Abstract
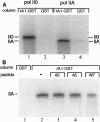
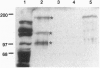
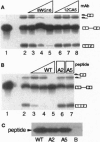
Although transcription and pre-mRNA processing are colocalized in eukaryotic nuclei, molecules linking these processes have not previously been described. We have identified four novel rat proteins by their ability to interact with the repetitive C-terminal domain (CTD) of RNA polymerase II in a yeast two-hybrid assay. A yeast homolog of one of the rat proteins has also been shown to interact with the CTD. These CTD-binding proteins are all similar to the SR (serine/arginine-rich) family of proteins that have been shown to be involved in constitutive and regulated splicing. In addition to alternating Ser-Arg domains, these proteins each contain discrete N-terminal or C-terminal CTD-binding domains. We have identified SR-related proteins in a complex that can be immunoprecipitated from nuclear extracts with antibodies directed against RNA polymerase II. In addition, in vitro splicing is inhibited either by an antibody directed against the CTD or by wild-type but not mutant CTD peptides. Thus, these results suggest that the CTD and a set of CTD-binding proteins may act to physically and functionally link transcription and pre-mRNA processing.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bartolomei M. S., Halden N. F., Cullen C. R., Corden J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988 Jan;8(1):330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar S., Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993 Dec 25;21(25):5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Issner R., Kim J., Mccaw P., Sharp P. A. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995 Oct;1(8):852–865. [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Nickerson J. A., Issner R., Penman S., Sharp P. A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994 Nov;127(3):593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D. B., Du L., van der Zee S., Warren S. L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995 Apr;129(2):287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Sharp P. A. Transcription initiation complexes and upstream activation with RNA polymerase II lacking the C-terminal domain of the largest subunit. Mol Cell Biol. 1990 Oct;10(10):5562–5564. doi: 10.1128/mcb.10.10.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagas P. M., Corden J. L. Structural studies of a synthetic peptide derived from the carboxyl-terminal domain of RNA polymerase II. Proteins. 1995 Feb;21(2):149–160. doi: 10.1002/prot.340210209. [DOI] [PubMed] [Google Scholar]
- Chevray P. M., Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J. L., Cadena D. L., Ahearn J. M., Jr, Dahmus M. E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M. E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995 Apr 4;1261(2):171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- Desai D., Gu Y., Morgan D. O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992 May;3(5):571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fu X. D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995 Sep;1(7):663–680. [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A. L. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993 Apr;18(4):117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- Heinrichs V., Baker B. S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995 Aug 15;14(16):3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Gralla J. D. RNA polymerase II phosphorylation: uncoupling from GAL4-VP16 directed open complex formation and transcription in a reconstituted system. Nucleic Acids Res. 1994 Nov 25;22(23):4958–4962. doi: 10.1093/nar/22.23.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. E., Dahmus M. E. The photoactivated cross-linking of recombinant C-terminal domain to proteins in a HeLa cell transcription extract that comigrate with transcription factors IIE and IIF. J Biol Chem. 1995 Oct 6;270(40):23390–23397. doi: 10.1074/jbc.270.40.23390. [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Dahmus M. E. The major late promoter of adenovirus-2 is accurately transcribed by RNA polymerases IIO, IIA, and IIB. J Biol Chem. 1989 Feb 25;264(6):3169–3176. [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994 May 20;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Garcia-Blanco M. A., Manley J. L. Protein-protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature. 1994 Mar 10;368(6467):119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994 Mar 31;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- LeMaire M. F., Thummel C. S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990 Nov;10(11):6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford G. L., Yamagata K., Kaufmann W. E., Barnes C. A., Sanders L. K., Copeland N. G., Gilbert D. J., Jenkins N. A., Lanahan A. A., Worley P. F. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995 Feb;14(2):433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. RNA processing. Splicing in space. Nature. 1994 Dec 22;372(6508):727–728. doi: 10.1038/372727a0. [DOI] [PubMed] [Google Scholar]
- Moore M. J., Sharp P. A. Site-specific modification of pre-mRNA: the 2'-hydroxyl groups at the splice sites. Science. 1992 May 15;256(5059):992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Neugebauer K. M., Stolk J. A., Roth M. B. A conserved epitope on a subset of SR proteins defines a larger family of Pre-mRNA splicing factors. J Cell Biol. 1995 May;129(4):899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- O'Brien T., Hardin S., Greenleaf A., Lis J. T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994 Jul 7;370(6484):75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Kruger W., Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991 Mar 22;64(6):1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- Roth M. B., Zahler A. M., Stolk J. A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991 Nov;115(3):587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D. G., Oettinger M. A., Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989 Dec 22;59(6):1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Seipel K., Georgiev O., Gerber H. P., Schaffner W. C-terminal domain (CTD) of RNA-polymerase II and N-terminal segment of the human TATA binding protein (TBP) can mediate remote and proximal transcriptional activation, respectively. Nucleic Acids Res. 1993 Dec 11;21(24):5609–5615. doi: 10.1093/nar/21.24.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H., Conaway J. W., Conaway R. C. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993 May 27;363(6427):371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Sisodia S. S., Sollner-Webb B., Cleveland D. W. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987 Oct;7(10):3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N. E., Steinberg T. H., Aronson D. B., Burgess R. R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989 Jul 5;264(19):11511–11520. [PubMed] [Google Scholar]
- Usheva A., Maldonado E., Goldring A., Lu H., Houbavi C., Reinberg D., Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992 May 29;69(5):871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- West M. L., Corden J. L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995 Aug;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis J. T., Xiao H., Greenblatt J., Friesen J. D. The upstream activator CTF/NF1 and RNA polymerase II share a common element involved in transcriptional activation. Nucleic Acids Res. 1994 Jun 11;22(11):1966–1973. doi: 10.1093/nar/22.11.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992 May;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zehring W. A., Greenleaf A. L. The carboxyl-terminal repeat domain of RNA polymerase II is not required for transcription factor Sp1 to function in vitro. J Biol Chem. 1990 May 25;265(15):8351–8353. [PubMed] [Google Scholar]
- Zehring W. A., Lee J. M., Weeks J. R., Jokerst R. S., Greenleaf A. L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P., Manley J. L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5' splice sites. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]