Abstract
A quantitative understanding of the advantages of nanoparticle-based drug delivery vis-à-vis conventional free drug chemotherapy has yet to be established for cancer or other disease despite numerous investigations. Here, we employ first-principles cell biophysics, drug pharmaco-kinetics and drug pharmaco-dynamics to model the delivery of doxorubicin (DOX) to hepatocellular carcinoma (HCC) tumor cells and predict the resultant experimental cytotoxicity data. The fundamental, mechanistic hypothesis of our mathematical model is that the integrated history of drug uptake by the cells over time of exposure, which sets the cell death rate parameter, and the uptake rate are the sole determinants of dose response relationship. A universal solution of the model equations is capable of predicting the entire, nonlinear dose response of the cells to any drug concentration based on just two separate measurements of these cellular parameters. This analysis reveals that nanocarrier-mediated delivery overcomes resistance to free drug because of improved cellular uptake rates, and that dose response curves to nanocarrier mediated drug delivery are equivalent to those for free-drug, but “shifted to the left,” i.e., lower amounts of drug achieve the same cell kill. We then demonstrate the model’s general applicability to different tumor and drug types, and cell-exposure time courses by investigating HCC cells exposed to cisplatin and 5-fluorouracil, breast cancer MCF-7 cells exposed to DOX, and pancreatic adenocarcinoma PANC-1 cells exposed to gemcitabine. The model will help in the optimal design of nanocarriers for clinical applications and improve the current, largely empirical understanding of in vivo drug transport and tumor response.
Keywords: Drug delivery, mathematical modeling, mesoporous silica nanoparticle, pharmacokinetics-pharmacodynamics model, protocells
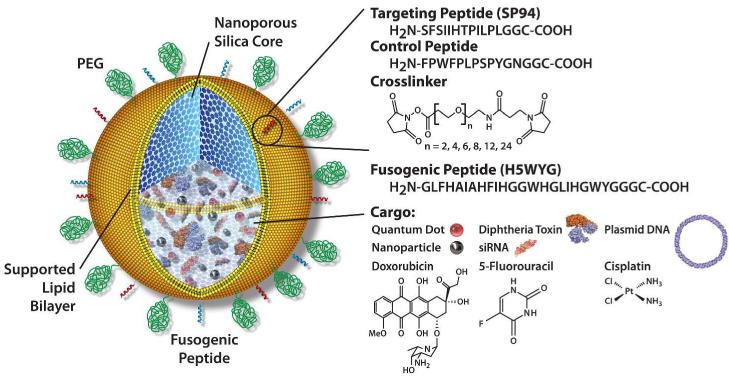
Stable nanoparticles capable of specifically binding to cancer cells and delivering high doses of therapeutic compounds could be transformational for cancer therapy by more efficiently delivering drugs into cancer cells, while simultaneously reducing toxic side effects in healthy cells and tissues.1-3 An ideal targeted nanoparticle drug carrier, or “nanocarrier,” should have: (i) the capacity for carrying high levels of multiple diverse molecular cargos, e.g., small molecules, drugs, siRNAs, peptides, and imaging agents; (ii) the ability to reside in the blood for extended periods without elimination by the immune or excretory systems; and (iii) the specificity for binding only to targeted disease cells (either endothelium or epithelium), while avoiding normal, healthy cells. To achieve these combined features, we have recently developed a modular, composite nanocarrier termed a “protocell” (Fig. 1).4-6 Protocells synergistically combine the advantages of liposomes (low inherent toxicity, immunogenicity, and long circulation times) and porous nanoparticles (stability and an enormous capacity for simultaneous delivery of multiple cargos). We have demonstrated that protocells have a 100-fold greater specificity in target-cell binding than equivalent fluid-phase liposomes.4 Furthermore, using a model of hepatocellular carcinoma (HCC), we demonstrated that protocells carrying a cocktail of doxorubicin (DOX), 5-FU, and cisplatin were so potent that a single targeted protocell could kill a multidrug-resistant hepatocellular carcinoma cell in vitro, while sparing normal hepatocytes.4
Figure 1.
Protocell Design. Nanoporous silica cores are loaded with multiple drug cargos by adsorption to the high surface area silica matrix. The drug-loaded core is then enveloped by a single lipid bilayer, which is further functionalized with: 1) polyethylene glycol (PEG) to reduce nonspecific interactions with its environment; 2) targeting peptides to direct protocell binding to specific cells; and 3) pH-responsive peptides which cause disruption of endosomes and the bilayer coating upon particle internalization into acidic intracellular compartments, allowing drug delivery into the cytosol of the target cell.
Here, using a mathematical modeling framework based on first principles of drug and cell mass conservation, with biophysical parameters describing cellular uptake rates of the drug/nanoparticle and cellular death rates, we accurately predict dose response of HCC to DOX, 5-FU, and cisplatin administered as free drugs or loaded into protocells, and provide mechanistic understanding of the observed increased efficiency and efficacy of targeted nanoparticle-based delivery compared to free drug. In particular, delivery of DOX using protocells was found to kill higher numbers of cells in a colony than the delivery of free DOX due to increased uptake rates by cells in nanocarrier-mediated delivery and mitigation of drug loss by MDR efflux pumps such as P-glycoprotein (Pgp).
Integrative research studying cellular response to both free and nanoparticle-delivered chemotherapeutic drugs has so far focused on one particular aspect of response, such as the dose response curve or the pharmacokinetics-pharmacodynamics of a particular drug.7-10 The area under the concentration versus time curve (AUC) has been the major predictor of anti-cancer agent effects on cell death. The survival of cells relative to controls, when plotted against either the extracellular AUC or CaT (where C is concentration of drug, T, exposure time and a is a constant dependent on tumor type), yields a nonlinear, sigmoidal curve that can be typically described by the Hill model.7 In line with these phenomenological approaches, many ad-hoc modifications have been made to the Hill model to describe dose response curves obtained from in vitro cytotoxicity experiments, including examining the shape of the concentration.versus. time curve and exploring how cell damage is affected or determined by drug accumulation within the cells.10, 11 However, the Hill-type models do not allow one to predict how features like the duration of drug exposure (a crucial factor in determining the shape of the dose response curve) affect cell kill. To address this problem, various mechanistic models, including exponential kill models9 and other types of models7, 8, 12-16 that rely on compartmental models of drug action, have been developed. However, these models require measurements of many biological properties of tumor cells and drugs (e.g., cell cycle phase specificity, cell cycle time, and the level of drug resistance), making them difficult to use in predicting dose response curves.
Here, we use an integrated experimental and mathematical modeling approach, which builds on our prior efforts,17, 18 to study and quantify in vitro tumor drug response. Using experimental cytotoxicity data, we develop a simple yet mechanistic mathematical model from first principles, coupling the cell and drug dynamics, and fit this model to the data to obtain parameters describing cellular uptake of—and response to—the drug. We demonstrate that the cell death rate is a universal mechanistic, predictable function of the time-integral of drug exposure, replacing unnecessarily complicated and ad-hoc phenomenological models of cell death described above.8, 9, 16 Furthermore, after calibrating the model using just two drug concentration data points, we accurately predict the nonlinear dose response curves for all drug concentrations and for both types of delivery methods, i.e., free drug and drug-loaded protocells, to both drug sensitive and resistant HCC cell types. We then demonstrate the general applicability of the model formulation to different cell/drug types, delivery vehicles, and time courses, where these differences are simply and uniquely accounted for by different values of two fundamental parameters, i.e., cell death and drug uptake rates.
Results and Discussion
Mathematical model
We performed a set of monolayer in vitro assays (see Materials and Methods) where the dynamics of viable cells (change in viable population of cells over time due to drug uptake) and drug (change in drug concentration over time due to uptake by cells) were inter-dependent. Thus, we developed a mathematical model, from first principles of cell and drug mass conservation, which describes the dynamics of the viable population of cells as a function of drug concentration and the history of drug uptake by the cells. See SI Text for details and formulation. Below, we report the solutions describing the changes in viable cell population and drug concentration over time for the three scenarios considered in the experiments.
Scenario 1: Continuous drug-exposure model
Defining the dimensionless variables, , , , Eqs. S1-S4 (SI Text) leads to semi-analytical implicit solutions for drug and cell concentrations and as a function of time :
| [1a,b] |
σ0 and n0 are initial drug and cell concentrations; λ the specific rate of uptake of drug (by a unit concentration of cells). The dimensionless constant is the ratio between the characteristic time scales associated with drug uptake by the cells and cell death.
Scenario 2: Discontinuous drug-exposure model
If drug exposure in the above scenario is discontinued at , Eq. 1 is only valid for , i.e., while drug is delivered continuously to the cells. It is straightforward to demonstrate that, for , Eq. 1 can be replaced by (Eqs. S5, S6, SI Text):
| [2a,b] |
and are the concentrations of viable cells and of drug, calculated from Eq. 1 at . Eq. 2 describes apoptosis of cells in the colony at an exponential rate set by the total uptake of drug that has occurred up to time .
Scenario 3: Constant drug-concentration model
For experimental conditions simulating constant drug concentration , Eqs. S7, S8 lead to the solution (SI Text):
| [3a,b] |
is total concentration of drug taken up from time 0 to time .
Analysis of Coupled Cell and Drug Dynamics
Asymptotic behaviors of Eq. 1 can be found for short and long drug exposure times, respectively (in dimensional variables):
| [4a] |
| [4b] |
σ∞ is the drug concentration value as time t goes to infinity and drug uptake by the cells is completed, and is calculated from Eq. 1b with n = 0. Based on the model assumptions, Eq. 4 reveals that the cells initially uptake drug, thus decreasing drug concentration over time at an exponential rate λ · n0. As the drug concentration tends to a constant long-time value σ∞, cells begin to die. At large times, cell death is exponential, with death rate λA · (σ0−σ∞) linearly proportional to the total amount of drug σ0−σ∞ that the cells have taken up (total uptake). Similar cell viability dynamics occur for discontinuous (Eq. 2) and constant (Eq. 3) drug exposure.
Analysis of Cellular Biology Parameters
The parameter values obtained from the fits of the mathematical model to the experimental data (Fig. 2; Materials and Methods) are reported in Fig. 3 vs. initial DOX concentration σ0 in the medium for drug sensitive and resistant HCC cell lines (Table S1, SI Text). Because the drug is packaged at high concentration within the protocell carrier, the effective uptake rates λ · n0 of drug by the cells are higher for protocell-based delivery than for free drug delivery (Fig. 3A) at the same total amount of drug administered in the medium. The uptake rates are independent (within error) of initial DOX concentration σ0 (and roughly on cell line) for both methods of delivery. The total amount of DOX, i.e., σ0−σ∞, taken up by the cells (inset) increases as expected with initial DOX concentration σ0, and is higher for protocell-based delivery than free-drug delivery.
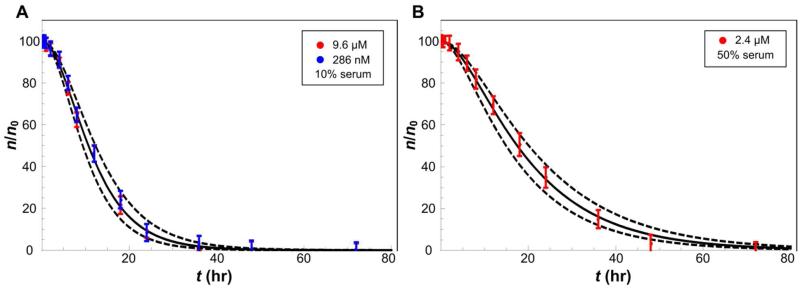
Figure 2.
Sample fits of Eq. 1 (solid curves) to cytotoxicity data (symbols: fraction of viable cells n/n0 with S.D.) at different drug concentrations, used to calculate mean values of drug-uptake rate and cell-death rate parameters. (A) Drug resistant (red; initial DOX concentration σ0 = 9.6 μM) and sensitive (blue; σ0 = 286 nM) HCC cell lines exposed to free DOX. (B) Drug resistant HCC cells (red; σ0 = 2.4 μM) exposed to DOX-loaded protocells. Additional fits to upper and lower bounds of the experimental measurements (dashed) were used to calculate standard deviations of the parameters (Tables S1,S2; SI Text).
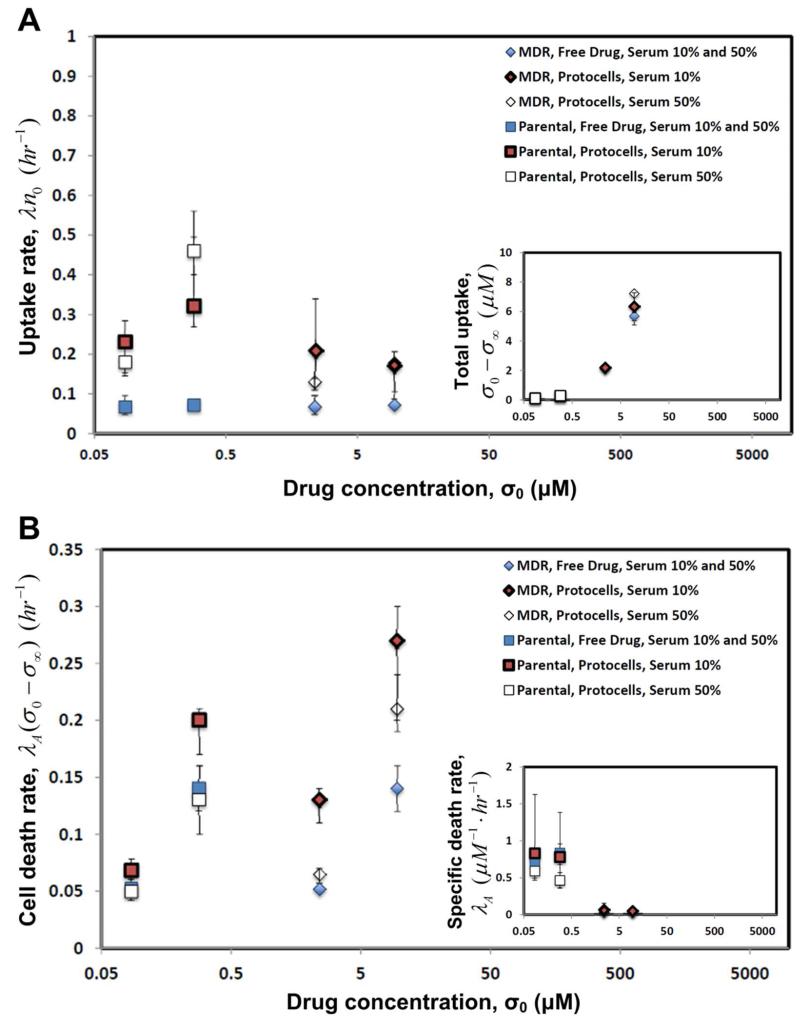
Figure 3.
DOX-loaded protocell delivery (red and white symbols with S.D.) leads to higher uptake rates and thus higher rates of cell death compared to free DOX (blue). (A) Uptake rate λ · n0 by drug-resistant (MDR, diamonds) and sensitive (parental, squares) HCC cell lines. Inset: total uptake of drug σ0 − σ∞, i.e., the difference between initial and long-time drug concentrations in the medium. (B) Long-time cell death rate λA · (σ0 − σ∞), established in the colony after total drug uptake σ0 − σ∞ has occurred. Inset: specific death rate λA, i.e., the death rate per unit concentration of drug uptaken. The parameters were calculated by fitting Eq. 1 to the time-dependent cytotoxicity data (e.g., see Fig. 2). For protocells, initial DOX concentration σ0 is the product of the concentration of protocells in the medium times the average concentration of drug in protocells. σ0 = 2.4 and 9.6 μM (MDR cells), and 0.0857 and 0.286 μM (sensitive).
The long-time death rates λA · (σ0−σ∞) for the drug sensitive and resistant cell lines (Fig. 3B) increase with initial DOX concentration σ0 for both methods of delivery. The protocell delivery of DOX leads to higher death rates than those corresponding to free-drug delivery, especially for MDR cells. The results also suggest that for the cells and drug considered here, serum percentage has a negative effect on death rate, which could be a result of protein association with the supported lipid bilayer (Fig. 1) of the protocell causing modest leakage of DOX. Moreover, the specific death rates λA for both drug resistant and sensitive cell lines are independent (within error) of the method of delivery and initial DOX concentration σ0 (inset). This supports the model hypothesis (Eq. S3, SI text) that a linear long-time death rate λA · (σ0−σ∞) proportional to the total uptake of drug is sufficient to capture the behavior of the cells, at least within the range of drug concentrations studied here, without the use of unnecessarily complicated models of cell death.8, 9, 16 Altogether, these results imply that the protocell-based delivery is more efficient and effective at killing cells than free-drug.
Test of the Time-Dependent Model
As a first test of model predictivity, we investigated the effect of the duration of cell exposure to DOX on cell death (Fig. 4) by using experimental data corresponding to delivery for 4 hours only to MDR HCC cell lines via both free DOX and DOX-loaded protocells at two initial DOX concentrations σ0. We applied the mathematical model Eq. 2 with td = 4 hr. We kept the values of the parameters λ · n0 and λA the same as previously calculated from the corresponding continuous DOX-exposure experiments (Fig. 3; Table S1, SI Text), since according to the model assumptions these parameters are intrinsic properties of the cells independent of the time-delivery protocol. The curves predicted by the mathematical model agree (Fig. 4) with the data corresponding to 4 hr drug exposure (coefficients of determination R2 = 0.988 and 0.982 for the free DOX and DOX-loaded 4 hour-exposure curves at an initial DOX concentration σ0 = 92.5 nM; and R2 = 0.995 and 0.973 for σ0 = 298 nM), thus validating the model assumptions and equations. When drug is delivered for 4 hours only, cells begin to regrow with a doubling time of mitosis of about 25–32 hr while at the same time the kill effect of the drug declines; thus, we do not expect the parameters of the model to accurately predict cell viability values for times longer than this doubling time. Consistent with the mathematical model, continuous delivery leads to higher total uptake, i.e., σ0−σ∞ > σ0−σ(4 hr), because σ(4 hr) > σ∞, and thus higher death rates and total death are achieved. This result is consistent with the observation that the uptake time scale 2 hr < (λ · n0)−1 < 15 hr, and thus after 4 hours the drug uptake process is typically far from being completed.
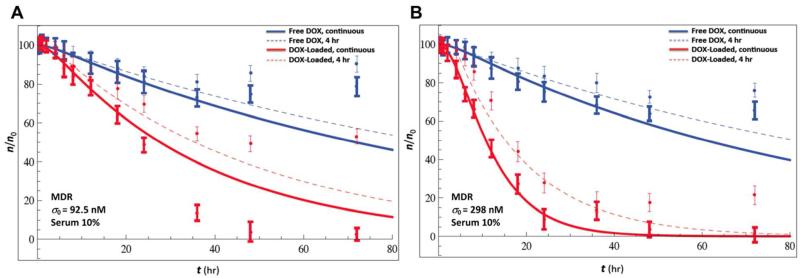
Figure 4.
Continuous delivery of DOX over time (thick symbols and solid lines) leads to higher long-time death rates compared to delivery for 4 hours (thin symbols and dashed lines). Percentage of viable cells n/n0 versus time t (symbols with S.D.) for free DOX (blue) and DOX-loaded protocells (red) delivered for 4 hours and continuously for 72 hours to MDR HCC cell lines; 10% serum; Eq. 1 (thick lines) and Eq. 2 (dashed lines). Initial DOX concentrations σ0 = 0.0925 μM (A) and 0.298 μM (B). The mathematical model, with parameters calibrated from the continuous-drug exposure experiments, is predictive of the viability corresponding to the 4-hr exposure experiments. The increase in viability observed at long times (symbols) is due to resumed proliferation and not accounted for in the mathematical model of cell kill.
General Applicability of the Model
We tested the generality of the model by applying it to continuous time-exposure experiments of HCC cells to different drug types, i.e., cisplatin (cis) and fluorouracil (5-FU). The long-time death rates λA · (σ0−σ∞) and specific death rates λA for cis and 5-FU were obtained for both parental and MDR HCC cell lines at a single value of initial drug concentration σ0, and are independent of delivery methods and cell line (within error) (Table S2, SI Text). The uptake rates of drug by the cells λ · n0 and total amount of drug taken up by the cells σ0−σ∞ for cis and 5-FU are also roughly insensitive to delivery method and cell line (within error). Therefore, in vitro, there is no apparent advantage in using protocells to deliver cis and 5-FU to HCC cells in contrast to the case for DOX (Fig. 3). These observations are consistent with the MDR cell line remaining sensitive to cis and 5-FU, which are not substrates for P-glycoprotein (P-gp; a transporter that transports substrates across extra- and intracellular membranes), and with the fact that cis and 5-FU administered in dimethyl sulfoxide (DMSO) can diffuse readily into cells. However this DMSO delivery approach cannot be used in vivo or in human patients, so it is reasonable to expect that there may be an advantage of using protocell-mediated delivery in a clinical setting. Note finally that the model correctly predicts lower uptake for these drugs than for DOX when protocells are used, which is consistent with the observation that the former drugs are loaded at lower concentration than DOX in the protocells.
We then tested applicability to different cell types, by revisiting our in vitro experiments17 with continuous delivery of free DOX to MCF-7 breast cancer cell lines. By fitting the numerical solution of Eq. 1 at t = 96 hr to the dose response data at several initial drug concentrations σ0 for MDR and parental MCF-7 cells (Fig. S1A, SI Text), we obtained the parameter values: λ · n0 = 0.1 hr−1 and λA = 0.08 hr−1 μM−1 (MDR); λ · n0 = 0.07 hr−1 and λA = 0.4 hr−1 μM−1 (parental). The coefficients of determination for the fits were R2 = 0.98 (MDR) and 0.94 (parental). These parameter values are similar to those found for HCC cells (parental) and larger for MDR MCF-7 than MDR HCC, which is consistent with the observation that smaller values of σ0 are necessary to achieve the same kill in MDR MCF-7 than MDR HCC cells.
Finally, we tested the model on a different cell type and drug, i.e., on human pancreatic adenocarcinoma cells continuously exposed to free gemcitabine, as well as to constant gemcitabine concentration (Methods). By fitting the numerical solution of Eq. 1 (continuous exposure) and Eq. 3 (constant exposure) at t = 96 hr (Fig. S1B, SI Text), we found the parameter values: λ · n0 = 0.028 hr−1 and λA = 0.74 hr−1 μM−1 (continuous); λ · n0 = 0.025 hr−1 and λA = 0.668 hr−1 μM−1 (constant). The coefficients of determination for the fits were R2 = 0.94 (continuous) and 0.99 (constant). The similarity of parameter values is expected because these values are a property of the cell/drug combination and should be independent of time protocol of delivery. Also, the larger specific death rates found in these experiments are consistent with the observation that cell kill is achieved at smaller initial drug concentrations.
Prediction of Dose Response Curves
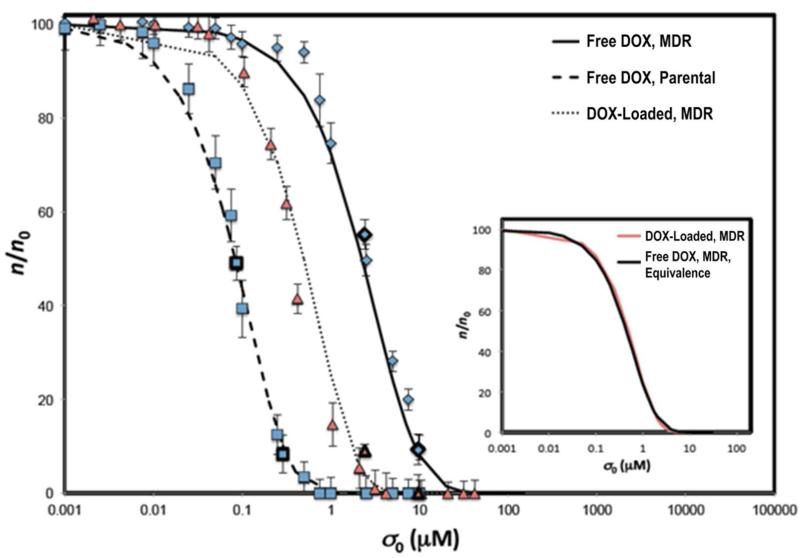
We used the average values of the specific death rate λA and uptake rate λ· n0 calibrated (Figs. 2,3; Table S1, SI Text) from the time-dependent data of HCC cell viability at two values of initial drug concentration σ0, and predicted theoretical dose-response curves spanning the full range of drug concentrations by numerically integrating Eq. 1 up to t = 24 hr (Methods). A comparison of the theoretical and experimental dose response curves, i.e., fraction of viable cells n/n0 at t = 24 hr versus initial drug concentration σ0, for free DOX and DOX-loaded protocells is reported in Fig. 5 (coefficients of determination R2 = 0.987, 0.991, and 0.996 for MDR cell line with free DOX, DOX-loaded protocells, and parental cell line with free DOX). The parameter values used were: λA = 0.051 (hr·μM)-1 and λ · n0 = 0.19 hr−1 (MDR with DOX-loaded protocells); λA = 0.025 (hr·μM)-1 and λ · n0 = 0.069 hr−1 (MDR with free DOX); λA = 0.769 (hr·μM)-1 and λ · n0 = 0.069 hr−1 (parental with free DOX). The ability to accurately predict the entire dose response curves from parameter fits based on using just two experimental concentration values, despite the nonlinear nature of drug response and without incorporating complicated kinetic arguments to describe cell death, provides further support to the mathematical model assumptions.
Figure 5.
Mathematical model predicts dose-response curves. Numerical integration of Eq. 1 at t = 24 hr (solid, dashed and dotted curves) and experimental cytotoxicity data (symbols with S.D.): free DOX (blue squares: parental; diamonds: MDR); DOX-loaded protocells (red). R2 = 0.987 (MDR, free DOX), 0.991 (MDR, DOX-loaded protocells), and 0.996 (parental, free DOX). For prediction of each dose-response curve, a single value of drug uptake rate λ · n0 and specific cell death rate λA parameters was calculated (Methods; Table S1, SI Text) by fitting Eq. 1 to time-course viability data at two drug concentrations (dark-outlined symbols), and then averaging the two resulting values for each parameter. Inset: “equivalence” between free-DOX and DOX-loaded-protocell dose-response is demonstrated by superposing least-square fits (R2 = 0.999) through the two data sets (by rescaling the free-drug concentration by a factor = 1/5 determined empirically).
“Equivalence” between free-drug and protocell dose-response curves
We further validated the mathematical model by demonstrating equivalence between free DOX and DOX-loaded HCC dose response (Fig. 5, inset), by superposing least-square fits through the two sets of dose-response data (rescaling the initial drug concentration σ0 for the free-drug case by a factor of roughly 1/5 determined empirically). This remarkable result validates the mathematical model prediction (Methods) that differences in drug response corresponding to different experimental conditions of drug uptake and cell death can be accounted for through different values of the associated rate parameters in the model (Fig. 3), while the fundamental functional relationships, e.g., Eq. 1 between dose and response, universally apply to all cell and drug types, delivery time-courses, and delivery vehicles, i.e., free drug vs. protocells. For the case where DOX-loaded protocell and free DOX delivery are compared, the results in Fig. 5 imply that the delivery of DOX using protocells is more efficient (because of higher cellular uptake rates) and affords a kill equal to the kill achieved by delivery of free DOX at a higher total concentration or at a later time.
Model implications
We demonstrated the general applicability and predictive power of the mathematical model presented here by applying it to different cell and drug types, and to both free and nanocarrier-mediated drug delivery. Actually, this general modeling approach based on conservation laws has been successfully applied to the prediction of patient-specific response to drug and of response to immunotherapy in mice.19, 20 To date, extensive research has been conducted to study the drug delivery capabilities of nanoparticles compared to the traditional methods of delivery.2, 21, 22 It has been shown that nanoparticles loaded with drug are often superior in reaching intended targets than other approaches. Here we sought to understand the mechanisms underlying how the delivery of DOX and other drugs to HCC tumor cells using the protocell drug delivery vehicle results in more efficient and effective cell death than traditional free-DOX delivery. We found that this outcome is primarily due to the fact that the protocell delivery allows the cells to uptake drug at higher rates, which then leads to faster rates of death. The initial concentration of DOX σ0 necessary to obtain the same amount of cell kill is less for protocell-mediated delivery compared to free-DOX delivery. This warrants the use of protocells as a viable alternative in the clinical setting. The ability of the protocells to circumvent or overcome the drug resistant pumps5 within the cells and to be loaded with extremely high concentrations of DOX allows them to outperform free drug and other nanocarriers.
The two model parameters, i.e., the integrated history of drug uptake by the cells over time of exposure and the uptake rate, sum up all higher-order phenomena (such as endosomal escape efficiency of the protocells etc.) that potentially play a role in determining drug response. Accordingly, these phenomena are not directly modeled in the mathematical formulation, as the two parameters are instead directly measured from “calibration” experiments as described throughout (see especially Fig. 2 and associated text). We have also demonstrated that (see e.g., Fig. 3), as a result of a number of higher-order phenomena, accounted for by different values of the two parameters, nanoparticle-based delivery leads to higher uptake-rates and thus higher cell kill than free-drug delivery. Direct incorporation of higher-order mechanisms could in principle lead to an “ab-initio” predictive model that would potentially not require calibration. Furthermore, the protocell platform incorporates two pH-sensitive triggers that help to very efficiently deliver drug into the cytoplasm. First, acidification of the endosome destabilizes the supported lipid bilayer allowing drug escape and, second, the fusogenic peptide incorporated in the supported lipid bilayer serves as a proton sponge leading to endosome swelling and disruption. Together these triggers help to rapidly deliver the drug into the endosome so that endosomal escape efficiency is not in any way rate limiting, and does not need to be specifically accounted for in the model as presented here.
Conclusions
Over seventy years of chemotherapy research have resulted in phenomenological explanations of cell death typically confined to specific cell and drug types, but have failed to provide a unified mechanistic understanding of how underlying biophysical processes affect drug uptake and cellular death rates. We present a universal solution of first-principle conservation equations of cell and drug mass, incorporating the fundamental biological hypothesis that the history of drug uptake by the cells is the sole determinant of death rates. This general mathematical model quantifies and predicts in vitro dose response of different human tumor types to a variety of drugs, elucidates the advantages of targeted nanoparticle-based drug delivery directly to cancer cells, and will help in the rational design of nanocarriers and drug dosing regimens.
Materials and Methods
Experimental Determination of Dose-Response and Time-Dependent Viability in HCC cells by Free Drug and Drug-Loaded Protocells
Detailed materials and methods that include procedures used to synthesize drug-loaded, SP94-targeted protocells have been published in the Supplementary Information section of C. Ashley, et al.5 Parental Hep3B (ATCC cat. no. HB-8064) were grown according to manufacturer instructions. MDR was induced via exposure to increasing concentrations of DOX in 24-hr intervals (25, …, 100, 150, …, 500, 600, …, 900 nM; 1, 1.5, …, 3, 4, and 5 μM), interspersed with 48-hr recovery periods, during which cells were incubated in complete growth medium without DOX.23
Dose-response curves for parental and MDR Hep3B, when exposed to free and protocell-encapsulated DOX, cisplatin, and 5-FU, were determined by seeding 6-well plates with 106 cells/well, allowing cells to adhere overnight, and then exposing cells to initial concentrations σ0 = 0.1-10,000 nM of drug in phenol red-free DMEM containing 10% or 50% FBS for 24 hours at 37°C. Here varying levels of serum were used to assess the stability of the protocells in complex media representative of in vivo environments. Cells that remained adherent were dislodged from the well using a cell scraper, centrifuged (4000 rpm, 2 minutes) to remove excess drug, and stained with SYTOX® Green nucleic acid stain and Alexa Fluor® 647-labeled annexin V. The numbers of viable (double-negative) and non-viable (single- or double-positive) cells were determined via flow cytometry (FACSCalibur); SYTOX® Green fluorescence was excited by the 488-nm laser and collected in the FL1 channel, while Alexa Fluor® 647 fluorescence was excited by the 633-nm laser and collected in the FL3 channel (670-nm long pass filter). The concentrations of drug necessary to kill 50% (LC50) and 90% (LC90) of cells were determined from the dose-response curves.
Time-response curves were then determined by continuously incubating cells with the LC50 or LC90 values of drug for 30 min to 72 hr (continuous drug exposure). Discontinuous drug-exposure time-response curves were constructed by exposing cells to free or protocell-encapsulated drug up to td = 4 hr and then incubating them in fresh medium for 30 min to 72 hr. Cell viability was determined at various times points as described above. Parental and MDR Hep3B exposed to 1 μM of CsA for 72 hours to reverse any Pgp-mediated resistance24 were used as controls. The time-dependent viability and doubling rates of untreated cells were used to normalize all viability data.
Experimental Determination of Apoptosis in PANC-1 cells by Free Gemcitabine
Cells were plated at a density of 5000 cells/well of a 96-well plate (Corning). Twenty-four hr after plating, the medium was replaced and gemcitabine was given at increasing concentrations σ0 = 0, 3.1, 6.2, 12.5, 25, 50, 100, 500, and 1000 nM. Additionally, two experimental groups were tested at the same time. One group received only an initial dose of gemcitabine at one of the above concentrations (continuous drug exposure). Another group received one of the above concentrations of gemcitabine every 24 hours, simulating a relatively “constant” exposure. Cell viability was determined with the MTS assay, according to manufacturer’s protocol (Promega), at t = 1, 2, 24, 48, 72, and 96 hr after initial gemcitabine exposure. All experiments were repeated three times.
We also used previously published data17 of continuous DOX exposure of human breast cancer MCF-7 cells (both parental and MDR) to test the mathematical model. For these cells, as well as for the human pancreatic cancer PANC-1 cells exposed to gemcitabine as described below, dose-response data revealed the existence of a subpopulation of < 10% of cells who were intrinsically resistant to the drugs used and survived at all drug concentrations applied. In the mathematical model analysis and for the purpose of comparing model results to in vitro viability data, these cells were removed from the analysis.
Mathematical Model of Coupled Cell and Drug Dynamics
The mathematical model from fundamental mass conservation considerations coupled with cytotoxicity monolayer data for protocells and free drug quantifies the advantages of the former nanocarrier drug delivery platform to tumor cells.
Equivalence of dose-response curves
The drug concentration variable can be numerically eliminated from Eqs. 1a,b to obtain dose-response curves, i.e., cell viability as function of initial drug concentration (through the parameter Φ) and time: . The functional form of F cannot be expressed analytically. This simple result demonstrates “equivalence” of dose-response curves corresponding to different values of drug uptake and cell death rate parameters particularly when these differences are due to different cell/drug types or delivery mechanisms, i.e., nanocarriers vs. free drug. Despite these differences, dose response is a universal function solely of dimensionless time and the parameter Φ. Similar considerations apply to the other time-exposure courses considered (Eqs. 2,3).
Calculation of drug concentration and cell viability
Here, the solutions of Eq. 1 were calculated by numerical integration of the corresponding differential Eqs. S1-S4 (SI Text), using Mathematica (routine “NDSolve”),25 and then fit to the cytotoxicity time-dependent data for continuous drug exposure of HCC cells at two different initial drug concentrations σ0 for each cell type thus calibrating the death and uptake rate parameters Φ and λ · n0, as shown in examples in Fig. 2, where by fitting both the mean and the upper/lower bounds of the range of viability measurements at each concentration produced estimates of sensitivity of the model parameters (Fig. 3; Tables S1,S2, SI Text). Similar fits were performed for continuous drug-exposure to different drug types (SI Text).
Time dependent curves were also produced and compared to the discontinuous drug-exposure experiments with HCC cells by evaluating Eq. 2 using the same parameter values found from the fits described above for continuous exposure (Fig. 4).
To predict dose-response curves for HCC cells (Fig. 5), the parameter values found at two concentrations by the fits to the continuous drug-exposure data described above were then simply averaged; Eq. 1 was numerically evaluated at time t = 24 hr for all concentrations σ0 to produce the curves in Fig. 5. A graph was then constructed by rescaling drug concentration to superpose dose-response data corresponding to free-drug and protocell delivery to demonstrate equivalence of the dose-response curves (Fig. 5, inset). Eqs. 1 and 3 were also directly fitted at t = 96 hr to dose-response data for MCF-7 breast cancer cell lines exposed continuously to DOX and to dose-response data for pancreatic adenocarcinoma PANC-1 cell lines exposed both continuously and to a constant concentration of gemcitabine, respectively, to produce the curves in Fig. S1, SI Text.
Supplementary Material
Acknowledgements
P Dogra, A Day (Cristini lab). Support: NIGMS K12GM088021 (JP); NSF Grant DMS-1263742, CTO PSOC-1U54CA143837, TCCN-1U54CA151668, USC PSOC-1U54CA143907, ICBP-1U54CA149196 (VC, ZW); NSF SBIR 1315372, the Victor and Ruby Hansen Surface Professorship in Molecular Modeling of Cancer (VC); Methodist Hospital Research Institute (EJK, VC); the Anne Eastland Spears Fellowship (EJK); the Roadmap for Medical Research under grant NIH PHS 2 PN2 EY016570B (CJB), NCI 1U01CA151792-01(ECC, CJB), DOE BES Materials Science and Engineering Program (ECC, CJB), Sandia National Laboratories LDRD (ECC, CEA, CJB), NIEHS 1U19ES019528-01, NSF EF-0820117 (CEA, CJB); the President Harry S. Truman Fellowship in National Security Science and Engineering at Sandia National Laboratories (CEA).
Footnotes
Conflicts of interest. The authors declare that no competing interests exist.
Author contributions. V.C. developed the drug uptake hypothesis and mathematical model. V.C. and C.J.B. conceived and designed research. J.P., C.E.A., E.C.C., C.J.B., and V.C. performed research. J.P., C.E.A., Z.W., T.B., E.K., C.J.B., and V.C. analyzed data. Z.W., C.J.B., and V.C. wrote the paper.
Supporting Information Available. A detailed description of the model derivation and additional results are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chidambaram M, Manavalan R, Kathiresan K. Nanotherapeutics to Overcome Conventional Cancer Chemotherapy Limitations. J Pharm Pharm Sci. 2011;14:67–77. doi: 10.18433/j30c7d. [DOI] [PubMed] [Google Scholar]
- 2.Davis ME, Chen Z, Shin DM. Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin VP. Recent Advances with Liposomes as Pharmaceutical Carriers. Nat Rev Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 4.Ashley CE, Carnes EC, Epler KE, Padilla DP, Phillips GK, Castillo RE, Wilkinson DC, Wilkinson BS, Burgard CA, Sewell RM, et al. Delivery of Small Interfering RNA by Peptide-Targeted Mesoporous Silica Nanoparticle-Supported Lipid Bilayers. ACS Nano. 2012;6:2174–2188. doi: 10.1021/nn204102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, et al. The Targeted Delivery of Multicomponent Cargos to Cancer Cells by Nanoporous Particle-Supported Lipid Bilayers. Nat Mater. 2011;10:389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Stace-Naughton A, Jiang X, Brinker CJ. Porous Nanoparticle Supported Lipid Bilayers (Protocells) as Delivery Vehicles. J Am Chem Soc. 2009;131:1354–1355. doi: 10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Kareh AW, Secomb TW. A Mathematical Model for Cisplatin Cellular Pharmacodynamics. Neoplasia. 2003;5:161–169. doi: 10.1016/s1476-5586(03)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Kareh AW, Secomb TW. Two-Mechanism Peak Concentration Model for Cellular Pharmacodynamics of Doxorubicin. Neoplasia. 2005;7:705–713. doi: 10.1593/neo.05118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner SN. A Mechanistic, Predictive Model of Dose-Response Curves for Cell Cycle Phase-specific and -nonspecific Drugs. Cancer Res. 2000;60:1417–1425. [PubMed] [Google Scholar]
- 10.Levasseur LM, Slocum HK, Rustum YM, Greco WR. Modeling of the Time-Dependency of In Vitro Drug Cytotoxicity and Resistance. Cancer Res. 1998;58:5749–5761. [PubMed] [Google Scholar]
- 11.Lankelma J, Fernández Luque R, Dekker H, Pinedo HM. Simulation Model of Doxorubicin Activity in Islets of Human Breast Cancer Cells. Biochim Biophys Acta. 2003;1622:169–178. doi: 10.1016/s0304-4165(03)00139-9. [DOI] [PubMed] [Google Scholar]
- 12.El-Kareh AW, Secomb TW. A Mathematical Model for Comparison of Bolus Injection, Continuous Infusion, and Liposomal Delivery of Doxorubicin to Tumor Cells. Neoplasia. 2000;2:325–338. doi: 10.1038/sj.neo.7900096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanga S, Sinek JP, Frieboes HB, Ferrari M, Fruehauf JP, Cristini V. Mathematical Modeling of Cancer Progression and Response to Chemotherapy. Expert Rev Anticancer Ther. 2006;6:1361–1376. doi: 10.1586/14737140.6.10.1361. [DOI] [PubMed] [Google Scholar]
- 14.Sinek J, Frieboes H, Zheng X, Cristini V. Two-Dimensional Chemotherapy Simulations Demonstrate Fundamental Transport and Tumor Response Limitations Involving Nanoparticles. Biomed Microdevices. 2004;6:297–309. doi: 10.1023/B:BMMD.0000048562.29657.64. [DOI] [PubMed] [Google Scholar]
- 15.Sinek JP, Sanga S, Zheng X, Frieboes HB, Ferrari M, Cristini V. Predicting Drug Pharmacokinetics and Effect in Vascularized Tumors using Computer Simulation. J Math Biol. 2009;58:485–510. doi: 10.1007/s00285-008-0214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliaz RE. Determination and Modeling of Kinetics of Cancer Cell Killing by Doxorubicin and Doxorubicin Encapsulated in Targeted Liposomes. Cancer Res. 2004;64:711–718. doi: 10.1158/0008-5472.can-03-0654. [DOI] [PubMed] [Google Scholar]
- 17.Frieboes HB, Edgerton ME, Fruehauf JP, Rose FR, Worrall LK, Gatenby RA, Ferrari M, Cristini V. Prediction of Drug Response in Breast Cancer using Integrative Experimental/Computational Modeling. Cancer Res. 2009;69:4484–4492. doi: 10.1158/0008-5472.CAN-08-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Ven AL, Wu M, Lowengrub J, McDougall SR, Chaplain MA, Cristini V, Ferrari M, Frieboes HB. Integrated Intravital Microscopy and Mathematical Modeling to Optimize Nanotherapeutics Delivery to Tumors. AIP Adv. 2012;2:11208. doi: 10.1063/1.3699060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascal J, Bearer EL, Wang Z, Koay EJ, Curley SA, Cristini V. Mechanistic Patient-Specific Predictive Correlation of Tumor Drug Response with Microenvironment and Perfusion Measurements. Proc Natl Acad Sci U S A. 2013;110:14266–14271. doi: 10.1073/pnas.1300619110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das H, Wang Z, Niazi MK, Aggarwal R, Lu J, Kanji S, Das M, Joseph M, Gurcan M, Cristini V. Impact of Diffusion Barriers to Small Cytotoxic Molecules on the Efficacy of Immunotherapy in Breast Cancer. PLoS One. 2013;8:e61398. doi: 10.1371/journal.pone.0061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic Nanoparticles for Drug Delivery in Cancer. Clin Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari M. Frontiers in Cancer Nanomedicine: Directing Mass Transport through Biological Barriers. Trends Biotechnol. 2010;28:181–188. doi: 10.1016/j.tibtech.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minko T, Kopeckova P, Kopecek J. Chronic Exposure to HPMA Copolymer-Bound Adriamycin Does Not Induce Multidrug Resistance in a Human Ovarian Carcinoma Cell Line. J Controlled Release. 1999;59:133–148. doi: 10.1016/s0168-3659(98)00186-2. [DOI] [PubMed] [Google Scholar]
- 24.Tong AW, Su D, Mues G, Tillery GW, Goldstein R, Klintmalm G, Stone MJ. Chemosensitization of Human Hepatocellular Carcinoma Cells with Cyclosporin A in Post-Liver Transplant Patient Plasma. Clin Cancer Res. 1996;2:531–539. [PubMed] [Google Scholar]
- 25.Wolfram Research, Mathematica Version 8.0, Mathematics and Algorithms. 2008 http://www.wolfram.com/learningcenter/tutorialcollection/MathematicsAndAlgorithms/MathematicsAndAlgorithms.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.