI+] prion.20-24 The frequency of the cross-seeding was proposed to be much less efficient than self-seeding, and to depend on the conformation of the specific [PIN+] prion variant used.19,25 Another possibility also proposed21,26 is that the [PIN+] prion aggregate binds to a prion inhibitor, titrating it away from the cytosol resulting in the more frequent conversion to the prion state.
Support for the cross-seeding model comes from in vitro demonstrations that prion fibers can promote the aggregation of heterologous prion protein.5,23,27 Furthermore, sonication of the fibers not only increases its ability to self-seed, but also enhances its ability to promote the aggregation of heterologous protein. This suggests that cross-seeding is occurring at the growing fiber ends. In addition, another group showed that low level expression of a fusion of Sup35 and Rnq1 dramatically increases the de novo appearance of [PSI+] in the presence of [PIN+].28 This can be explained by the cross-seeding model since the Rnq1 in the fusion is expected to join the [PIN+] aggregate thereby bringing the soluble Sup35 in the fusion in contact with [PIN+], where it can be cross-seeded.
Support for the chaperone titration model has come from the findings that amyloid aggregates bind to and titrate chaperones away from the cytosol.29-31 It is also possible that both mechanisms contribute to the increased de novo appearance of the heterologous prion.
In our recent article we investigated why some [PIN+] variants cause the appearance of [PSI+] more efficiently than other [PIN+] variants in terms of the cross-seeding model.5 We hypothesized that the cross-seeding event contains two steps: (1) binding of soluble Sup35 to the [PIN+] seed and (2) conversion of the bound Sup35 to the prion state (see Fig. 1). If the major difference between the efficiency of the [PIN+] variants were due to differences in the efficiency of binding of Sup35 to the [PIN+] cross-seed (step 1) we reasoned that the different [PIN+] variants would be immune-captured with Sup35 with different efficiencies. This was not the case. Furthermore the number of [PIN+] seeds per cell characteristic for each [PIN+] variant, as determined by a genetic method,32 also did not correlate with their efficiency of [PSI+] induction (Table 1). Taken together, this indicates that [PIN+] variant-specific differences are not caused by differences in the ability of Sup35 to bind to the growing fiber ends of [PIN+] aggregates.
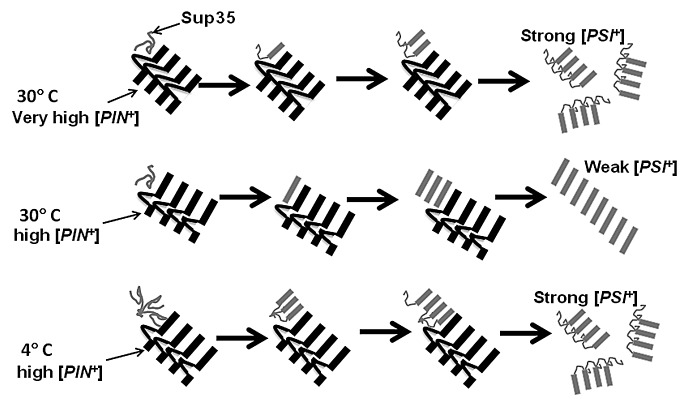
Figure 1. Model showing preferential seeding of specific [PSI+] variants by [PIN+] variants at different temperatures. Very high and high [PIN+] prion domains are cartooned as parallel in register β sheets (black bars) with non-β sheet loops in variant-specific positions. The Sup35 prion domain (gray) is preferentially cross seeded into parallel in register β sheets38 (gray bars) by different lengths of β sheet regions available in high and very high [PIN+]. In the left, Sup35 is shown to first bind to the [PIN+] aggregates (step 1). Only then does the [PIN+] amyloid region convert the Sup35 into amyloid (step 2). Due to the interference of its non-β sheet loop, very high [PIN+] preferentially seeds strong [PSI+], with a small β-sheet core which is vulnerable to sheering and the production of the smaller aggregates characteristic of strong [PSI+]. High [PIN+], which has a larger uninterrupted β-sheet region, preferentially seeds weak [PSI+] with a large β-sheet core. At 4 °C, soluble Sup35 is shown forming a nucleus that interferes with the initial β sheet conversion even when seeded by high [PIN+]. This results in the formation of a smaller β sheet-core and the induction of strong [PSI+]. Evidence for the in vitro formation of such a Sup35 nucleus at 4 °C has been presented.35
We also examined chaperones known to affect the propagation of prions, but found no differences in the levels of the chaperones Hsp104, Sis1, or Ssa1 associated with the different [PIN+] variant aggregates.33 This fails to support the titration model.
[PIN+] Variant-Specific Information is Transmitted to Newly Induced [PSI+] In Vivo
The above results suggested to us that the distinctions between the [PIN+] variants may reflect their different abilities to convert bound Sup35 into the prion conformation (step 2). If this were true, we reasoned that the [PIN+] variants may not only differ in the efficiency with which they convert soluble Sup35 to a prion, but may also differ in their preference to convert Sup35 into particular [PSI+] variants. Indeed, we found this to be the case. More weak than strong [PSI+] variants were induced in the presence of low and high [PIN+] variants, while the reverse was true in the presence of very high [PIN+] (Table 1 and Fig. 1).
To determine if the [PIN+] variant aggregates themselves were sufficient to control the spectrum of [PSI+] variants induced, we set up an in vitro system. As expected, low, high, and very high [PIN+] aggregates made in vitro promoted the conversion of soluble Sup35 to amyloid. However, when the in vitro seeded Sup35 amyloid was transformed back into [psi–] yeast,34 where the [PSI+] variant induced could be scored, we found no preference for induction of weak vs. strong [PSI+] by any of the [PIN+] variants. Clearly other cellular factors not present in the in vitro system were necessary for the variant-specificity of cross-seeding seen in vivo.
Addendum: Temperature Effects Transmission of Variant-specific Information
One environmental factor that controls the specificity of seeding in vitro is temperature. Incubation of soluble Sup35 at 4 °C in vitro in the absence of any seed, has been shown to promote the appearance of strong [PSI+], while incubation at higher temperatures preferentially causes the appearance of weak [PSI+].34 In contrast, once fibers are formed, they seed the formation of fibers with their variant specificity despite the incubation temperature.
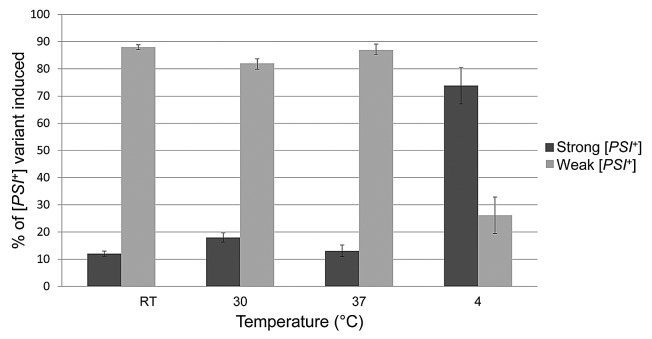
Here were show that temperature also exerts a strong effect in vivo on the variants of [PSI+] preferentially induced in the presence of high [PIN+] (Figs. 1 and 2). Weak [PSI+] appeared preferentially when cells overexpressing Sup35 were incubated at room temperature, 30°C, or 37°C. However, strong [PSI+] appeared preferentially when induction was at 4 °C.
Figure 2. Temperature affects the type of [PSI+] variant induced in vivo. [PSI+] was induced de novo in yeast strain 74D-69439 carrying the m.d. high [PIN+] variant by overexpressing the prion and middle domains of Sup35 fused to GFP under the control of a copper promoter from plasmid pCup1-SUP35NM-GFP.40 Induction was in plasmid selective glucose medium supplemented with 50 μM copper at room temperature (RT), 30 °C and 37 °C for 48 h, and at 4 °C for one week. To score for the variant of [PSI+] induced we made use of the nonsense suppressible allele, ade1–14 present in our yeast strain. Induced cells were plated and grown on complex media (YPD41). The standard colony color assay for suppression of ade1–14 was used: [psi–] colonies are red, weak [PSI+] colonies are pink, strong [PSI+] colonies are white.7 These colors reflect the level of readthrough of the ade1–14 nonsense mutation caused by the appearance of [PSI+] and inactivation of the Sup35, translational release factor. Red sectoring colonies were subcloned before scoring.
It remains to be determined if temperature (a) exerts its effect indirectly by changing the concentration of chaperones and other cellular proteins, (b) directly alters the conformation of the [PIN+] seed, or (c) alters the conformation of soluble Sup35 as depicted in Figure 1 and previously proposed to explain the in vitro results.35 Whatever the case, it is clear that both intra- and extracellular factors influence the frequency of induction of specific heterologous prion variants. While the work shown here is specific for effects of temperature, other stresses on the cell such as oxidative stress my also influence the preference for the appearance of specific prion variants. Since variants have different effects on the cells, the environment could select for variants that promote survival.
Discussion
The data presented in our recent article5 supports the cross-seeding model and suggests that variant differences occur at the second step of cross-seeding. Indeed, different [PIN+] variants appeared identical in the efficiency of the first step in cross-seeding, the binding of soluble Sup35 to [PIN+] fiber. Furthermore, cross-seeding efficiencies did not reflect differences in the average number of [PIN+] prion seeds per cell characteristic for the variant. Rather, the second step in cross-seeding, the actual conversion of the bound Sup35 to an amyloid conformation, is implicated as the mechanism of [PIN+] variant differences. In this case the shape of the cross-seeding [PIN+] variant would be intrinsically more or less capable of converting bound heterologous Sup35 into the [PSI+] prion. This is further supported by our finding that the [PIN+] prion variant used as the cross-seed affects the kinds of [PSI+] variant preferentially induced. In contrast, since none of the chaperones tested were preferentially titrated into one vs. another [PIN+] variant, there is no evidence supporting the titration model.
Interestingly, the environment also influences the type of variant formed in vivo. We demonstrated this in this addendum by altering the incubation temperature. Other cellular stressors may also influence the type of prion variant induced. If so the environment could preferentially induce prions that provide an advantage in the stress condition.
These findings are likely to be relevant to the de novo formation of amyloid aggregates of specific proteins associated with other neurodegenerative diseases.2-4 Whether amyloid aggregates of a single protein can always form distinct variants associated with different disease characteristics remains to be determined. Evidence already hints that disease associated amyloid aggregates can cross-seed the de novo appearance of amyloid aggregates associated with a different disease.36,37
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Grant R01GM056350 to SWL. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cellularlogistics/article/25698
References
- 1.Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–61. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- 2.Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto C. Transmissible proteins: expanding the prion heresy. Cell. 2012;149:968–77. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Münch C, Bertolotti A. Propagation of the prion phenomenon: beyond the seeding principle. J Mol Biol. 2012;421:491–8. doi: 10.1016/j.jmb.2011.12.061. [DOI] [PubMed] [Google Scholar]
- 5.Sharma J, Liebman SW. Exploring the basis of [PIN+] variant differences in [PSI+] induction. J Mol Biol. 2013;425:3046–59. doi: 10.1016/j.jmb.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 7.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–86. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–91. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–45. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman DA, Wickner RB. The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet. 2013;9:e1003257. doi: 10.1371/journal.pgen.1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–43. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 12.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–8. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- 13.Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. A size threshold limits prion transmission and establishes phenotypic diversity. Science. 2010;330:680–3. doi: 10.1126/science.1197785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–7. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 15.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheibel T, Kowal AS, Bloom JD, Lindquist SL. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr Biol. 2001;11:366–9. doi: 10.1016/S0960-9822(01)00099-9. [DOI] [PubMed] [Google Scholar]
- 17.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–52. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–85. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–9. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–19. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–82. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 22.Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–82. doi: 10.1016/j.jmb.2006.10.069. [PIN+] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–9. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derkatch IL, Liebman SW. Prion-prion interactions. Prion. 2007;1:161–9. doi: 10.4161/pri.1.3.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liebman SW, Bagriantsev SN, Derkatch IL. Biochemical and genetic methods for characterization of [PIN+] prions in yeast. Methods. 2006;39:23–34. doi: 10.1016/j.ymeth.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–94. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 27.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–87. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 28.Choe YJ, Ryu Y, Kim HJ, Seok YJ. Increased [PSI+] appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryot Cell. 2009;8:968–76. doi: 10.1128/EC.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–43. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Hong JY, Derkatch IL, Liebman SW. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 2013;9:e1003236. doi: 10.1371/journal.pgen.1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, et al. PolyQ Proteins Interfere with Nuclear Degradation of Cytosolic Proteins by Sequestering the Sis1p Chaperone. Cell. 2013;154:134–45. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–72. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 35.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6:225–30. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morales R, Estrada LD, Diaz-Espinoza R, Morales-Scheihing D, Jara MC, Castilla J, et al. Molecular cross talk between misfolded proteins in animal models of Alzheimer’s and prion diseases. J Neurosci. 2010;30:4528–35. doi: 10.1523/JNEUROSCI.5924-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundmark K, Westermark GT, Olsén A, Westermark P. Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: Cross-seeding as a disease mechanism. Proc Natl Acad Sci U S A. 2005;102:6098–102. doi: 10.1073/pnas.0501814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci U S A. 2008;105:2403–8. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–4. doi: 10.1126/science.7754373. [psi+] [DOI] [PubMed] [Google Scholar]
- 40.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- 41.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-V. [DOI] [PubMed] [Google Scholar]