Abstract
Meiotic recombination enables the reciprocal exchange of genetic material between parental homologous chromosomes and ensures faithful chromosome segregation during meiosis in sexually reproducing organisms. This process relies on the complex interaction of DNA repair factors, and many steps remain poorly understood in mammals. Here we report the identification of MEIOB, a meiosis-specific protein, in a proteomics screen for novel meiotic chromatin-associated proteins in mice. MEIOB contains an OB domain with homology to one of the RPA1 OB folds. MEIOB binds to single-stranded DNA and exhibits 3’ to 5’ exonuclease activity. MEIOB forms a complex with RPA and with SPATA22, and these three proteins colocalize in foci that are associated with meiotic chromosomes. Strikingly, chromatin localization and stability of MEIOB depends on SPATA22 and vice versa. Meiob-null mouse mutant exhibits a failure in meiosis and sterility in both sexes. Our results suggest that MEIOB is required for meiotic recombination and chromosomal synapsis.
Meiosis, the hallmark of gametogenesis in sexually reproducing organisms, creates haploid germ cells from diploid progenitors and ensures their genetic diversity through recombination. Physical links between homologous chromosomes permit the formation of crossovers required for allele recombination and also facilitate accurate chromosome segregation at the first meiotic cell division1,2. Meiotic recombination is initiated at DNA double-strand breaks (DSBs) generated by SPO113; these DSBs become resected from the 5’ end, yielding extended 3’ single-stranded DNA (ssDNA) overhangs4. Replication protein A (RPA) coats ssDNA, forming nucleoprotein filaments that prevent degradation and secondary structure formation. Subsequent invasion of the 3’ single strand into the duplex of the homologue is mediated by DMC1 and RAD51. Strand invasion causes the displacement of one strand of the homologue, resulting in D-loop formation5. The invading 3’ end primes new DNA synthesis, such that D-loop extension toward the second end allows for the capture of the 3’ ssDNA end of the other break (second end). End ligation leads to the formation of double Holliday junctions (dHJs). While early steps of meiotic recombination (DSB formation, end resection, strand invasion, and D-loop formation) have been relatively well characterized, the intermediate steps of this process such as second end capture are poorly defined.
RPA is a conserved ssDNA-binding heterotrimer complex of RPA1, RPA2, and RPA36. The four oligonucleotide-binding (OB) folds of RPA1 mediate ssDNA-binding of the RPA complex7. RPA is essential for many aspects of DNA metabolism, notably DNA replication, DNA repair, and meiotic recombination. During meiotic recombination, RPA binds to various ssDNA intermediates, including resected 3’ ssDNA and the D-loop. Studies of mutant alleles of yeast RPA1 have demonstrated that RPA is required for meiotic recombination8. RPA is indispensible for mouse development; a homozygous point mutation in Rpa1 causes early embryonic lethality9.
Here we report the identification of MEIOB, a meiosis-specific protein with homology to one OB fold of RPA1, in a systematic proteomics screen for meiotic chromatin-associated proteins. We find that mouse MEIOB has evolved to fulfill an essential function in meiosis.
Results
Proteomics screen for meiotic chromatin-associated proteins
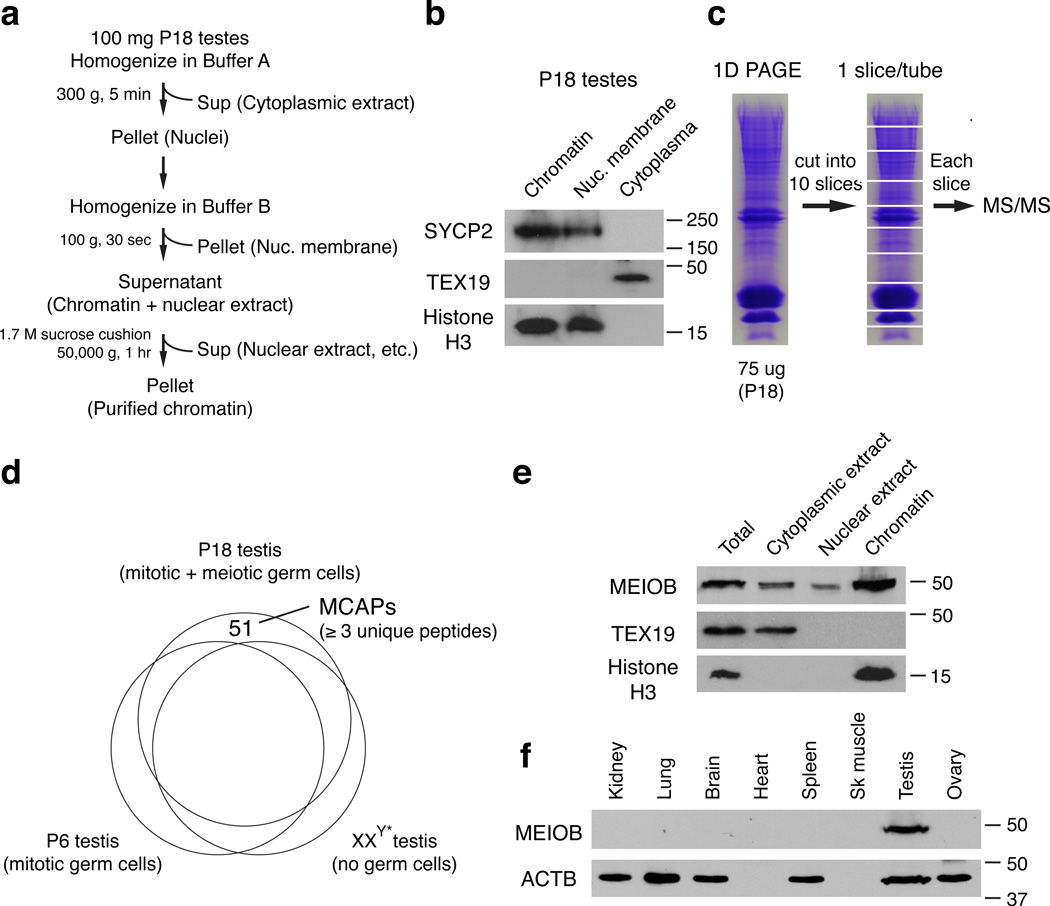
To systematically identify proteins that are associated with meiotic chromosomes, we isolated chromatin from postnatal day 18 (P18) testes, which lack post-meiotic germ cells and are thus highly enriched for meiotic germ cells. Our adaptation of a chromatin purification protocol for testis samples yielded highly pure chromatin (Fig. 1a)10,11. SYCP2, a synaptonemal complex protein associated with meiotic chromatin12, was present in the chromatin fraction, whereas the cytoplasmic germ cell-specific protein TEX19 was not detectable in the chromatin fraction by western blot analysis (Fig. 1b)13. To improve detection of proteins with low abundance, we separated chromatin-associated proteins from P18 testes by SDS-PAGE and analyzed 10 different size fractions by MS/MS (Fig. 1c). A total of 1300 proteins with at least one unique peptide were identified.
Figure 1. Proteomic identification of meiotic chromatin-associated proteins from mouse testes.
(a) Biochemical purification of chromatin from P18 testes. (b) Purity assessment of isolated meiotic chromatin by western blotting. SYCP2 is known to associate with meiotic chromatin. TEX19 is a cytoplasmic protein control. Histone H3 is a ubiquitous chromatin-associated protein. (c) Identification of chromatin-associated proteins by tandem mass spectrometry (MS/MS). The gel was cut into 10 slices, separating bands with abundant proteins from those with lower protein content. (d) Systematic identification of meiotic chromatin-associated proteins (MCAPs) by subtractive analysis of three proteomic datasets. Proteins represented with at least 3 unique peptides in P18 testes but absent from both P6 and XXY* testes were considered candidate meiotic chromatin-associated proteins. (e) Association of MEIOB with chromatin in P18 testes. Western blot analysis was performed using 20 µg protein from cytoplasmic extract, nuclear extract, and chromatin. TEX19 and histone H3 serve as cytoplasmic and chromatin controls, respectively. (f) MEIOB protein expression in adult mouse tissues. ACTB serves as a loading control. Note that heart and skeletal muscle contain little ACTB. Protein molecular mass standards are shown in kilodaltons.
In addition to meiotic cells, P18 testes contain mitotic germ cells and somatic cells (Fig. 1d). To identify proteins only associated with meiotic chromatin, we applied a subtractive approach, excluding chromatin-associated proteins present in mitotic cells. Using the same biochemical/proteomics protocol, we identified chromatin-associated proteins from postnatal day 6 mouse testes (P6), which contain spermatogonia but no meiotic cells14, and from XXY* testes, which lack all germ cells and thus contain only somatic cells15. By subtracting these two datasets from the P18 dataset, with a stringent cutoff of at least 3 unique peptides in P18 but none in P6 and XXY*, we identified 51 putative meiotic chromatin-associated proteins (Fig. 1d and Supplementary Table S1).
Identification of previously known meiotic chromatin-associated proteins confirmed the validity of our approach: 13 of 51 proteins identified are known to localize to meiotic chromatin (SYCP1, SYCP2, SYCP3, SYCE2, SMC1B, HORMAD1, H2AX, MDC1, TERF1, TDP1, PIWIL1, PIWIL2, and YBX2; Supplementary Table S1), including all three synaptonemal complex proteins. Furthermore, the genes encoding 19 proteins identified in our screen had been subjected to targeted deletion in mice: eleven mutants exhibited meiotic arrest, two showed post-meiotic spermiogenic arrest, four displayed embryonic lethality (with an unknown role in meiosis), and two were dispensable for fertility. The identification of these known meiosis factors validates our biochemical/proteomic approach and suggests that the other 32 proteins identified in our screen will likely be novel meiotic chromatin-associated proteins (Supplementary Table S1).
MEIOB is a novel meiosis-specific protein
To test whether the newly identified putative meiotic chromatin-associated proteins indeed localize to meiotic chromatin and function in meiosis, we focused on one of these uncharacterized proteins – MEIOB (Meiosis-specific with OB domains). MEIOB (470 aa) is predicted to contain an OB fold (aa 167–272) with limited homology to the DBD-B OB domain of RPA1 (Supplementary Fig. S1). MEIOB homologues are present in mammals, chicken, fugu fish, Drosophila, and Arabidopsis. Western blot analyses using specific antibodies raised against MEIOB confirmed its association with chromatin; MEIOB was also detectable in the cytoplasmic and soluble nuclear fractions of the testis (Fig. 1e). Abundantly expressed in the testis, MEIOB was not detectable in somatic tissues by western blot (Fig. 1f). MEIOB localized predominantly to the nuclei of spermatocytes in the testis (Supplementary Fig. S2). Therefore, MEIOB is a novel meiotic chromatin-associated protein.
MEIOB colocalizes with RPA in foci on meiotic chromosomes
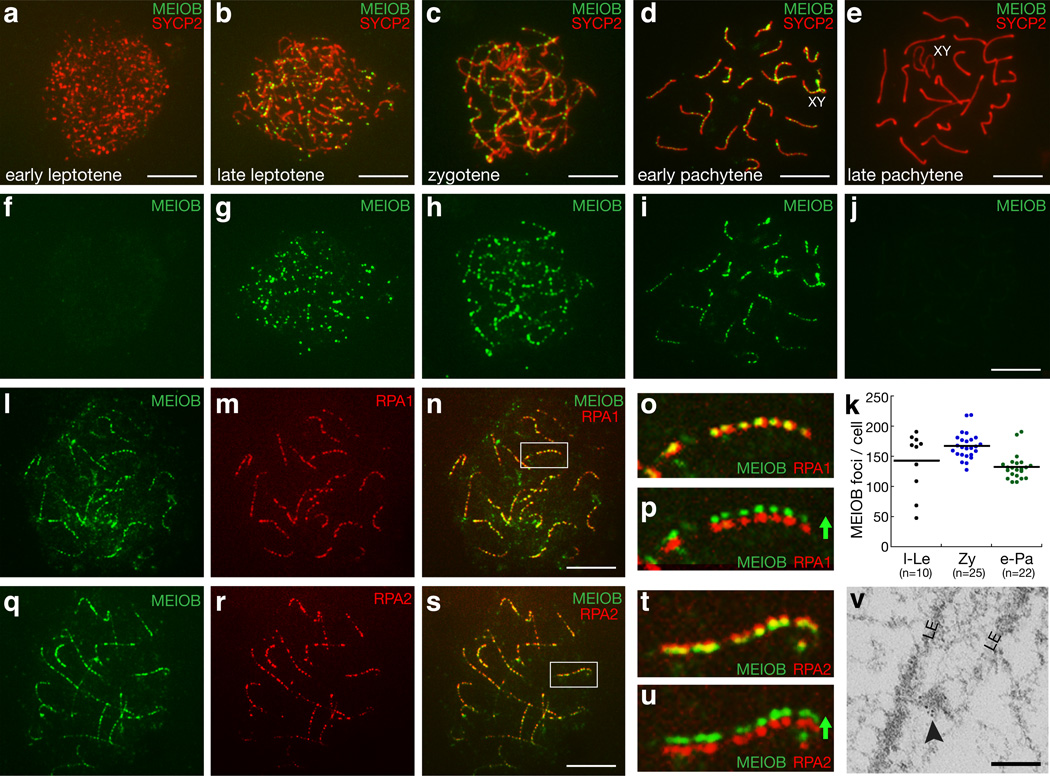
Immunostaining of spread nuclei of spermatocytes showed that MEIOB formed discrete foci on chromosomes; foci that exhibited a dynamic distribution pattern during meiosis (Fig. 2a–j). MEIOB foci were absent at the early leptotene stage (Fig. 2a), became first detectable on late leptotene chromosomes (Fig. 2b), formed arrays along synaptonemal complexes in zygotene and early pachytene spermatocytes (Fig. 2c, d), and disappeared in late pachytene spermatocytes (Fig. 2e). At a range of 100–200 per cell, MEIOB foci peaked at the zygotene stage (Fig. 2k). Notably, MEIOB foci were formed along the partially synapsed sex chromosomes at the pachytene stage (Fig. 2d). In addition, MEIOB formed foci on meiotic chromosomes in fetal oocytes (Supplementary Fig. S3), implying functional relevance of MEIOB for meiosis in both sexes. MEIOB was absent from the adult ovary (Fig. 1f), in which oocytes are at the diplotene stage. No focal staining was observed in Meiob−/− spermatocytes with the antibody used for these analyses, demonstrating that the antibody is specific for MEIOB and does not cross-react with RPA1 (Supplementary Fig. S4a).
Figure 2. MEIOB colocalizes with RPA in foci on meiotic chromosomes.
(a–e) Distribution pattern of MEIOB foci on chromatin of spermatocytes from early leptotene through late pachytene stages. (f–j) the same cells in panels a–e are shown with MEIOB labeling only. (k) Number of MEIOB foci in spermatocytes at successive stages of meiotic prophase I. Stages were determined by SYCP2 staining. l-Le, late leptotene; Zy, zygotene; e-Pa, early pachytene; n indicates the number of cells counted. The line indicates the average. (l–p) MEIOB colocalizes with RPA1 on the meiotic chromosomes of spermatocytes. The same spermatocyte is shown with MEIOB labeling only (l), RPA1 labeling only (m), and in a merged image (n). Enlarged views of the marked chromosome in panel n are shown in panels o and p (without or with offset channels). (q–u) MEIOB colocalizes with RPA2 on the meiotic chromosomes of spermatocytes. The same spermatocyte is shown with MEIOB labeling only (q), RPA2 labeling only (r), and in a merged image (s). Enlarged views of the marked chromosome in panel s are shown in panels t and u (without or with offset channels). (v) Electron microscopy with immunogold (6 nm particles, arrowhead) -labeled MEIOB reveals signal between the lateral elements (LE) in spermatocytes. Scale bars: a–s, 10 µm; v, 0.2 µm.
We next evaluated the potential interaction of MEIOB with other meiotic recombination related proteins and found that MEIOB co-localized with RPA, which forms foci on prophase I meiotic chromosomes16–18. Immunofluorescence analyses with antibodies specific for both the RPA1 and RPA2 subunits of RPA confirmed complete focal association with MEIOB (Fig. 2l–u). As previously shown, RPA foci gradually replace RAD51 foci during meiotic recombination, thus, only a subset of RPA foci overlap with RAD51 foci17. The relative distribution pattern of MEIOB and RAD51 foci is similar to that of RPA and RAD51 foci. Early leptotene spermatocytes contained a large number of RAD51 foci but lacked MEIOB foci (Supplementary Fig. S5a). In zygotene and pachytene spermatocytes (Supplementary Fig. S5b–g), the distribution of MEIOB and RAD51 foci appeared to be dynamic: some foci contained both proteins, some contained RAD51 only, and others contained MEIOB only. RAD51 and DMC1 always colocalize in foci on meiotic chromosomes19. As expected, MEIOB colocalized with DMC1 in only a subset of foci (Supplementary Fig. S6a–d’). In a subset of foci, MEIOB also colocalized with MSH4, a member of the ZMM group involved in meiotic recombination (Supplementary Fig. S6e–h’)20. Thus, MEIOB and RPA always colocalize, whereas MEIOB colocalizes with RAD51, DMC1, or MSH4 only in a subset of foci. Ultrastructural analysis by immunogold electron microscopy showed MEIOB signal in the electron dense nodule between lateral elements of the synaptonemal complex (Fig. 2v). These results suggest that MEIOB associates with RPA during meiotic recombination.
MEIOB binds to single-stranded DNA
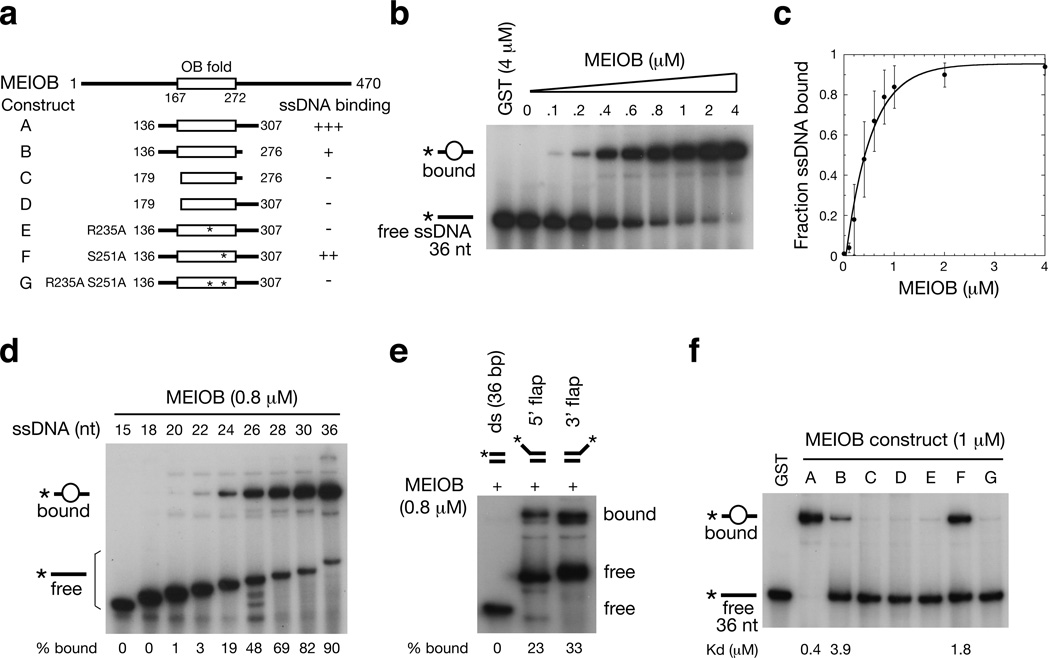
A central OB domain is the only predicted protein motif in MEIOB (Fig. 3a). OB folds are compact barrel-like structures that can recognize and interact with single-stranded nucleic acids or those with an unusual structure21,22. We expressed the central region of MEIOB including the OB-fold (aa 136–307; Fig. 3a, construct A) as a GST fusion protein and assessed its nucleic acid binding capacity. Truncated MEIOB (construct A) bound to ssDNA with an apparent Kd of 0.4 µM (Fig. 3b, c). Truncated MEIOB exhibited high affinity for single-stranded DNA at least 24 nt in length and lower affinity for shorter fragments (Fig. 3d). Truncated MEIOB also bound to dsDNA with 5’ or 3’ single-stranded flaps, but not to dsDNA with blunt ends (Fig. 3e). These experiments demonstrate that MEIOB is an ssDNA-binding protein.
Figure 3. MEIOB binds to ssDNA but not dsDNA.
(a) MEIOB contains an OB fold. All MEIOB constructs were expressed as GST fusion proteins. Only construct A (MEIOB central region) (aa 136–307) was used in experiments illustrated in panels b through e. The summary of ssDNA-binding activity is based on the results from panel f: +++, strong; ++, intermediate; +, weak; -, no binding. The Kd values are shown in panel f. (b) Truncated MEIOB binds to ssDNA. The ssDNA (d36GT) was 5’-labeled with 32P (indicated by asterisk). (c) Binding curve for truncated MEIOB-ssDNA (d36GT) from triplicate EMSA experiments (mean ± sd). (d) Binding of truncated MEIOB to ssDNA oligonucleotides of various lengths. All oligonucleotides [d(GT)n or d(GT)nG] were 5’ 32P-labeled. (e) Truncated MEIOB binds to 5’ and 3’ ssDNA flap (30-nt) but not dsDNA. One strand of the dsDNA and the 5’ flap DNA were 5’ 32P-labeled. The long strand in the 3’ flap DNA was 3’ 32P-labeled. (f) EMSA analysis of various MEIOB constructs depicted in panel A. GST was used as negative controls.
To identify essential elements of the ssDNA-binding domain of MEIOB, we expressed protein segments with additional N- and C-terminal truncations and point mutations (constructs B-G; Fig. 3a, f). We found that deletion of aa 277–307, located C-terminal to the OB-fold, resulted in a 10-fold reduction in the binding affinity (Kd for construct B: 3.9 µM), whereas deletion of aa 136–178 (construct C) abolished DNA-binding activity. Based on the crystal structure of human RPA123, several residues in the DBD-B OB domain are involved in DNA-binding; these are conserved in the MEIOB OB domain (Supplementary Fig. S1). Analysis of mutant protein segments with point mutations at such residues identified R235 of MEIOB as essential for DNA-binding. Residue S251 was important but not required for DNA-binding, as the S251A mutation reduced the binding affinity by 4.5 fold (Kd for construct F: 1.8 µM).
MEIOB exhibits ssDNA-specific 3’ to 5’ exonuclease activity
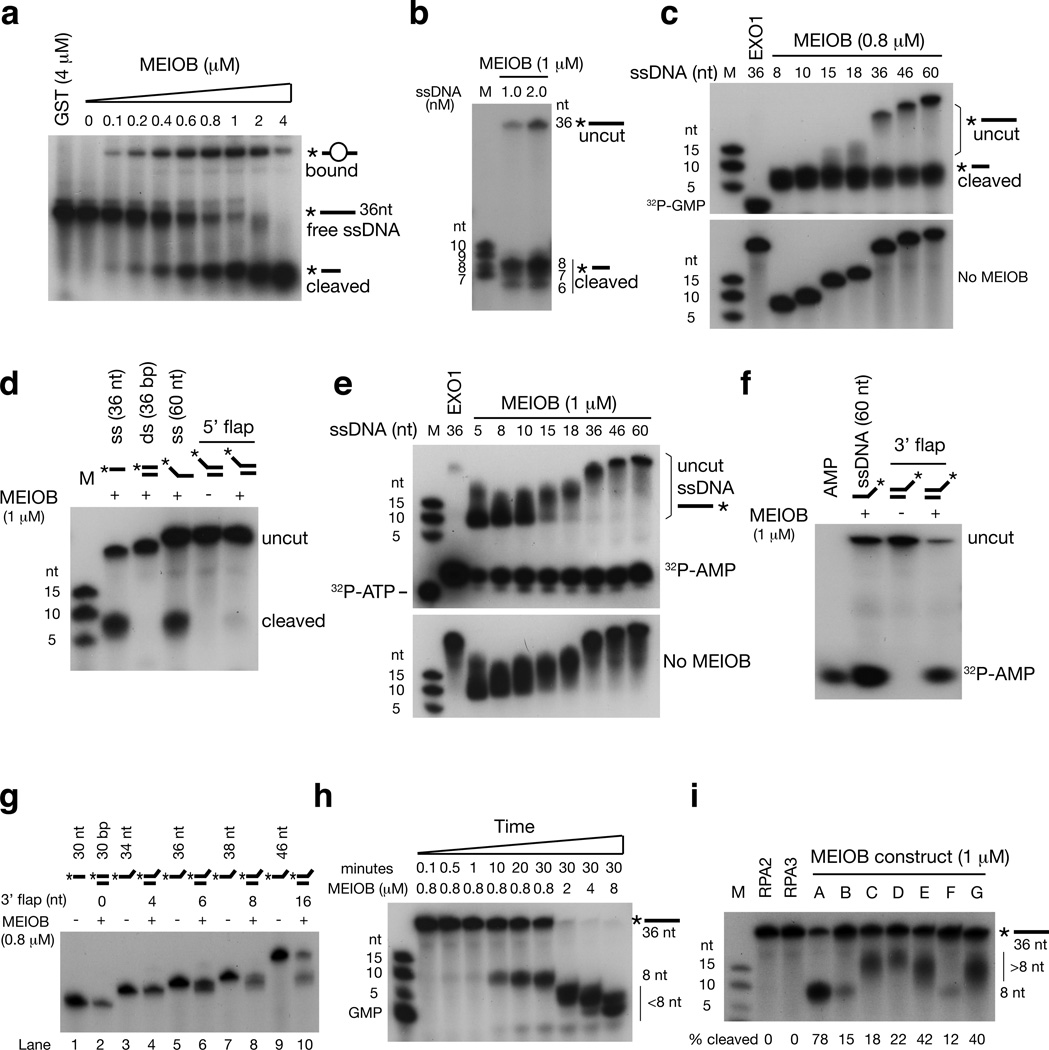
When the in vitro binding assays with 5’-32P-labeled ssDNA were performed in the presence of Mg2+, a cleaved product appeared proportionally to MEIOB concentration, revealing that truncated MEIOB (construct A in Fig. 3a) exhibits Mg2+-dependent nuclease activity (Fig. 4a). Truncated MEIOB cleaved ssDNA into oligonucleotides of 6 to predominantly 8 bases in length (Fig. 4b). Although its binding affinity to oligonucleotides of ≤ 18 nt was low (Fig. 3d), truncated MEIOB cleaved ssDNA fragments as short as 10–15 nt into 8-nt oligonucleotides (Fig. 4c). The generation of 5’-labeled 6-to-8-nt products indicated that truncated MEIOB has either endonuclease or 3’ exonuclease activity. Truncated MEIOB did not process 5’ flap ssDNA, showing that it lacks 5’ exonuclease or endonuclease activity (Fig. 4d). Rather, truncated MEIOB cleaved nucleotides from the 3’end of 3’-radiolabeled oligos (Fig. 4e), demonstrating that it is a 3’ exonuclease. The 3’ exonuclease activity of truncated MEIOB was robust for ssDNA of ≥18 nt and lower for shorter oligonucleotides (5–15 nt) (Fig. 4e), suggesting that the enzymatic activity of truncated MEIOB increases with stronger DNA binding. The ability of truncated MEIOB to digest 3’ flap DNA further validated its 3’ exonuclease activity (Fig. 4f). The exonuclease activity of truncated MEIOB on 3’ flap was similar to that on ssDNA. Truncated MEIOB digested 3’ flaps efficiently to ~8 nt, digested 6-nt and 8-nt 3’ flaps very inefficiently, but had no effect on 4-nt 3’ flaps (Fig. 4g). At 0.8 µM, truncated MEIOB digested ssDNA efficiently only to a length of 8 nt after 10 minutes of incubation (Fig. 4h). With a ten-fold excess (8 µM), truncated MEIOB further digested ssDNA to less than 5 nt or even completely (Fig. 4h). In conclusion, truncated MEIOB exhibits 3’ exonuclease activity specific for ssDNA in vitro. However, we can not exclude the possibility that in vitro biochemical analyses on truncated MEIOB proteins may not reflect precisely the activity of the full-length protein in vitro, in vivo, or in partner with other proteins.
Figure 4. MEIOB exhibits ssDNA-specific 3’ exonuclease activity.
Construct A (MEIOB central region, Fig. 3a) (aa 136–307) was used in experiments illustrated in panels a through h. (a) Truncated MEIOB exhibits nuclease activity in addition to its ssDNA-binding activity. (b) Size determination of 5’-labeled products generated by MEIOB. An 8-nt product was predominant. All products (panels b through i) were resolved on denaturing acrylamide gels. (c) Truncated MEIOB processes 5’ labeled oligonucleotides of various lengths. ExoI digests ssDNA completely. (d) Truncated MEIOB lacks nuclease activity on dsDNA and 5’ flap (30 nt) DNA substrates. (e) Truncated MEIOB exhibits 3’ exonuclease activity on 3’ labeled oligonucleotides of various lengths. EXO1 serves as a positive control. (f) Truncated MEIOB efficiently processes oligonucleotides with a 3’ flap (30 nt). (g) Extent of 3’ flap digestion by truncated MEIOB. (h) Time course and gradient analyses of truncated MEIOB nuclease activity. Note that MEIOB at higher concentrations (2 to 8 µM) produced shorter end products (<8 nt). (i) Nuclease activities of various MEIOB mutants shown in Fig. 3A. Recombinant GST-RPA2 and -RPA3 were used as negative controls. Percent (%) cleavage represents the average from three independent experiments.
We next tested the nuclease activity of various MEIOB mutants with truncations and point mutations (Fig. 3a). We found that all the mutant proteins exhibited much lower nuclease activities than construct A (Fig. 4i). Interestingly, similar to construct A, constructs B and F digested the 36 nt ssDNA to 8 nt. In contrast, constructs C, D, E, and G produced digested oligonucleotides longer than 8 nt, suggesting lower enzymatic processivity. Because constructs A, B, and F bind to ssDNA, but C, D, E, and G do not (Fig. 3f), these data suggest that the DNA-binding property of MEIOB might stimulate its enzymatic processivity.
MEIOB is required for meiosis and fertility in both sexes
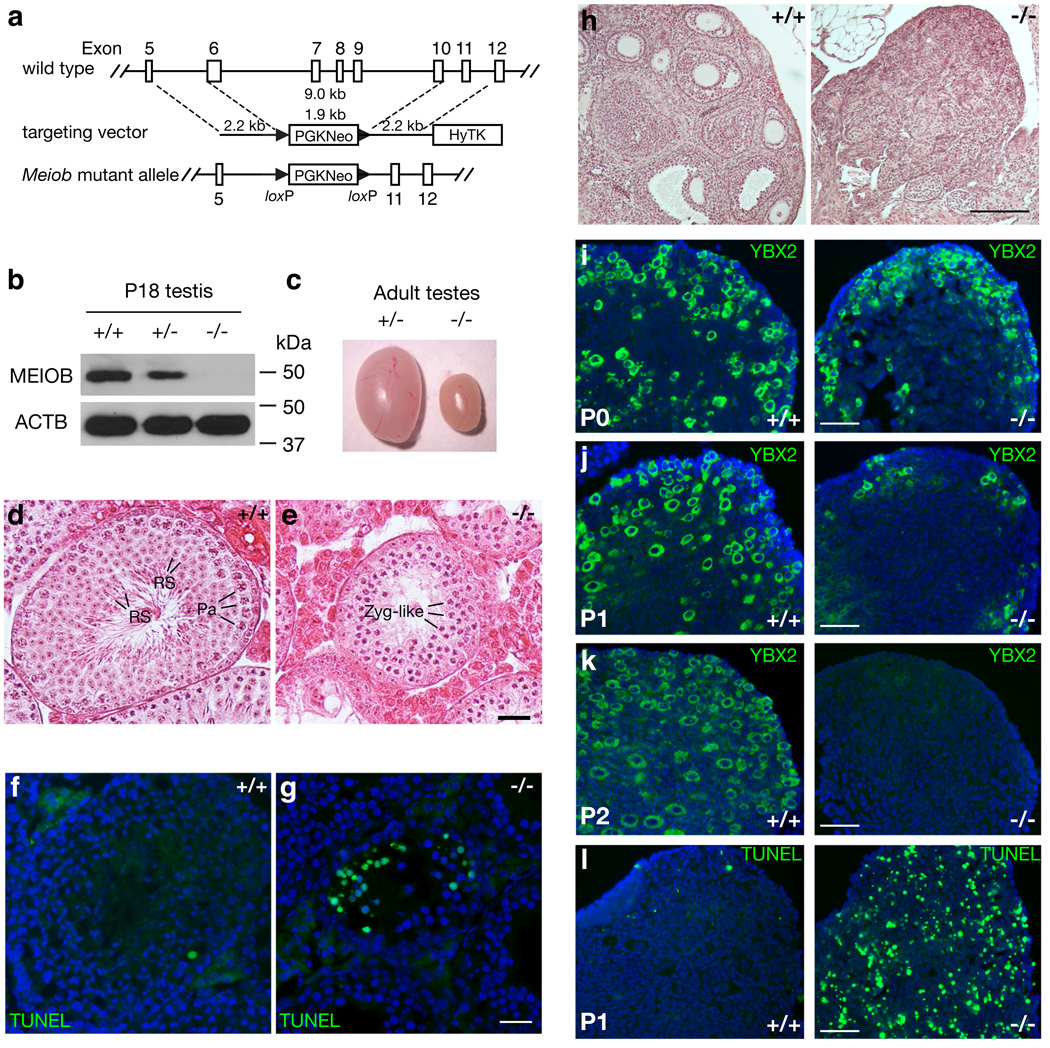
To uncover the requirement of MEIOB for meiosis, we disrupted the Meiob gene (Fig. 5a). Meiob−/− mice were viable and appeared to be healthy. Western blotting demonstrated the absence of MEIOB protein in Meiob−/− testes (Fig. 5b), confirming that the Meiob mutant was null. Interbreeding of heterozygous (Meiob+/−) mice yielded a normal Mendelian ratio (182:357:169) of Meiob+/+, Meiob+/−, and Meiob−/− offspring.
Figure 5. MEIOB is required for meiosis and fertility in mice of both sexes.
(a) Targeted inactivation of the Meiob gene. The mouse Meiob gene maps to Chromosome 17 and has 14 exons. Deletion of exons 6–10 (aa 111–293) is expected to cause a frame shift in the resulting transcript. (b) Absence of MEIOB protein in P18 Meiob−/− testis. (c) Significant size reduction in 8-wk-old Meiob−/− testis. (d, e) Histological analysis of 8-wk-old wild type and Meiob−/− testes. Abbreviations: Zyg, zygotene spermatocytes; Pa, pachytene spermatocytes; RS, round spermatids; ES, elongated spermatids. (f, g) TUNEL analysis of wild type and Meiob−/− seminiferous tubules. Note the large number of apoptotic cells (green) in Meiob−/− tubules. Scale bar (e, g), 25 µm. (h) Histological analysis of ovaries from adult wild type and Meiob−/− mice. Scale bar, 50 µm. (i–k) Progressive loss of oocytes in Meiob−/− ovaries. Frozen sections prepared from postnatal day 0, 1, and 2 wild type and Meiob−/− ovaries were immunolabeled with anti-YBX2 antibodies. (l) TUNEL analysis of P1 ovaries. Nuclear DNA (i–l) was counterstained with DAPI (blue). Scale bars (i–l), 50 µm.
Both Meiob−/− males and females were sterile. Disruption of Meiob resulted in dramatically reduced testis size (Fig. 5c). Testes from 8-wk-old Meiob−/− males weighed 70% less than wild type (54.5 ± 6.7 mg vs 184.0 ± 10.5 mg per pair; n=4) (Student’s t test, P < 0.0001). Histological analysis revealed complete meiotic arrest in germ cells of Meiob−/− adult males (Fig. 5d, e). In contrast to wild type seminiferous tubules that contained a full spectrum of germ cells (Fig. 5d), tubules from Meiob−/− testes completely lacked post-meiotic germ cells, with zygotene-like spermatocytes reflecting the most advanced germ cells (Fig. 5e). In addition, 3-wk-old Meiob−/− testis also exhibited a block in meiosis, showing that inaction of Meiob causes meiotic failure during the first wave of spermatogenesis and in adults (Supplementary Fig. S7). TUNEL assays showed that Meiob-deficient spermatocytes underwent apoptosis (Fig. 5g), suggesting that mutant spermatocytes are eliminated at the pachytene checkpoint24.
Ovaries from adult Meiob−/− female mice were small and devoid of oocytes of any developmental stage (Fig. 5h). To determine the time point of oocyte loss, we examined ovaries at different ages, using anti-YBX2 antibodies to identify oocytes25. Meiob−/− ovaries contained an abundant number of oocytes at birth (P0, Fig. 5i), but were almost devoid of oocytes by postnatal day 1 (P1; Fig. 5j), with complete lack of oocytes by day 2 (P2; Fig. 5k). TUNEL assays revealed that oocytes in Meiob−/− ovaries underwent massive apoptosis at P1 (Fig. 5l), which is in agreement with a complete oocyte loss within the first two days after birth. Complete early postnatal loss of oocytes is characteristic of recombination-defective (Dmc1, Msh5, or Atm) mouse mutants26. Therefore, MEIOB is essential for meiosis in both sexes.
MEIOB is required for chromosomal synapsis
To elucidate the molecular defects of Meiob-deficient meiotic germ cells, we monitored the process of chromosomal synapsis by assessing the distribution of proteins of the central (SYCP1) and axial/lateral elements (SYCP2) of synaptonemal complexes (Supplementary Fig. S8)12,27. In leptotene spermatocytes, the axial elements marked by SYCP2 began to form but SYCP1 filaments were not yet detectable (Supplementary Fig. S8a). At the zygotene stage, axial elements are normally fully formed and chromosomes progressively synapse. However, Meiob−/− spermatocytes exhibited severe defects in chromosomal synapsis (Supplementary Fig. S8b–h): 40% of Meiob−/− spermatocytes formed long axial elements, suggesting progression to the zygotene stage; however, these cells completely lacked synapsis (Supplementary Fig. S8b). Such cells were not observed in wild type males (Supplementary Fig. S8h). In wild type postnatal day 16 testes, ~65% of spermatocytes reached the pachytene stage. No normal pachytene spermatocytes were observed in Meiob−/− males (Supplementary Fig. S8h), but ~15% of Meiob−/− spermatocytes appeared to be at a pachytene-like stage, with shortened axial elements, suggesting chromosomal condensation, but no evidence for synapsis (Supplementary Fig. S8f, h). These results demonstrate that MEIOB is essential for chromosomal synapsis.
Meiob−/− mice exhibit defects in meiotic recombination
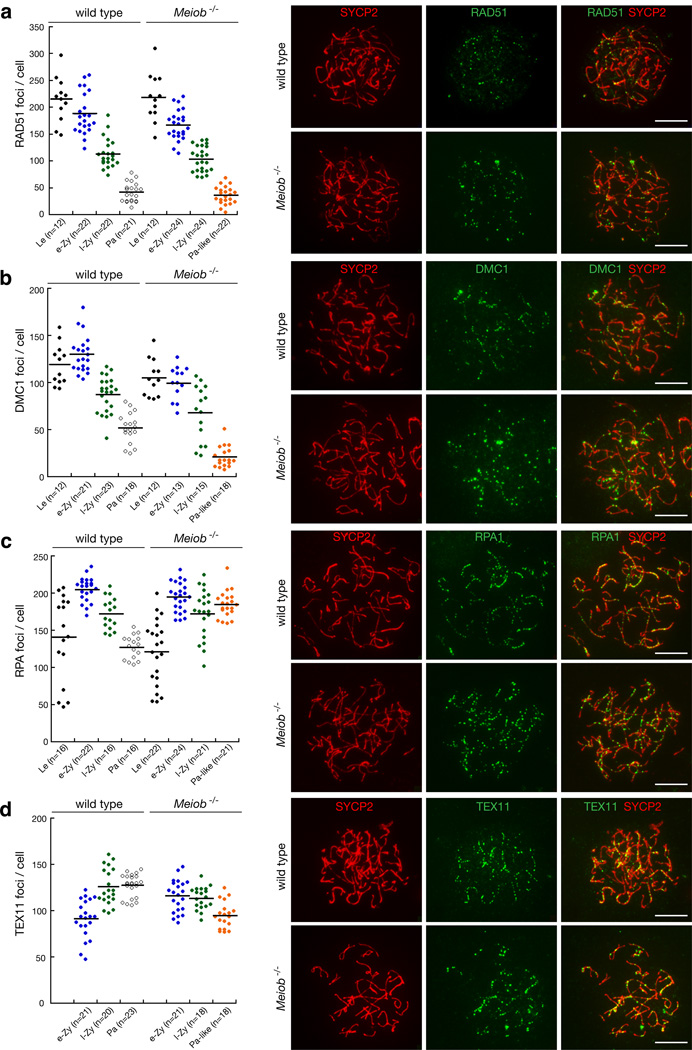
We next examined the process of meiotic recombination. During meiosis, SPO11-generated DSBs lead to phosphorylation of H2AX through the activation of the ATM kinase28–30. ATR is recruited to unsynapsed chromosomes and phosphorylates H2AX at the zygotene and pachytene stages31,32. Recombinases RAD51/DMC1 together with the HOP2-MND1 heterodimer direct strand invasion into the homologous chromosomes, resulting in D-loop formation33. RPA may bind to the D-loop, whereas ZMM proteins such as MSH4 and TEX11 facilitate the formation of crossovers20,34. These proteins form recombination nodules and can be visualized as foci on meiotic chromosomes35. Analysis of chromosome spreads from Meiob−/− spermatocytes revealed the presence of the majority of recombination factors assessed: γH2AX persisted on chromatin, and ATR formed foci on synaptonemal complexes (Supplementary Fig. S9). These results are in agreement with the severe synaptic defects in Meiob−/− spermatocytes (Supplementary Fig. S8). The initial numbers of RAD51 and DMC1 foci between wild type and Meiob−/− spermatocytes were similar at leptotene (Fig. 6a, b). The numbers of RAD51, DMC1, RPA, and TEX11 foci were also comparable between wild type and Meiob−/− spermatocytes at the zygotene stage (Fig. 6a–d). Interestingly, in the Meiob−/− pachytene-like spermatocytes, the number of RAD51 and DMC1 foci decreased as expected, but RPA foci persisted (Fig. 6c), suggesting that, in the absence of MEIOB, meiotic recombination fails after strand invasion, or RAD51/DMC1 filament is unstable and falls off, leading to replacement by RPA1.
Figure 6. MEIOB is essential for meiotic recombination.
Immunolabeling for proteins of the synaptonemal complex (SYCP2, lateral elements) and recombination nodules was performed on spread nuclei of spermatocytes from wild type and Meiob−/− testes at postnatal day 16. The meiotic stages of spermatocytes were determined based on the morphology of the synaptonemal complexes. Spermatocytes were categorized into the following groups: leptotene (Le), early-mid zygotene (e-Zy), late zygotene (l-Zy), and pachytene (Pa). Pachytene-like (Pa-like) spermatocytes contained prominent but short lateral elements (See supplementary Fig. S8f). Pa-like spermatocytes were present in Meiob−/− testes but absent in wild type testes. Dot plots for each DNA repair protein show the number of foci per cell; solid lines designate the average for each spermatocyte category. n, number of cells counted. Representative images of spermatocytes at early-mid zygotene stages are shown in separate channels and as merged images. (a) RAD51 foci. (b) DMC1 foci. (c) RPA foci. (d) TEX11 foci. Scale bars, 10 µm.
At the mid-late pachytene stage, MLH1 foci become apparent in normal spermatocytes, marking sites of crossover36,37. Meiob−/− spermatocytes did not progress to the mid-late pachytene stage and thus were devoid of MLH1 foci. The meiotic defects in fetal oocytes were similar to those observed in spermatocytes from Meiob−/− mice. However, a small percentage of Meiob−/− oocytes (7%) advanced to the pachytene stage with full synapsis by 17.5 days of embryonic development, whereas 93% of Meiob+/− oocytes developed to this stage. This difference with males may result from the possibility that Meiob−/− oocytes proceed in meiosis with some synapsis defects, which may be overcome with time, whereas mutant spermatocytes are rapidly eliminated upon synapsis defects. All Meiob−/− pachytene oocytes (n=30) were γH2AX-positive, indicating that DSBs were not repaired, and lacked MLH1 foci (Supplementary Fig. S10). Our results demonstrate that the markers of early steps of meiotic recombination (DSB formation, 5’ end resection, and strand invasion) are present in the absence of MEIOB, but that markers of subsequent steps of meiotic recombination are lacking. Our results suggest that defective chromosomal synapsis in Meiob−/− mice is likely attributed to defects in meiotic recombination.
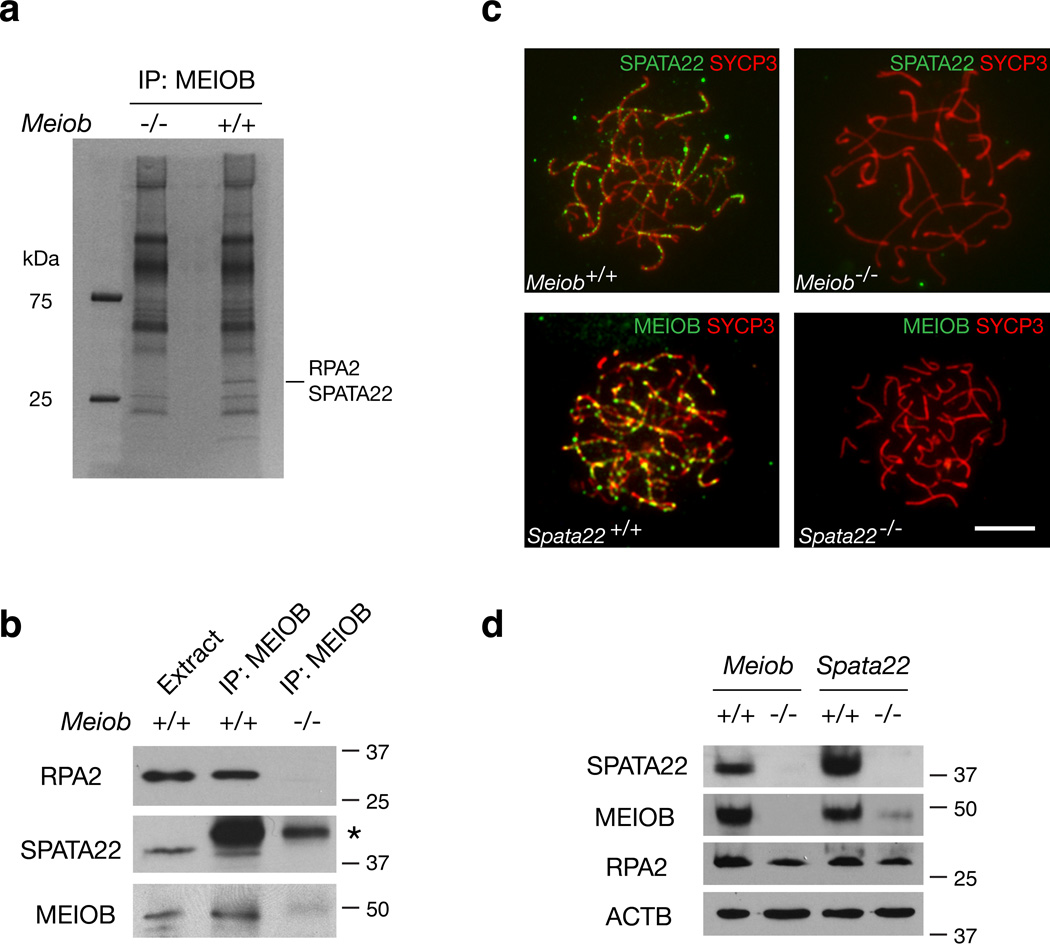
MEIOB forms complexes with RPA2 and SPATA22
To identify protein partners of MEIOB, we performed immunoprecipitation with testicular extracts using anti-MEIOB antibodies, and identified a single prominent band that was present in the immunoprecipitated proteins from wild type but absent from Meiob−/− testes (Fig. 7a). MS/MS analysis revealed that this band contained RPA2 and SPATA22, a meiosis-specific protein38. To verify the association of MEIOB with RPA2 and SPATA22, we performed co-IP experiments followed by Western blot analysis and confirmed that RPA2 and SPATA22 were present in the protein fraction immunoprecipitated with the anti-MEIOB antibody (Fig. 7b). We found that SPATA22, similar to MEIOB, formed discrete foci on meiotic chromosomes (Fig. 7c and Supplementary Fig. S4b). Specificity of the SPATA22 antibody was confirmed by the absence of foci in Spata22−/− spermatocytes (Supplementary Fig. S4b). In addition, SPATA22 colocalized with MEIOB in foci on meiotic chromosomes from spermatocytes (Supplementary Fig. S11). These studies demonstrate that MEIOB forms protein complexes with RPA and with SPATA22 in the testis.
Figure 7. MEIOB forms complexes with RPA2 and SPATA22.
(a) Identification of MEIOB-associated proteins from P18 testes by immunoprecipitation and mass spectrometry. The gel was stained with SYPRO Ruby. (b) Co-immunoprecipitation of MEIOB with RPA2 and SPATA22 from testicular protein extracts. The asterisk indicates a non-specific band. (c) Inter-dependent localization of MEIOB and SPATA22 on meiotic chromosomes from spermatocytes. Spata22−/− refers to Spata22repro42/repro42 mice38. Scale bar, 10 µm. (d) Western blot analysis of MEIOB and SPATA22 proteins in testes from wild type, Meiob−/−, and Spata22−/− mutant mice.
Inter-dependent localization of MEIOB and SPATA22
RPA foci were formed on Meiob-null meiotic chromosomes (Fig. 6c), indicating that the interaction of RPA with meiotic chromatin is independent of MEIOB. In stark contrast, SPATA22 foci were absent in Meiob-null spermatocytes, and MEIOB foci were absent in Spata22-null spermatocytes (Fig. 7c), suggesting that MEIOB and SPATA22 can interact with meiotic chromosomes only in the presence of each other. Reciprocal interdependence between these two proteins was further supported by the observation that SPATA22 protein was absent in Meiob−/− testes and MEIOB protein nearly absent in Spata22−/− testes (Fig. 7d). These results also indicate that the stability of these two proteins depends on their co-expression and interaction. Consistent with this hypothesis, the Meiob mutant phenocopies the meiotic failure observed in Spata22-null male and female mice38. SPATA22 has no recognizable domains, whereas MEIOB binds to ssDNA and exhibits 3’ exonuclease activity. These results suggest that MEIOB may function as the catalytic subunit of this novel meiotic recombination complex that requires SPATA22 as an essential co-factor.
Discussion
Genetic and cytological studies in diverse organisms from yeast to humans have yielded a wealth of knowledge on meiosis. However, more than in other model organisms, approaches to gain insight into mammalian meiosis have been hampered by various obstacles. Here, we have developed a biochemical and proteomics strategy to systematically identify meiotic chromatin-associated proteins. Our strategy has proven highly successful, identifying synaptonemal complex components, proteins critical for meiotic recombination, and factors involved in chromatin modification. In this report, we have characterized the essential function of one novel meiotic chromatin-associated protein – MEIOB. Furthermore, these proteomic screen and functional studies set the stage for future characterization of other novel putative meiotic chromatin-associated proteins (Supplementary Table S1).
Many key steps of meiotic recombination such as second-end capture are poorly understood. Biochemical and genetic studies have shown that yeast RAD52 plays a key role in second end capture39–42. In biochemical assays, mammalian RAD52 also mediates second end capture43. However, RAD52-deficient mice display no abnormalities in viability and fertility, suggesting that RAD52 is not essential for recombination in mice, and that other genes may functionally substitute RAD52 in mice44.
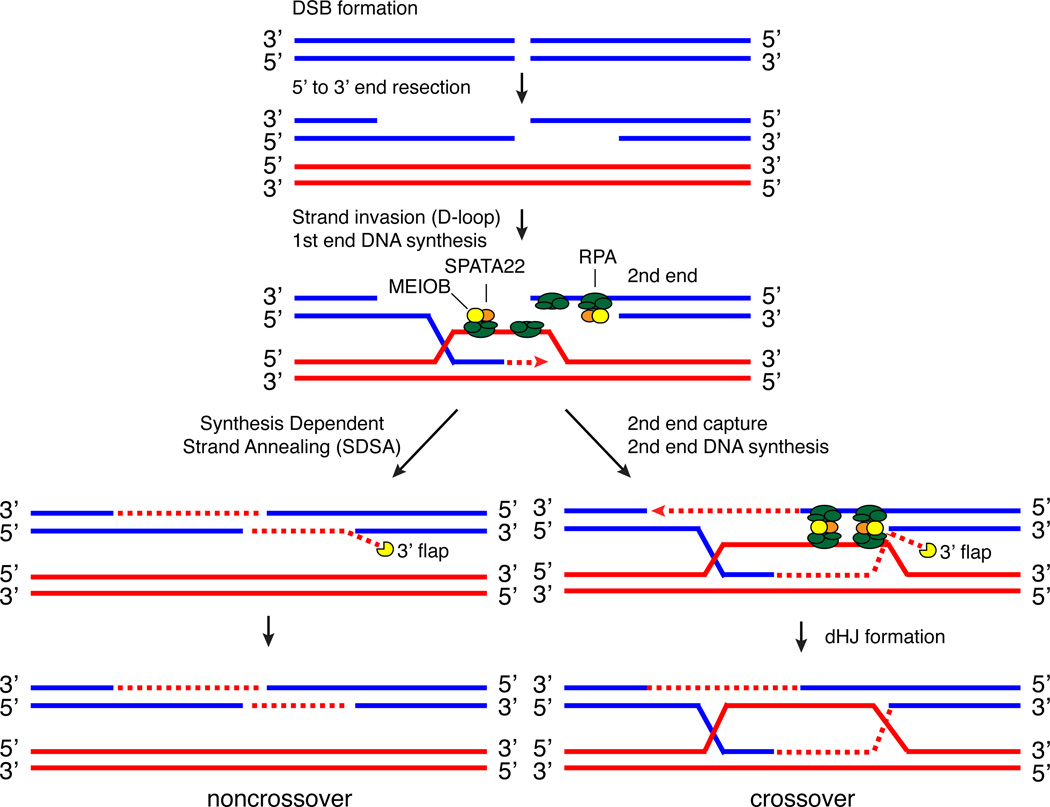
Our results support a model in which MEIOB and its co-factor SPATA22 may mediate second-end capture during mammalian meiotic recombination (Fig. 8). MEIOB is not required for the early steps of meiotic recombination, including DSB formation, end resection, and strand invasion. However, crossovers are never formed in Meiob-deficient germ cells and, in normal pachytene spermatocytes, the disappearance of MEIOB foci precedes the appearance of crossovers. These results indicate that MEIOB is likely to function at the time point after strand invasion (Fig. 8). Similar to RAD52, MEIOB binds to ssDNA and interacts with RPA. Therefore, in our model, we propose that MEIOB mediates second-end capture through interactions with the RPA proteins, which coat both the D-loop and the resected second end. Second end capture is a prerequisite to the dHJ pathway (Fig. 8)45. The severe synaptic defects and persistence of γH2AX caused by loss of MEIOB suggest that MEIOB is globally required for DSB repair during meiosis.
Figure 8. Proposed model for MEIOB function during meiotic recombination.
Key steps of meiotic recombination are illustrated. We propose that the RPA-MEIOB-SPATA22 complex coats both the D-loop and the ssDNA of the second end. In this model, the interaction between RPA and MEIOB-SPATA22 mediates second end capture. Most DSBs are repaired through the SDSA (synthesis dependent strand annealing) pathway, in which the D-loop collapses back to its sister chromatid. After second end capture, intermediates continue to form double Holliday junctions, which are resolved into either crossovers or non-crossovers. We propose that MEIOB removes the 3’ flaps resulting from the first end DNA synthesis in both dHJ and SDSA pathways.
We postulate that MEIOB may digest the 3’ ssDNA flap from the first end after annealing of second end. After strand invasion, the first end primes DNA synthesis to extend the D-loop towards the second end. Subsequent to second end capture, the first end needs to be ligated to the resected strand of the second end. The first end-primed DNA synthesis could progress beyond the end point of the resected strand of the second end, such that a 3’ ssDNA flap forms after annealing of the D-loop with the single-stranded second end or after synthesis dependent strand annealing (SDSA) (Fig. 8). The timing of DSB end resection and first end-primed DNA synthesis is different, thus, generation of a gap or 3’ flap is expected after second end annealing. Whereas a gap can be filled by DNA synthesis, the repair of a 3’ flap requires different mechanisms. MEIOB may bind to and remove the 3’ flap through its nuclease activity, such that the ends can be ligated seamlessly without insertions. If 3’ flap is not processed, DNA helicases may act upon this intermediate and destabilize it. In vivo, RPA may inhibit MEIOB activity on 3’ ends of ssDNA, thereby blocking promiscuous degradation. It is also possible that binding partners such as RPA and SPATA22 may modulate the nuclease activity and substrate specificity of MEIOB in vivo.
In Drosophila and Arabidopsis, RPA1-related proteins (DmHDM and AtRPA1A) have evolved to regulate crossover formation, but neither is essential for meiotic recombination46,47. Although certain mitotic DNA repair proteins (such as RPA and RAD51) are utilized during meiotic recombination, many species have developed meiosis-specific repair machinery. One such factor is DMC1, a meiosis-specific paralogue of RAD5148–50. Yeast meiotic recombination only requires the D-loop forming activity of DMC1, while RAD51 plays a supporting role51,52. DMC1 has evolved to initiate and engage the first inter-homolog chromosome interaction unique to meiosis. Our results suggest that MEIOB might have evolved to drive another inter-homolog interaction unique to meiosis – capture of the second homolog end.
Methods
Biochemical purification of chromatin
We optimized previously reported chromatin purification protocols (Fig. 1a)10,11 as follows: 100 mg testis tissue was homogenized in 1 ml Buffer A (250mM sucrose, 10mM Tris HCl pH 8.0, 10mM MgCl2, 1mM EGTA, 1× protease inhibitor cocktail III) and centrifuged at 300 g to separate nuclei (pellet) from cytoplasmic extract (supernatant). The nuclei were homogenized in Buffer B (Buffer A + 0.1% Triton X-100, 0.25% NP-40) and centrifuged briefly at 100 g to remove nuclear membrane particles (pellet), which contain some chromatin (discarded). The chromatin-containing supernatant was layered over a 1.7 M sucrose cushion and centrifuged at 50,000 g for one hour in an ultracentrifuge to remove soluble nuclear proteins and remnant cytoplasmic proteins/organelles. The pellet (chromatin) was washed three times, resuspended in Tris buffer, sonicated, and boiled in an equal volume of 2xSDS PAGE buffer to dissolve all chromatin-associated proteins (~200 µg).
Proteomics screen and mass spectrometry
75 µg chromatin-associated proteins from each testis sample were separated on a 10% SDS-PAGE gel (Fig. 1c) and stained with Coomassie blue. The gel lane was cut into 10 slices, digested with trypsin, and subjected to HPLC followed by tandem mass spectrometry (MS/MS) at the University of Pennsylvania Proteomic Core Facility. All spectra from 10 MS/MS runs (one run/slice) were pooled and analyzed using the Scaffold 2.6 software. A total of ~1300 proteins were identified from each testis sample. Meiotic chromatin-associated proteins were identified by electronic subtraction using Scaffold 2.6 software as follows: meiotic chromatin-associated proteins = P18 testes – (P6 testes + XXY* testes) (Fig. 1d).
Recombinant proteins
All recombinant GST-fusion proteins were expressed in E. coli (strain BL21) using the pGEX4T-1 vector. The cDNAs encoding mouse MEIOB, mouse RPA2, and mouse RPA3 were amplified by PCR from testis cDNAs and subcloned into the pGEX4T-1 vector. Point mutations in MEIOB were generated by PCR-based mutagenesis. All constructs were verified by sequencing. For induction, 3 ml of LB medium (100 µg/ml ampicillin) was inoculated with a single colony, incubated for 6 hours at 37°C, and used to inoculate 50 ml LB medium (100 µg/ml ampicillin) at 1:1000 dilution. After overnight incubation, 50 ml culture was transferred to 600 ml fresh LB medium with ampicilin and continued to incubate at 37°C till an OD600 of ~1.0. IPTG was added to a final concentration of 0.2 mM and the culture was incubated with shaking (180 rpm) at 18°C for 3 hours. Bacteria were pelleted by centrifugation at 4°C, resuspended in 20 ml cold DPBS buffer (Gibco 14190-136), and sonicated on ice. After centrifugation at 12,000 g for 20 minutes at 4°C, the supernatant was transferred to a new tube. 300 µl of glutathione-Sepharose 4B beads (pre-treated with cold PBS) was added to the lysate and incubated with continuous mixing for 2 hours at 4°C. The beads were pelleted by centrifugation for 1 minute and washed 9 times with 10 ml ice-cold PBS each. Proteins were eluted with 1ml elution buffer (10 mM glutathione, 50 mM Tris-HCl, pH 8.0) for 6 times and the eluents were pooled. The eluted proteins were concentrated using the Pierce 20K MWCO concentrators (Cat. No. 89886A, Thermo Scientific).
Electrophoretic mobility shift and nuclease assays
All biochemical assays were performed with purified recombinant GST-MEIOB proteins expressed in E. coli. Nuclease assays were performed for 30 min at room temperature with 1 µM (or otherwise indicated) recombinant MEIOB protein and 2 nM 5’ or 3’ end-labeled oligonucleotides (Supplementary Table S2) in binding buffer (5x: 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, 0.25 µg/µl poly(dI-dC):poly(dI-dC), 20% glycerol; Promega). The EMSA binding assays were performed at room temperature for 30 minutes in the same buffer as for the nuclease assays except with 10 mM EDTA. DNA-protein complexes and DNA products were separated on 10% native or 25% denaturing acrylamide gels, and PhosphorImager radiographs were quantified using ImageQuant software.
Generation of polyclonal antibodies
The 6xHis-MEIOB (245–466 aa) fusion protein was expressed in E. coli using the pQE-30 vector, affinity purified with Ni-NTA resin, and used to immunize rabbits and guinea pigs (Cocalico Biologicals Inc.), yielding antisera UP2327 (rabbit) and UP-GP 99 (guinea pig). Affinity purified antibodies were used for immunofluorescence and western blotting analyses. Other antibodies used for western blotting are ACTB (1:7500, Cat. No. A5441, clone AC-15, Sigma), histone H3 (1:2500, Cat. No. 05-499, clone 6.6.2, Millipore), and TEX19 (1:500)13.
Targeted inactivation of the Meiob gene
In the targeting construct, a 9-kb genomic DNA segment harboring exons 6–10 of Meiob was replaced with the neomycin selection cassette (Fig. 5a). The two homologous arms (2.2 kb each) were amplified from a Meiob-containing BAC clone (RP23-397O23) by PCR with high-fidelity DNA polymerase. The thymidine kinase negative selection marker was inserted adjacent to the right arm. V6.5 ES cells were electroporated with linearized pUP105/ClaI targeting construct, cultured in the presence of 350 µg/ml G418 and 2 µM ganciclovir, and screened by long-distance PCR12. Out of 384 double resistant ES cell clones, two homologously targeted clones were obtained and both yielded germline transmission of the Meiob mutant allele through chimeric mice. All offspring were genotyped by PCR. Wild type allele (514 bp) was assayed by PCR with primers AGAGCCTTCTTTCACAATG and TTTATCTGCCCAGGACTAC. The mutant allele (382 bp) was assayed by PCR with primers ACCTATACATTGGTGACCTTGA and CCTACCGGTGGATGTGGAATGTGTG. Mice were maintained and used for experimentation according to the guidelines of the Institutional Care and Use Committee of the University of Pennsylvania.
Statistics
Statistical analysis was performed with Student’s t test. The values were presented as mean ± s.d. (standard deviation).
Histological and surface nuclear spread analyses
For histological analysis, testes or ovaries were fixed in Bouin’s solution overnight, embedded with paraffin, and sectioned using a microtome. Sections were processed and stained with hematoxylin and eosin. For immunofluorescence analysis, testes or ovaries were fixed in 4% paraformaldehyde for 3 hours at 4°C, dehydrated in 30% sucrose, processed, and sectioned. TUNEL assays were carried out with the ApopTag Fluorescein In Situ Apoptosis Detection Kit (catalog no. S7110, Serologicals Corporation/Chemicon). For spread analysis, spermatocytes from postnatal day 16 testes were used unless noted otherwise. Testicular tubules or embryonic ovaries were extracted in a hypotonic treatment buffer. Cells were suspended in 0.1 M sucrose and were then spread on a thin layer of paraformaldehyde solution containing Triton X-10034. The following primary antibodies were used for immunofluorescence on spread nuclei: SYCP1 (1:50, Cat. No. ab15090, Abcam), SYCP212, SYCP3 (1:100, Cat. No. 611230, BD Biosciences), γH2AX (1:500, Cat. No. 16-202A, Clone JBW301, Millipore), ATR (1:200, Cat. No. PC538, EMD Biosciences), RPA1 (1:50, Cat. No. ab87272, Abcam), RPA2 (1:100, Cat. No. 2208S, clone 4E4, Cell Signaling Technology), RAD51 (1:50, gift from the late Peter Moens), DMC1 (1:50, Cat. No. sc-22768 (H-100), Santa Cruz Biotech), MLH1 (1:50, Cat. No. 550838, clone G168-15, BD Biosciences), MSH4 (1:50, gift from Chengtao Her)53, SPATA22 (1:50, Cat. No. 16989-1-AP, ProteinTech Group), and TEX11 (1:10)34.
Co-immunoprecipitation
Co-IP was performed with postnatal day 18 testes using anti-MEIOB antibodies (Fig. 7a). Testes (200 mg) were homogenized in 1 ml RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM DTT, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA) with 1x protease inhibitor cocktail III. Immunoprecipitated proteins were run on a 4–15% gradient SDS-PAGE gel and stained with SYPRO Ruby (Bio-Rad). The protein band exclusive to the wild type testis sample was subjected to mass spectrometry.
For co-IP experiments followed by western blotting (Fig. 7b), 200 mg of postnatal day 18 testes (wild type or Meiob−/−) were homogenized in 1 ml RIPA buffer plus 1x protease inhibitor cocktail III on ice, and 4 ml cold RIPA buffer with 1x protease inhibitor cocktail III and 90 U benzonase/ml was added to the lysate. The lysate was incubated on a rocking platform at room temperature for 2 hours, followed by centrifugation at 16,100 g for 15 minutes at 4°C. The supernatant was transferred to a new tube and centrifuged again at 100,000 g for one hour at 4°C. The supernatant was used for immunoprecipitation with anti-MEIOB antibodies followed by western blotting. Full scans of key western blots are provided in Supplementary Fig. S12.
Supplementary Material
Acknowledgments
We thank B. Kaufman for advice on biochemical purification, C.X. Yuan for help with mass spectrometry, R. Schultz for the YBX2 antibody, the late P. Moens for the RAD51 antibody, C. Her for the MSH4 antibody, J. Kolesar for help with ImageQuant analysis, Z. Deng and S. Kasowitz for help with sequence alignments, and S. Eckardt for help with manuscript preparation. NIH grants GM089893 and GM076327 supported P.J.W.; HD42137 supported M.A.H.; funds from Midwestern University supported SL; and the Priority Program 1384 of the Deutsche Forschungsgemeinschaft supported R.B.
Footnotes
Author contributions
M.L. and F.Y. carried out most experiments and data analysis; N.A.L. performed blastocyst injection and generated chimeric mice; J.L., S.L., and M.A.H. contributed with SPATA22-related experiments; R.B. performed the electron microscopy experiment; P.J.W. conceptualized the proteomics screen, analyzed data and wrote the manuscript. All the authors were involved in the discussion and commented on the manuscript.
Competing financial interest statement: The authors declare no competing financial interests.
References
- 1.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Gerton JL, Hawley RS. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 2005;6:477–487. doi: 10.1038/nrg1614. [DOI] [PubMed] [Google Scholar]
- 3.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 4.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 6.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 7.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soustelle C, Vedel M, Kolodner R, Nicolas A. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 2002;161:535–547. doi: 10.1093/genetics/161.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu DS, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, et al. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, et al. The Ubiquitin Ligase Ubr2, a Recognition E3 Component of the N-End Rule Pathway, Stabilizes Tex19.1 during Spermatogenesis. PLoS One. 2010;5:e14017. doi: 10.1371/journal.pone.0014017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarrey JR. In: Cell and Molecular Biology of the Testis. Desjardins C, Ewing L, editors. New York: Oxford University Press; 1993. pp. 58–89. [Google Scholar]
- 15.Hunt PA, Eicher EM. Fertile male mice with three sex chromosomes: evidence that infertility in XYY male mice is an effect of two Y chromosomes. Chromosoma. 1991;100:293–299. doi: 10.1007/BF00360527. [DOI] [PubMed] [Google Scholar]
- 16.Plug AW, et al. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell. Sci. 1998;111(Pt 4):413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- 17.Moens PB, et al. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell. Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 18.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell. Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 19.Tarsounas M, Pearlman RE, Moens PB. Meiotic activation of rat pachytene spermatocytes with okadaic acid: the behaviour of synaptonemal complex components SYN1/SCP1 and COR1/SCP3. J. Cell. Sci. 1999;112(Pt 4):423–434. doi: 10.1242/jcs.112.4.423. [DOI] [PubMed] [Google Scholar]
- 20.Kneitz B, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 24.Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, et al. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Giacomo M, et al. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. U. S. A. 2005;102:737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries FA, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Capetillo O, Liebe B, Scherthan H, Nussenzweig A. H2AX regulates meiotic telomere clustering. J. Cell Biol. 2003;163:15–20. doi: 10.1083/jcb.200305124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J. Cell. Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 30.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner JM, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 32.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 33.Petukhova GV, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 34.Yang F, et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 36.Edelmann W, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 37.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 38.La Salle S, et al. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 2012;86:45. doi: 10.1095/biolreprod.111.095752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Rijkers T, et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svendsen JM, Harper JW. GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24:521–536. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joyce EF, Tanneti SN, McKim KS. Drosophila hold'em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics. 2009;181:335–340. doi: 10.1534/genetics.108.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osman K, Sanchez-Moran E, Mann SC, Jones GH, Franklin FC. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 2009;28:394–404. doi: 10.1038/emboj.2008.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida K, et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 50.Pittman DL, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 51.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bishop DK. Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell. Cycle. 2012;11:4105–4106. doi: 10.4161/cc.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Her C, Wu X, Griswold MD, Zhou F. Human MutS homologue MSH4 physically interacts with von Hippel-Lindau tumor suppressor-binding protein 1. Cancer Res. 2003;63:865–872. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.