Abstract
Introduction
Quantification of plasma viral load (PVL) is used to monitor disease progression in SIV-infected macaques. This study was aimed at optimizing of performance characteristics of the quantitative PCR (qPCR) PVL assay.
Methods
The PVL quantification procedure was optimized by inclusion of an exogenous control Hepatitis C Virus armored RNA (aRNA), a plasma concentration step, extended digestion with proteinase K, and a second RNA elution step. Efficiency of viral RNA (vRNA) extraction was compared using several commercial vRNA extraction kits. Various parameters of qPCR targeting the gag region of SIVmac239, SIVsmE660 and the LTR region of SIVagmSAB were also optimized.
Results
Modifications of the SIV PVL qPCR procedure increased vRNA recovery, reduced inhibition and improved analytical sensitivity. The PVL values determined by this SIV PVL qPCR correlated with quantification results of SIV-RNA in the same samples using the “industry standard” method of branched-DNA (bDNA) signal amplification.
Conclusions
Quantification of SIV genomic RNA in plasma of rhesus macaques using this optimized SIV PVL qPCR is equivalent to the bDNA signal amplification method, less costly and more versatile. Use of heterologous aRNA as an internal control is useful for optimizing performance characteristics of PVL qPCRs.
Keywords: MIQE, qPCR, PCR inhibition, AIDS, plasma viral load (PVL)
Introduction
Plasma viral load (PVL) is widely used to assess the status and progression of simian immunodeficiency virus (SIV) infections in nonhuman primates (NHP) [6, 27, 37]. Quantitative polymerase chain reaction (qPCR) is commonly used for determination of PVL [5, 12, 21, 28, 35, 40]. Plasma is a challenging specimen for quantification of viral RNA due to the presence of PCR inhibitors and RNAses, and removing such inhibitors improved analytical sensitivity for detection of viral RNA [9, 10, 23, 26]. Inadequate preparation of plasma, suboptimal vRNA extraction efficiency, and poor primer and probe design also each contribute to inaccurate quantification of viral copy number in plasma.
The purpose of this study was to improve sensitivity and reproducibility of SIV PVL qPCR through optimizing various multiple steps of SIV PVL qPCR (sample preparation, cDNA synthesis, and amplification) and by using heterologous armored RNA (aRNA). Armored RNA (aRNA) contains a short RNA viral sequence packaged in an Escherichia-coli bacteriophage MS2 protein coat to represent a synthetic target virus [17, 34]. Known concentrations of aRNA can be spiked into test specimens to control for yields in sample preparation and reverse transcription [15, 16, 38]. Commercially-available aRNAs have been used in human diagnostics for quality assurance in viral load determination, including human immunodeficiency virus (HIV) [4, 13, 17–20, 22, 31, 33, 34, 41, 42]. To date however, aRNA has not been used for quality control in quantification of SIV RNA [5].
Our overall approach was to optimize SIV PVL qPCR assay for (SIVmac239, SIVsmE660 and SIVagmSAB) using aRNA and an internal positive control (IPC). The aRNA for Hepatitis C Virus (HCV) was chosen as the IPC because HCV does not infect NHPs to confound data interpretation [1]. The results from the qPCR were correlated with the branched-DNA (bDNA) signal amplification and hybridization. This study was performed and results presented in compliance with the “minimum information for publication of quantitative real-time PCR experiments” guidelines (MIQE; http://www.rdml.org/miqe) [8, 24, 25, 36].
Materials and Methods
Animals and collection of plasma
Rhesus macaques (Macaca mulatta) were housed at the Tulane National Primate Research Center, Covington, LA. All procedures using these animals were performed by the clinical veterinary staff under the guidance of veterinarians and approved by the Institutional Animal Care and Use Committee (IACUC) of Tulane University in compliance with the AALAS “Policy on the Human Care and Use of Laboratory Animalscv”
Blood specimens from SIV-infected or uninfected rhesus macaques were collected in EDTA anti-coagulant and centrifuged at 1,500 × g for 10 minutes at 4°C. Plasma was aspirated, ailquoted into cryovial tubes, and stored at −80°C until extraction of viral RNA (vRNA).
Reference PVL test
For reference of comparison between the modified PVL qPCR assay procedures described here, one set of all initial plasma samples tested in the study also was evaluated by the SIV RNA 4.0 bDNA signal amplification nucleic acid probe assay (Siemens Diagnostic, Berkeley, CA) as described [3, 29, 32]. The bDNA lower limit of detection (LOD) was 125 SIV RNA copies / ml of plasma.
Plasma sample preparation
In selected experiments where indicated, armored RNA (aRNA) of Hepatitis C Virus genotype 2b (HCV-2b; cat. #42010, Asuragen; Austin, TX) containing a short RNA sequence packaged in an E. coli bacteriophage MS2 protein coat was used as the IPC to monitor RNA recovery and quantification [17, 34]. For each assay, 10 ul aRNA were added to each plasma sample, and based on the manufacturer’s information, the copy numbers fell within the dynamic range of the HCV-2b calibration curve.
Plasma samples were assayed neat or centrifuged at 25,000 × g for 1 hour to pellet virus particles [12]. After centrifugation, the excess plasma supernatant was carefully removed from the virus pellet before continuing with the vRNA extraction protocol. The centrifugation step allows for quantification of virus from higher volumes of plasma when PVL is expected to be low. To inactivate nucleases and other contaminants, proteinase K (cat. #AM2548, Life Technologies; Carlsbad, CA) was added to neat plasma or virus pellet specimens at a final concentration of 2.5 mg/ml followed by incubation at 37°C for 30 min to inactivate further proteolysis [12].
vRNA isolation
Viral RNA (vRNA) was extracted from plasma using High Pure Viral RNA kit (cat. #11858882001, Roche; Indianapolis, IN) using the manufacture’s protocol with the modification that the elution volume was increased from 50 μl to 100 μl and supplemented with RNase Inhibitor (0.3 units/μl; cat. #N8080119, Life Technologies). The increased elution volume allowed for higher vRNA recovery from the supplied spin filter tubes containing glass fiber fleece. To further improve vRNA recovery, the eluted vRNA (100 μl) samples were subjected to a second RNA extraction/elution using the RNA Clean and Concentrator kit – 25 (cat. #R1018, Zymo Research; Irvine, CA) as per the manufacture’s protocol. The final elution volume of 50 μl was supplemented with RNase Inhibitor (0.8 units/μl; Life Technologies). The final vRNA was transferred in 15 μl and 8 μl volumes to two 0.2 ml PCR tubes, (cat. #27-104, Genesee Scientific; San Diego, CA) for the SIV target and IPC reverse transcription (RT) reactions, respectively, and stored at −80°C prior to cDNA synthesis. The remaining vRNA was stored in a 1.5 ml microfuge tube (cat. #AM12450, Life Technologies) at −80°C.
Synthesis of cDNA
Reverse transcription (RT) master mix was prepared containing MultiScribe™ Reverse Transcriptase (1.25 units/μl; cat. #4311235, Life Technologies), RNase Inhibitor (0.4 units/μl; Life Technologies), Buffer II (1× final; Life Technologies), MgCl2 (6.5 nM/μl; cat. #N8080130, Life Technologies), dNTPs (2 nM/μl final; cat. #N8080260, Life Technologies), and BSA as a carrier protein (0.5 mg/ml; cat. #AM2548, Life Technologies) for vRNA cDNA synthesis in a final reaction volume of 50 μl. BSA was included to mitigate PCR inhibition caused by endogenous plasma protease activity, to minimize adsorption of template onto the tube surface, and to reduce primer dimer formation [26]. The RT master mix was dispensed into the 0.2 ml tubes containing vRNA template using an X-stream electronic repeating pipette (Eppendorf; Hauppauge, NY). Two sets of RT reactions were performed using gene-specific primers at 400 nM per reaction. In the first reaction, 15 μl vRNA were incubated with SIV-specific primers and in the second set, 8 ul vRNA were incubated with specific IPC primers (Table 1). SIV primer annealing outside the target qPCR amplicon template was designed to recognize SIVmac239, SIVmac251, and SHIVs containing the SIVmac gag gene. SIV-specific qPCR reverse primers were used to distinguish SIVagmSAB and SIVsmE660. cDNA synthesis reactions were performed in a thermocycler (9700, Life Technologies) that included an RNA denaturing step of 95°C for 10 minutes, synthesis at 48°C for 45 minutes and a final incubation at 70°C for 5 minutes. The synthesized cDNA samples were stored at −30°C until the reaction.
Table 1.
Primer and hydrolysis probe sequences of the SIV and IPC targets.
| SIVmac239 | |
| Nested Primers for Calibration Curve Construction; | |
| MAC.GAG outer For | 5′ – AGGTTACGGCCCGGCGGAAAGAAAA – 3′ |
| MAC.GAG outer Rev | 5′ – CCTACTCCCTGACAGGCCGTCAGCATTTCTTC – 3′ |
| MAC.GAG inner For | 5′ – AGTACATGTTAAAACATGTAGTATGGGC – 3′ |
| MAC.GAG inner Rev | 5′ – CCTTAAGCTTTTGTAGAATCTATCTACATA – 3′ |
| qPCR Primers and Hydrolysis Probe; | |
| SIVmac239 RT Rev | 5′ – TATGGGGTTCTGTTGTCT – 3′ |
| SIVmac239 Forward | 5′ – AGGCTGCAGATTGGGACTTG – 3′ |
| SIVmac239 Reverse | 5′ – TGATCCTGACGGCTCCCTAA – 3′ |
| SIVmac239 Probe | 5′ – FAM ACCCACAACCAGCTCCACAACAAGGAC IABKFQ – 3′ |
| SIVagmSAB | |
| Nested Primers for Calibration Curve Construction; | |
| AGM.LTR outer For | 5′ – ACTGGGCGGTACTGGGAGTGGCTT – 3′ |
| AGM.LTR outer Rev | 5′ – AACTAAGGCAAGACTTTATTGAGG – 3′ |
| AGM.LTR inner For | 5′ – ATTGAGCCTGGGTGTTCTCT – 3′ |
| AGM.LTR inner Rev | 5′ – CAAGACTTTATTGAGGCAAT – 3′ |
| qPCR Primers and Hydrolysis Probe; | |
| SIVagmSAB Forward | 5′ – ATTGAGCCTGGGTGTTCTCT – 3′ |
| SIVagmSAB Reverse | 5′ – AAGACCTCACCAGAGTGCTCTTAG – 3′ |
| SIVagmSAB Probe | 5′ – FAM TAAGTCTGAACCAGCTTGAGCCTGGGTGTT IABKFQ – |
| SIVsmE660 | |
| Nested Primers for Calibration Curve Construction; | |
| E660.GAG outer For | 5′ – TTAGGTTACGGCCCAACGGAAAGA – 3′ |
| E660.GAG outer Rev | 5′ – ATACCCAGACCCTTGAGCACCAAT – 3′ |
| E660.GAG inner For | 5′ – TTAGGTTACGGCCCAACGGAAAGA – 3′ |
| E660.GAG inner Rev | 5′ – GCTGCGGGTGTTGTAAATCCCAAT – 3′ |
| qPCR Primers and Hydrolysis Probe; | |
| SIVsmE660 Forward | 5′ – GCAGAGACATCTAGTGGTGGAAAC – 3′ |
| SIVsmE660 Reverse | 5′ – GTAATTTCCTCCTCTGCCACTAGG – 3′ |
| SIVsmE660 Probe | 5′ – FAM AATGCCAGCAACAAGCAGACCAACAGCA IABKFQ – 3′ |
| HCV-2b (IPC) | |
| qPCR Primers and Hydrolysis Probe; | |
| HCV-2b Forward | 5′ – ACACTCCGCCATGAATCACT – 3′ |
| HCV-2b Reverse | 5′ – CTGTACGACACTCATACTAACGCC – 3′ |
| HCV-2b probe | 5′ – MAX TGTCTTCACGCAGAAAGCGTCTAGCCAT IABKFQ – 3′ |
Primers and hydrolysis probe design for SIV and internal positive control (IPC) targets
The primers and hydrolysis probes for viruses containing SIVmac gag were designed and selected according to parameters defined by Integrated DNA Technologies (IDT) and the Primer Express 3.0 software (Life Technologies). The primers and hydrolysis probes for SIVagmSAB, SIVsmE660 and HCV-2b were designed and selected according to parameters defined from using Primer Quest (IDT, Coralville, IA; http://www.idtdna.com/Primerquest/Home/Index). The specificity of all the oligonucleotides described in Table 1 was examined further by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). In addition, the secondary structure of all target amplicon sequences in the gag gene or the LTR was verified in silico with UNAfold (http://www.idtdna.com/UNAFold?). The target amplicon of primers designed for each virus is described as follows:
SIVmac239
The 73 bp oligonucleotide target amplicon for the TaqMan qPCR system is located between 758 – 832 bp within the gag region (130 – 1662 bp) of the SIVmac239 genome (Genbank accession # AY587015). This conserved target sequence enables quantification of SIVmac239, SIVmac251 and SHIV viruses containing SIVmac gag.
SIVagmSAB
The 93 bp oligonucleotide target amplicon for the TaqMan qPCR system is located between 758 – 832 bp within the LTR region (9270 – 10036 bp) of the SIVagmSAB genome (Genbank accession # U04005) [14]. The LTR sequence enables quantification of SIVagmSAB and derivatives.
SIVsmE660
The 94 bp oligonucleotide target amplicon for the TaqMan qPCR system is located between 1376 – 1469 bp within the gag region (1065 – 2588 bp) of SIVsmE543 (Genbank accession # U72748) and the SIVsmE660 clone of SIVsmE543. The gag sequence enables quantification of SIVsmE543, SIVsmE660, and derivatives.
HCV-2b aRNA (IPC)
The 70 bp oligonucleotide IPC amplicon for the TaqMan qPCR system is located within the 5′UTR gene of HCV genotype 2b (http://www.asuragen.com/pdfs/Updated06Jul12_AR%20HCV2b%2042010.pdf). The sequence enables quantification for the HCV-2b aRNA positive control sequence.
The melting temperatures of the qPCR desalted primers and hydrolysis probes were 60°C and 70°C, respectively. Hydrolysis probe sequences were based on the absence of more than four identical nucleotide stretches, absence of G at the 5′ end that could contribute to quenching, and a melting temperature 10° C higher than that of the primers [7]. The fluorescence reporter dye at the 5′ end was FAM (6-carboxyfluorescein) for all of the SIV target hydrolysis probes. MAX (IDT equivalent to VIC®) was used for the IPC (HCV-2b) hydrolysis probe and a black hole quencher dye was applied at the 3′ end of Iowa black FQ quencher (IABkFQ) as a non-fluorescent chromophore, for all hydrolysis probes.
Exogenous Calibration Curves
Amplicons from ssRNA of test viruses were prepared to generate the exogenous calibration curves. EDTA-preserved blood was obtained at necropsy from experimentally-infected rhesus macaques exhibiting high viremia with SIVmac239 (animal CL41), SIVagmSAB (animal JT08), and SIVsmE660 (animal BI87). Extracted and purified vRNA was used for nested RT-PCR targeting the respective sequences to create a linearized DNA templates for SIVmac239 (836 bp), SIVagmSAB (273 bp), and SIVsmE660 (606 bp). Nested RT-PCR amplifications for both the first RT-PCR and second PCR were performed in 50 μl volume reactions containing 400 ng of forward and reverse primers for amplification using a 9700 Thermocycler (Life Technologies). The RT-PCR master mix and PCR master mix were dispersed into the 0.2 ml tubes containing vRNA or cDNA, respectively. In the RT-PCR, 5 μl of vRNA was assayed with a 1 Step RT-PCR Kit (cat. #210212; Qiagen, Valencia, CA) with the outer primer pairs. Reverse transcription included an RNA denaturing step of 95°C for 30 min and synthesis at 50°C for 15 min, followed by 40 cycles of amplification at 94°C for 30 sec, 60–65°C for 45 sec, and 72°C for 1 min, with a final reaction at 72°C for 10 min. In the PCR test, 5 μl of cDNA was assayed in Master Mix (1× final; cat. #M7502; Promega, Madison, WI) and included the inner primer sets. The hot start PCR incubation program was 91°C for 1 min followed by 40 cycles at 94°C for 30 sec, 60–65°C for 45 sec, 72°C for 1 min and a final step at 72°C for 10 min.
HCV-2b RNA was released from MS2 pseudocapsids by heating to 70°C for 3 minutes. The HCV-2b RNA transcript was converted to cDNA by a single RT-PCR reaction (described above) and amplified with a further 40 PCR reactions (described above) to create linearized DNA templates. The nested RT-PCR (SIV targets) and PCR (HCV-2b) products were visualized after electrophoresis in 1.5% low melt agarose (cat. #16520050, Life Technologies) gels and then purified with Zymoclean™ Gel DNA Recovery Kit (cat. #D4001, Zymo Research) according to the manufacturer’s instructions. A T7 promoter sequence (5′ – GGATCCTAATACGACTCACTAT – 3′) was added to the forward qPCR primers of each amplicon to construct this ssRNA exogenous calibration curve. The RNA calibration curve was produced by in vitro transcription using the T7 RNA polymerase with T7 RiboMAX Express Large Scale RNA Production System (cat. #P1320, Promega). RNA transcript concentrations were calculated using an Epoch Microplate Spectrophotometer (BioTek; Winooski, VT).
The exogenous calibration curves were generated by plotting Cq values against log-transformed concentrations of serial tenfold (log) dilutions of target nucleic acid from purified single-stranded RNA (ssRNA) transcripts of the complete qPCR target amplicons. The calibration curves were used for interpolating quantitative information of the target in an unknown sample and were used to determine the dynamic detection range of the targets [11, 39]. In addition, the exogenous calibration curves were assayed to define qPCR reaction sensitivity or the limit of detection (LOD) as “the lowest amount of analyte in a sample that can be detected” [2, 30]. The LOD was based on three or more replicates and calculated using GenEx 5 (www.multid.se).
Quantification of SIV PVL by qPCR
SIV target cDNA (14 μL in a 96-well format or 5.6 μl in a 384-well microtiter plate format) and exogenous control cDNA (5 μL and 2 μl for 96- and 384-well microtiter plate formats, respectively) were assayed in duplicate. TaqMan Gene Expression Master Mix (cat. #4369510, Life Technologies), amplification primers (250 nM) and the hydrolysis probe (900 nM; Life Technologies) were added using an X-stream repeating pipette (Eppendorf). Reaction volumes were 50 μl in 96-well optical plates (cat # N8010560, Life Technologies) or 20 μl in 384-well optical plates (cat #4309849, Life Technologies). Microtiter plates were covered with optical film (cat. #4311971, Life Technologies) vortexed (MixMate; Eppendorf), and pulse centrifuged (Centrifuge 5430; Eppendorf). Amplification and detection of target sequence fluorescence probe signals were detected on 7900HT (96-well format) or QuantStudio 12K Flex (384-well format) Sequence Detectors (Life Technologies) using a program of 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Data were captured and analyzed with Sequence Detector Software (Life Technologies). Viral copy numbers were determined by plotting Cq values obtained from unknown (i.e. test) samples against the exogenous calibration curves generated from known amounts of in vitro-transcribed RNA and further normalized from known copies of spiked aRNA to determine the number of virus copies per ml of initial specimen sample or test dilution, as applicable. The baseline was adjusted from 15 to 10 Cq based on the 108 copy number of the calibration curve intersecting with the threshold line at 12 (+/− 0.5) cycles.
Assay Quality Control
To control for assay variation, validation of results, IPC quantification consistency, and reagent stability, replicates of an SIV-positive control plasma sample also were analyzed in parallel with every set of test samples. SIV-positive control samples also were used to verify that the PCR master mix and reagents were prepared correctly to produce amplification of the target nucleic acid. Negative control plasma samples were included for processing with every set of test samples to monitor cross contamination. A non-amplification control (NAC) was processed with the RT reactions to ensure absence of template contamination. A non-template control (NTC) was included in the qPCR reactions to ensure that there was no cross contamination between reactions.
Statistical analysis
GraphPad InStat and Prism programs for Windows (GraphPad Software, San Diego California USA, www.graphpad.com) were used to perform statistical analyses and graph results, respectively. Analyses used included linear regression, Analysis of Covariance (ANCOVA) to compare linear slopes and intercepts, and Student’s t Test or ANOVA to compare means between two or more groups, respectively. Efficiency (E) was calculated as E = 10(−1/slope) from the slope of the standard curve and percent efficiency (E%) = (E – 1 ) × 100.
Results
The purpose of this study was to improve quantification of SIVmac, SIVagm, and SIVe660 in plasma by qPCR using modification outlined in Figure 1. Initial steps to improve RNA purification included comparison of commercial RNA extraction kits, introduction of proteinase K and an additional elution step to remove potential inhibitors and increase RNA recovery. For normalization and quality control, aRNA was introduced to the specimen and a centrifugation step was added to increase the volume of plasma that could be assayed to concentrate virus and thus increase assay analytical sensitivity. Commercially-available reverse transcriptases, addition of BSA as a carrier, and increasing MgCl2 concentrations also were tested for improving cDNA synthesis. Results from the virus standard exogenous calibration curves were further analyzed to define sensitivity, repeatability, and quality assurance of the overall SIV PVL qPCR assay
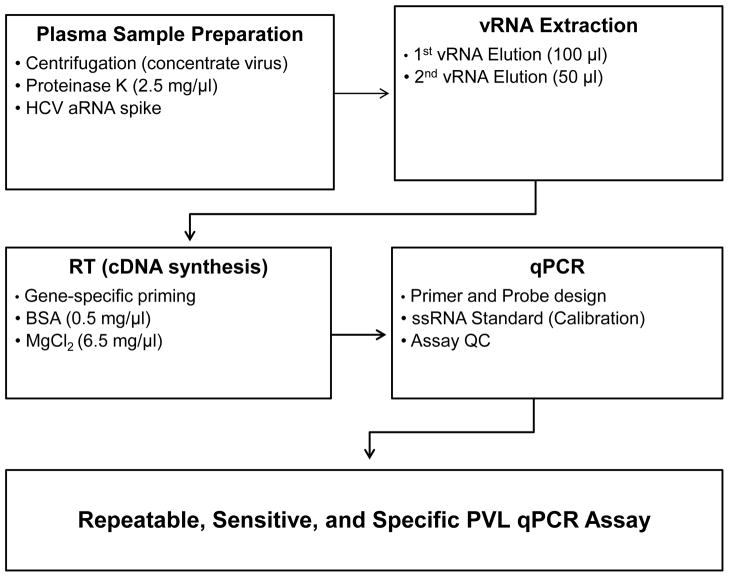
Figure 1.
Outline of assay for SIV PVL quantification. Modifications applied for plasma sample preparation, virus RNA extraction, reverse transcription to synthesize cDNA, and qPCR are described in detail in the Materials and Methods Section. The resulting optimal conditions are described in the Results.
vRNA extraction
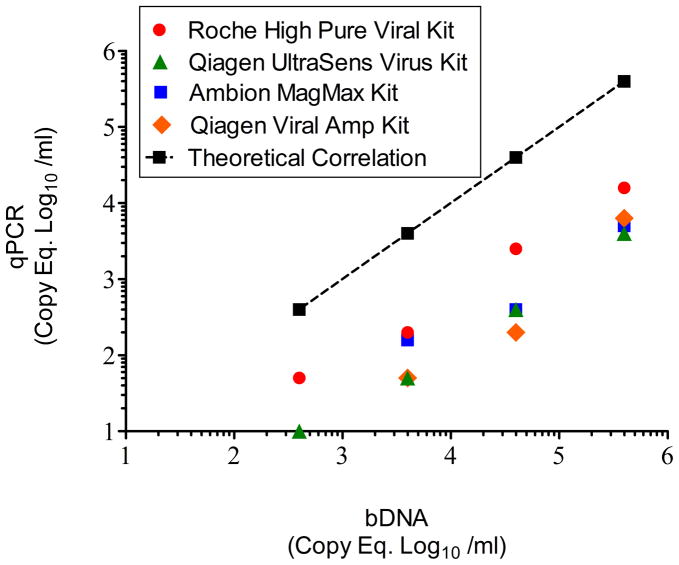
The High Pure Viral RNA kit (Roche), Viral Amp kit (Qiagen; Valencia, CA), Ultrasens Viral kit (Qiagen), and MagMAX Viral kit (Life Technologies) were compared for vRNA extraction using the manufacture’s procedure from serially-diluted plasma recovered from rhesus macaques infected with SIVmac239 (animal ID # CN79) followed by RT and qPCR prior to implementation of modification. The qPCR results from each dilution were plotted on the y axis against bDNA PVL (Siemens Healthcare) results on the x axis (Fig. 2). There was no significant difference in copy equivalents (Eq) at each dilution between the commercial extraction kits. Use of the Roche High Pure Viral Kit, however, consistently produced the higher recovery and was used in the following experiments. The results also demonstrated that the unmodified RT and qPCR procedure using the Roche High Pure Viral Kit produced results below the theoretical correlation of equality with the bDNA results.
Figure 2.
Comparison of vRNA extraction kits. SIVmac239-viremic plasma was evaluated by bDNA. Serial 10-fold dilutions of the plasma were then subjected to the four commercially-available vRNA extraction kits. The the log10-transformed qPCR SIV copy equivalents (Eq) per ml at each dilution were then plotted against the theoretical bDNA values (based on the initial neat plasma SIV PVL). The theoretical linear regression for equal correlation with bDNA was plotted (dotted line). Although the highest recovery was observed with the Roche High Pure Viral kit, there was no significant differences in the slopes or axis intercepts between the four extraction kits by ANCOVA analysis. By ANCOVA, the linear regression and lines comparing the unmodified qPCR using the Roche High Pure Viral (extraction) kit and bDNA were parallel (i.e. no significant difference in slopes) but the linear elevation (intercept) of the qPCR results was significantly lower (P < 0.0001) than the bDNA results.
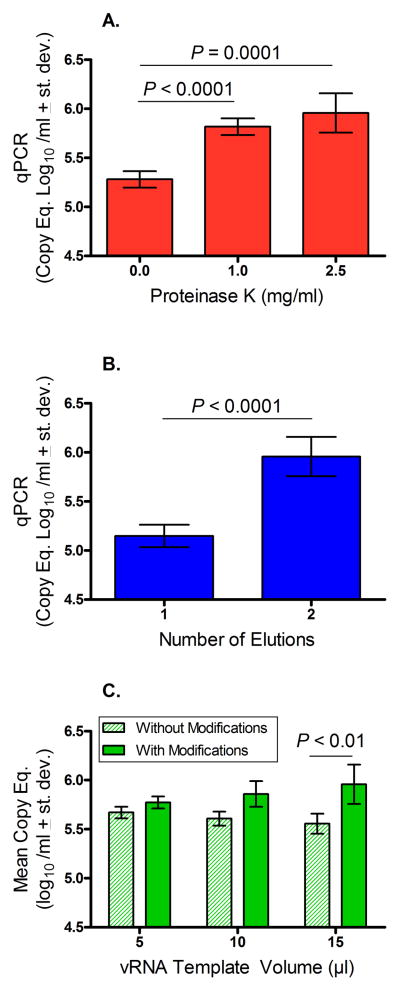
In an attempt to further improve extraction of vRNA for RT and qPCR, proteinase K was added to SIVmac239 viremic plasma specimens. The results demonstrated that a significantly higher PVL was detected by qPCR after incubation with Proteinase K at either 1.0 or 2.5 mg/ml compared to untreated plasma samples (P < 0.001; P = 0.0001, respectively; Fig 3A).
Figure 3.
Effects of modifications applied to vRNA extraction and isolation. Panel A. Addition of Proteinase K at 1.0 and 2.5 mg / ml to SIVmac239-containing plasma followed by extraction using the Roche High Pure Viral Kit, cDNA synthesis (using 15 μl vRNA template), and qPCR produced significantly higher SIV PVL results compared to untreated plasma specimens (P < 0.0001, P = 0.0001, respectively). Panel B. Viral RNA recovered after a second elution step that was reverse trasnscribed to cDNA followed by qPCR produced significantly higher SIV PVL results compared to using vRNA from the single elution step in the Roche High Pure Viral Kit (P < 0.0001). Panel C. Viremic plasma was subjected to both Proteinase K digestion and a second elution step to purify vRNA. Different volumes of vRNA template from the modified versus unmodified extraction methods were reverse transcribed for cDNA synthesis followed by qPCR. Although not significant at lower volumes of vRNA, addition of 15 μl of vRNA to the RT followed by qPCR resulted in a significantly higher SIV PVL recovery after application of the dual modification extraction procedure than from the non-modified extraction (P< 0.01). Student’s t Test was used to compare means between groups.
The extracted vRNA then was analyzed using a NanoDrop 1000 (Wilmington, DE) and the spectral profile based on published information (http://nanodrop.com/Library/T042-NanoDrop-Spectrophotometers-Nucleic-Acid-Purity-Ratios.pdf) indicated possible contamination with residual guanidine that could interfere with downstream qPCR. To overcome this possibility, the eluted vRNA from the High Pure Viral RNA kit was subjected to a second elution using the RNA Clean & Concentrator −25 kit (Zymo Research). The purified vRNA was analyzed again on the NanoDrop and no guanidine contamination appeared to remain (data not shown). This additional elution step also increased the concentration of vRNA and reduced the elution volume from 100 μl to 50 μl. The results in Fig 3B show that the additional RNA elution step also significantly improved the PVL results as measured by qPCR (P < 0.0001). The two modifications of treating the plasma virus specimen with Proteinase K at 2.5 mg/ml and the second elution procedure were then applied together (Fig 3C). There were no significant differences in mean copy equivalents of SIV PVL when comparing the modified versus non-modified extraction protocol after adding 5 or 10 μl of vRNA template to the RT followed by qPCR. Interestingly, however, the combined modification significantly increased SIV PVL recovery by qPCR (P < 0.01) after adding 15 μl of vRNA template to the RT suggesting that this method reversed the effects of PCR inhibitors that may be carried over when a higher volume of vRNA template is used for amplification by qPCR.
Synthesis of cDNA
To optimize the synthesis of cDNA from vRNA during the RT step, different sources of reverse transcriptase were compared using the SIVmac239 ssRNA standard template. There was no significant difference observed between the seven enzymes (Table 2). For the subsequent experiments, the Multiscribe enzyme (Life Technologies) was used because the instructions for this enzymes use a one-step protocol compared to the other enzymes that required at least one additional step. Addition of BSA as a protein carrier stabilizer and increasing the concentration of MgCl2 to 6.5 nM / μl during the RT procedures also improved the downstream qPCR results compared to RT reactions that lacked BSA and used MgCl2 at the lower concentration of 5.5 nM / μl (data not shown).
Table 2.
Comparison of reverse transcription (RT) enzymes by qPCR efficiency
| RT enzyme | Vendor | E (%) | Slope | y-intercept | R2 |
|---|---|---|---|---|---|
| MultiScribe | Life Technologies | 101 | −3.29 | 39.76 | 0.9992 |
| 101 | −3.29 | 39.10 | 0.9994 | ||
| 101 | −3.30 | 39.30 | 0.9998 | ||
| 101 | −3.29 | 38.56 | 0.9998 | ||
| High Capacity | Life Technologies | 112 | −3.06 | 38.89 | 0.9960 |
| HIV (AMV) | Life Technologies | 101 | −3.31 | 38.88 | 0.9995 |
| Super Script III | Life Technologies | 104 | −3.24 | 38.40 | 0.9989 |
| Transcriptor | Roche | 104 | −3.22 | 38.55 | 0.9997 |
| Smart MMLV | CloneTech | 103 | −3.26 | 38.66 | 0.9996 |
| Smart Scribe | CloneTech | 104 | −3.24 | 38.40 | 0.9989 |
Accommodation for testing higher plasma volumes
The commercial viral RNA extraction kit protocol uses 0.2 ml of plasma. To improve sensitivity of viral detection, especially when SIV PVL is likely to be low, a centrifugation step was included so that virus could be concentrated from higher volumes of plasma. To test this, serial dilutions of 0.5 ml and 1.5 ml volumes of SIV-containing plasma were subjected to an initial centrifugation of 25,000 × g for one hour to pellet the virus. After removing the supernatants, the virus pellets in a final volume of 0.2 ml were extracted for vRNA and subjected to RT and qPCR for comparison with bDNA results. Linear regression and comparison of lines by ANCOVA indicated that there was no significant difference in viral copy levels between plasma input volumes of 1.5 ml versus 0.5 ml (P = 0.7599; Fig. 4). In addition, the results demonstrated that centrifugation of plasma increased sensitivity of detection by at least one tenfold dilution using the higher initial plasma volume of 1.5 ml compared to 0.5 ml.
Figure 4.
Effect of increased plasma volume and centrifugation to concentrate SIV. Viremic plasma from SIVmac239-infected plasma was serially diluted ten-fold and volumes of 0.5 ml and 1.5 ml were subjected to centrifugation prior to vRNA extraction, cDNA synthesis and qPCR. SIV copy equivalents (Eq) per ml of each dilution were compared against the bDNA copy Eq. By linear regression, results of SIV PVL exhibited high goodness of fit (R2) values using 0.5 ml and 1.5 ml initial volumes of viremic plasma that were statistically significant. By ANCOVA, there was no significant difference in slope or intercept between linear regression values of results in serially diluted plasma and application of 0.5 ml versus 1.5 ml plasma volumes to the extraction procedure indicating that increased plasma volume and centrifugation retain qPCR output fidelity.
Addition of an exogenous aRNA control
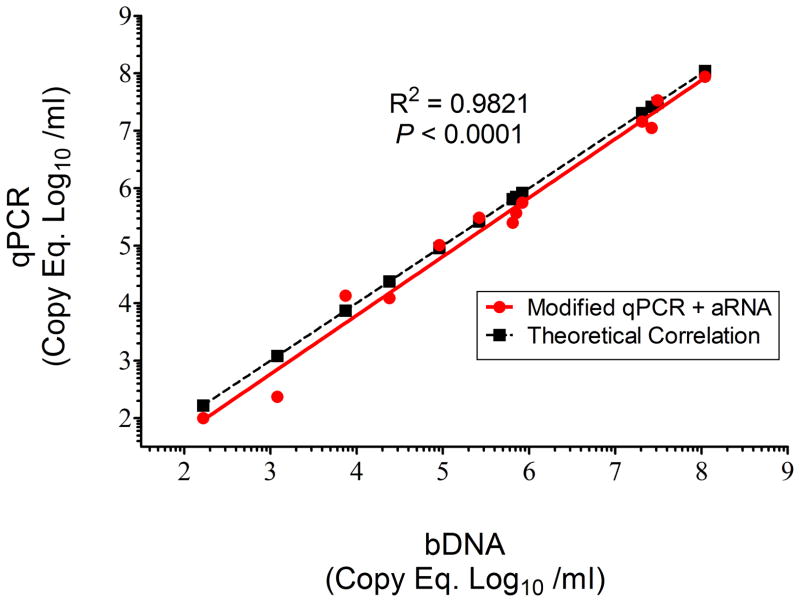
For quality control a HCV aRNA was added to neat plasma prior to initial centrifugation and nucleic acid extraction to quantify vRNA recovery, distinguish true target negatives from PCR inhibition and account for any inefficiency that may occur during each step in the protocol. To test this, a known concentration of aRNA based on the lot information provided by the manufacturer, was added to 13 different plasma samples from rhesus macaques infected with SIVmac239 (as the target virus) followed by vRNA extraction and RT using the modifications described above followed by qPCR. The results were correlated to bDNA PVL levels and as shown in Fig 5, a high goodness of fit was demonstrated (R2 = 0.9821; P < 0.0001) between these two methods. By ANCOVA, there was no significant difference between slopes of the linear regression of the modified plus aRNA qPCR results compared to the theoretical correlation of equality linear regression of bDNA results (P = 0.58).
Figure 5.
Addition of HCV aRNA for quality assurance. The aRNA was added to serial ten-fold dilutions of SIVmac239-containing plasma followed by vRNA extraction, RT cDNA synthesis and qPCR. SIV copy Eq /ml of each dilution were determined and plotted against the bDNA-determined SIV PVL. The results demonstrated a high goodness of fit (R2 = 0.9821; P < 0.001) after addition of aRNA. Furthermore, theoretical concordance with bDNA was plotted (dotted line) and no significant differences were measured between bDNA and the qPCR plus aRNA procedures (P = 0.58).
Exogenous Calibration Curves
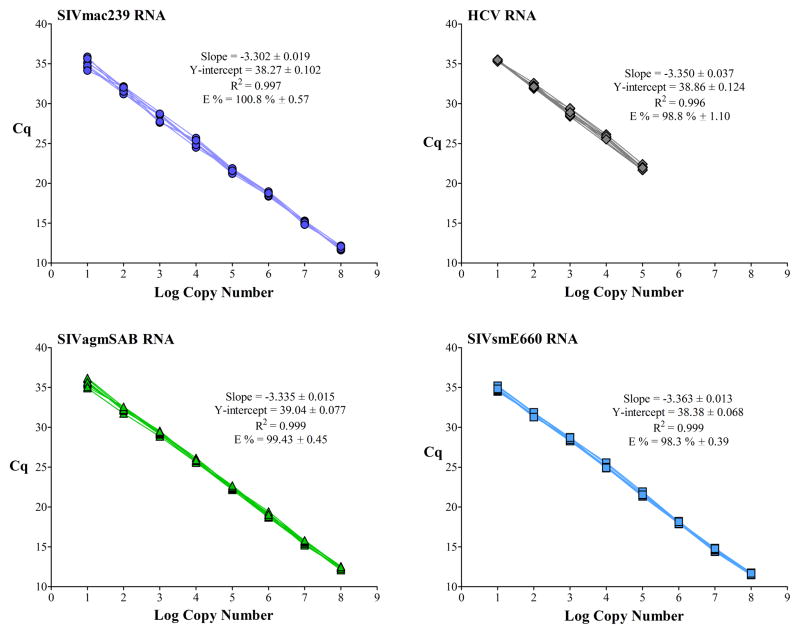
Serial ten-fold dilutions of amplicon stocks produced from ssRNA transcripts of the HCV aRNA, SIVmac239, SIVagmSAB, and SIVsmE660 were used to generate the four calibration curves ranging from 101 to 108 log copy numbers for interpolation of results from test specimens. Results of the calibration curves demonstrated highly reproducible slopes with a mean PCR efficiency (E %) of 100.2% (n = 121). Intra-assay variation or repeatability of eight replicate exogenous calibration curves for each target were analyzed by linear regression plots to verify the overall performance of each calibration assay (Fig 5). There were no significant differences in between any sets of linear regression coefficients within each amplicon set and R2 goodness of fit values were statistically significant (P < 0.0001) within each calibration curve. The coefficient of variance (CV) percent values were below 2% for each of the exogenous calibration curves further corroborating reproducibility within each amplicon calibration curve (Table 3).
Table 3.
Coefficient of variance (CV) of the exogenous calibration curves.
| CV (%) 1
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Virus | 108 | 107 | 106 | 105 | 104 | 103 | 102 | 101 |
|
|
|
|||||||
| MAC | 1.5% | 1.0% | 1.2% | 1.1% | 1.8% | 1.9% | 1.9% | 1.7% |
| HCV | n.a.2 | n.a. | n.a. | 1.5% | 1.4% | 1.6% | 0.8% | 0.6% |
| AGM | 1.3% | 1.4% | 1.4% | 0.7% | 0.9% | 0.8% | 0.8% | 1.2% |
| E660 | 0.9% | 1.1% | 0.8% | 0.8% | 1.0% | 0.7% | 0.6% | 0.8% |
CV % values were determined from eight replicates of each viral amplicon in serial ten-fold dilutions of the exogenous calibration curves.
n.a.; not applicable
Assay repeatability and sensitivity
To test the repeatability of the qPCR procedure, aRNA was spiked into two plasma specimens from viremic rhesus macaques infected with SIVmac239 and assayed multiple times. Results in Table 4 show that the PVL results exhibited low standard deviations and CV % values indicating high repeatability or consistency within each sample test. No significant differences were observed between the qPCR versus bDNA results (data not shown) further indicating that the modified qPCR method is comparable to bDNA analysis.
Table 4.
Assay reproducibility of the RT and qPCR procedure1.
| Plasma Sample | n | Mean viral log10 Copies (Eq / ml ± st. dev.) | CV (%) |
|---|---|---|---|
| 1 | 109 | 7.2 ± 0.247 | 3.4 |
| 2 | 81 | 7.4 ± 0.236 | 3.2 |
Plasma specimens also were analyzed by bDNA and the PVL results were not significantly different from the results by RT and qPCR.
The LOD or sensitivity of the assay was calculated to be 3.5 copies per reaction using GenEx 5 (www.multid.se) based on 3 or more replicates. Serially-diluted plasma samples in initial plasma volumes of 1.5 ml or 0.5 ml that were concentrated by centrifugation prior to vRNA extraction also were tested and compared to define assay sensitivity in relation to input volume of plasma. The lower limits of quantification (LOQ) were calculated as 28 and 83 viral copies eq/ml from 1.5 ml and 0.5 ml of input plasma volumes, respectively.
Discussion
This report describes the optimization of RT and qPCR for quantification of SIV in rhesus macaque plasma specimens in accordance with MIQE guidelines (30–33). Modifications that improved analytical sensitivity and reversed the effects of potential PCR inhibitors for extracting vRNA using commercial kits included centrifugation to concentrate virus, addition of Proteinase K, and a second elution step to increase vRNA recovery. Incorporation of an exogenous aRNA controlled for extraction and RT in the final qPCR. The RT reaction also was improved by addition of BSA protein carrier that helped stabilize the reaction and increased MgCl2. The calibration curves for each of the amplicons generated from ssRNA virus targets further demonstrated repeatability and high efficiency to enable reliable interpolation of SIV PVL results.
Studies utilizing RT and qPCR typically include a reference gene target to normalize gene expression such as a constitutive “housekeeping” gene that is expressed at relatively constant levels under all conditions. In human clinical studies that apply RT and qPCR for measuring viral levels in plasma, aRNA has been used instead for quality assurance to provide improved analytical sensitivity, specificity, and technical reproducibility [4, 13, 18, 22]. The results presented here shows that aRNA designed for human diagnostics can be used to improve quality control in studies performed in nonhuman primates, as well. This study demonstrated that the addition of the aRNA as an IPC at a known concentration to plasma is a useful diagnostic tool that can be applied to the quantification of plasma viral load of SIV in NHP plasma.
This modified qPCR plus aRNA applies the same volume (0.5ml) of concentrated virus from plasma as used in the commercially-available bDNA procedure. Addition of aRNA allowed for quality control during the stages of vRNA extraction, RT, and qPCR to enable accurate viral load quantification. Virus-specific qPCR primers and hydrolysis probes for each of the SIV target amplicons also contributed to ensure high PCR efficiencies (E %) that improved assay sensitivity and precise assessment of viral loads. The RT and qPCR method described here improved the LOD to 28 viral copy Eq / ml by allowing for concentration of virus from a higher initial plasma volume of 1.5 ml compared to the LOD of 83 viral copy Eq / ml from 0.5 ml plasma. This is important in studies requiring accurate and sensitive quantification of SIV when low copy numbers are anticipated.
The current study applied MIQE guidelines to establish and optimize vRNA extraction, RT and qPCR detection of SIV levels in NHP plasma samples compared with the industry standard bDNA testing. These considerations included analytical sensitivity, specificity, and repeatability. Advantages of the qPCR include target specificity and time efficiency since the qPCR can be completed in one day. The principles included in this modified qPCR using aRNA should be beneficial for optimizing and quantifying additional SIV stains as well as other RNA viruses. In addition, the principles outlined in this study have been applied to the quantification of viral RNA and proviral DNA targets in tissues and cells during experimental SIV infection of NHP. With further improvements, this method is expected to provide the foundation to apply digital PCR (dPCR) to further increase analytical sensitivity of the assay to detecting a single copy of a target.
Figure 6.
Calibration curves for three strains of SIV and the HCV aRNA. Amplicons from ssRNA templates of SIVmac239, SIVagmSAB, SIVsmE660, and HCV (aRNA) at concentrations ranging from 101 – 108 copies were assayed by qPCR to generate calibration curves. Eight replicate sets were tested for each amplicon and the results demonstrated repeatability based on high efficiency (E%) within the recommended range of 90 – 105 % and high goodness of fit above 0.98 from testing the amplicons in a range of at least 6 logs of dilution [24].
Acknowledgments
The authors gratefully acknowledge funding from the NIH grant OD011104 to the Tulane National Primate Research Center.
References
- 1.Abee CR. Nonhuman Primates in Biomedical Research. Amsterdam: Elsevier Academic Press; 2012. [Google Scholar]
- 2.Armbruster DA, Tillman MD, Hubbs LM. Limit of detection (LQD)/limit of quantitation (LOQ): comparison of the empirical and the statistical methods exemplified with GC-MS assays of abused drugs. Clinical chemistry. 1994;40:1233–1238. [PubMed] [Google Scholar]
- 3.Baumeister MA, Zhang N, Beas H, Brooks JR, Canchola JA, Cosenza C, Kleshik F, Rampersad V, Surtihadi J, Battersby TR. A sensitive branched DNA HIV-1 signal amplification viral load assay with single day turnaround. PLoS One. 2012;7:e33295. doi: 10.1371/journal.pone.0033295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beld M, Minnaar R, Weel J, Sol C, Damen M, van der Avoort H, Wertheim-van Dillen P, van Breda A, Boom R. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. Journal of clinical microbiology. 2004;42:3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry N, Herrera C, Cranage M. Detection, quantification, and characterisation of HIV/SIV. Methods in molecular biology. 2011;665:133–160. doi: 10.1007/978-1-60761-817-1_9. [DOI] [PubMed] [Google Scholar]
- 6.Brown CR, Czapiga M, Kabat J, Dang Q, Ourmanov I, Nishimura Y, Martin MA, Hirsch VM. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. Journal of virology. 2007;81:5594–5606. doi: 10.1128/JVI.00202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin SA. A-Z Quantitative PCR. La Jolla, CA: International University Line; 2004. [Google Scholar]
- 8.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 9.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. Journal of biomolecular techniques : JBT. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler DP, Wagnon CA, Bolton H., Jr Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Applied and environmental microbiology. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chervoneva I, Hyslop T, Iglewicz B, Johns L, Wolfe HR, Schulz S, Leong E, Waldman S. Statistical algorithm for assuring similar efficiency in standards and samples for absolute quantification by real-time reverse transcription polymerase chain reaction. Analytical biochemistry. 2006;348:198–208. doi: 10.1016/j.ab.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 12.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. Journal of medical primatology. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 13.Dingle KE, Crook D, Jeffery K. Stable and noncompetitive RNA internal control for routine clinical diagnostic reverse transcription-PCR. Journal of clinical microbiology. 2004;42:1003–1011. doi: 10.1128/JCM.42.3.1003-1011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diop OM, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin MC. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. Journal of virology. 2000;74:7538–7547. doi: 10.1128/jvi.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics : JMD. 2008;10:203–211. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donia D, Divizia M, Pana A. Use of armored RNA as a standard to construct a calibration curve for real-time RT-PCR. Journal of virological methods. 2005;126:157–163. doi: 10.1016/j.jviromet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Dreier J, Stormer M, Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. Journal of clinical microbiology. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drosten C, Seifried E, Roth WK. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. Journal of clinical microbiology. 2001;39:4302–4308. doi: 10.1128/JCM.39.12.4302-4308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotsch A, Schubert A, Bombis A, Wiedmann M, Zauke M, Schorling S. Nuclease-resistant single-stranded DNA controls for nucleic acid amplification assays. Journal of clinical microbiology. 2007;45:2570–2574. doi: 10.1128/JCM.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hietala SK, Crossley BM. Armored RNA as virus surrogate in a real-time reverse transcriptase PCR assay proficiency panel. Journal of clinical microbiology. 2006;44:67–70. doi: 10.1128/JCM.44.1.67-70.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, Ruprecht RM. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one-versus two-enzyme systems. AIDS research and human retroviruses. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Yang CM, Wang LN, Meng S, Deng W, Li JM. A novel real-time multiplex reverse transcriptase-polymerase chain reaction for the detection of HIV-1 RNA by using dual-specific armored RNA as internal control. Intervirology. 2008;51:42–49. doi: 10.1159/000119119. [DOI] [PubMed] [Google Scholar]
- 23.Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC research notes. 2008;1:70. doi: 10.1186/1756-0500-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson G, Nolan T, Bustin SA. Real-time quantitative PCR, pathogen detection and MIQE. Methods in molecular biology. 2013;943:1–16. doi: 10.1007/978-1-60327-353-4_1. [DOI] [PubMed] [Google Scholar]
- 25.Johnson GL, Bibby DF, Wong S, Agrawal SG, Bustin SA. A MIQE-compliant real-time PCR assay for Aspergillus detection. PloS one. 2012;7:e40022. doi: 10.1371/journal.pone.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and environmental microbiology. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EM, Chung HK, Livesay J, Suschak J, Finke L, Hudacik L, Galmin L, Bowen B, Markham P, Cristillo A, Pal R. Molecular methods for evaluation of virological status of nonhuman primates challenged with simian immunodeficiency or simian-human immunodeficiency viruses. Journal of virological methods. 2010;163:287–294. doi: 10.1016/j.jviromet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS research and human retroviruses. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- 29.Ling B, Veazey RS, Hart M, Lackner AA, Kuroda M, Pahar B, Marx PA. Early restoration of mucosal CD4 memory CCR5 T cells in the gut of SIV-infected rhesus predicts long term non-progression. Aids. 2007;21:2377–2385. doi: 10.1097/QAD.0b013e3282f08b32. [DOI] [PubMed] [Google Scholar]
- 30.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic acids research. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. Journal of clinical microbiology. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith SM, Holland B, Russo C, Dailey PJ, Marx PA, Connor RI. Retrospective analysis of viral load and SIV antibody responses in rhesus macaques infected with pathogenic SIV: predictive value for disease progression. AIDS Res Hum Retroviruses. 1999;15:1691–1701. doi: 10.1089/088922299309739. [DOI] [PubMed] [Google Scholar]
- 33.Sninsky JJ, Innis Michael A, Gelfand David H. Pcr Applications: Protocols for Functional Genomics. San Diego: Academic Press; 1999. [Google Scholar]
- 34.Stevenson J, Hymas W, Hillyard D. The use of Armored RNA as a multi-purpose internal control for RT-PCR. Journal of virological methods. 2008;150:73–76. doi: 10.1016/j.jviromet.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suryanarayana K, Wiltrout TA, Vasquez GM, Hirsch VM, Lifson JD. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS research and human retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 36.Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods. 2010;50:S1–5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Ten Haaft P, Verstrepen B, Uberla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. Journal of virology. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WalkerPeach CR, Winkler M, DuBois DB, Pasloske BL. Ribonuclease-resistant RNA controls (Armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clinical chemistry. 1999;45:2079–2085. [PubMed] [Google Scholar]
- 39.White HE, Hedges J, Bendit I, Branford S, Colomer D, Hochhaus A, Hughes T, Kamel-Reid S, Kim DW, Modur V, Muller MC, Pagnano KB, Pane F, Radich J, Cross NC, Labourier E. Establishment and Validation of Analytical Reference Panels for the Standardization of Quantitative BCR-ABL1 Measurements on the International Scale. Clinical chemistry. 2013 doi: 10.1373/clinchem.2012.196477. [DOI] [PubMed] [Google Scholar]
- 40.Whitney JB, Luedemann C, Bao S, Miura A, Rao SS, Mascola JR, Letvin NL. Monitoring HIV vaccine trial participants for primary infection: studies in the SIV/macaque model. Aids. 2009;23:1453–1460. doi: 10.1097/QAD.0b013e32832b43d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan S, Li J, Xu R, Wang L, Zhang K, Zhang R. Armored long RNA controls or standards for branched DNA assay for detection of human immunodeficiency virus type 1. Journal of clinical microbiology. 2009;47:2571–2576. doi: 10.1128/JCM.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, Ma Y, Zhao S, Yang N. Armored RNA as positive control and standard for quantitative reverse transcription-polymerase chain reaction assay for rubella virus. Archives of virology. 2007;152:219–224. doi: 10.1007/s00705-006-0839-3. [DOI] [PubMed] [Google Scholar]