Abstract
MicroRNAs (miRNAs) have emerged as key regulators in the pathogenesis of cancers where they can act as either oncogenes or tumor suppressors. Most miRNA measurement methods require total RNA extracts which lack critical spatial information and present challenges for standardization. We have developed and validated a method for the quantitative analysis of miRNA expression by in situ hybridization (ISH) allowing for the direct assessment of tumor epithelial expression of miRNAs. This co-localization based approach (called qISH) utilizes DAPI and cytokeratin immunofluorescence to establish subcellular compartments in the tumor epithelia, then multiplexed with the miRNA ISH, allows for quantitative measurement of miRNA expression within these compartments. We use this approach to assess miR-21, miR-92a, miR-34a, and miR-221 expression in 473 breast cancer specimens on tissue microarrays. We found that miR-221 levels are prognostic in breast cancer illustrating the high-throughput method and confirming that miRNAs can be valuable biomarkers in cancer. Furthermore, in applying this method we found that the inverse relationship between miRNAs and proposed target proteins is difficult to discern in large population cohorts. Our method demonstrates an approach for large cohort, tissue microarray-based assessment of miRNA expression.
Keywords: miRNA, ISH, Tissue Microarrays, miR-221
MicroRNAs (miRNAs) are short non-coding RNA molecules of 18–25 nucleotides that bind to target mRNAs and inhibit translation or promote degradation to ultimately down-regulate target protein expression (1). Each miRNA can potentially regulate hundreds of mRNAs, and it has been estimated that 30%–60% of all mRNAs are regulated by miRNAs playing roles in regulation of virtually every cellular process (2, 3). Improper miRNA regulation has been attributed to cancer (4) and scores of papers on miRNAs as prognostic and predictive biomarkers have been recently published. Since their discovery, the detection of miRNAs has been difficult, with most methods requiring total RNA extracts which lack critical spatial information. In situ hybridization (ISH), however, allows for the direct assessment of expression levels in tissue and, importantly, the evaluation of expression in malignant cells as well as stromal cells and invading lymphocytes.
Recent advances in ISH have enabled detection of miRNAs in formalin-fixed, paraffin embedded tissue (FFPE) using locked nucleic acid (LNA) modified probes and 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) fixation, and numerous studies have been published utilizing these techniques (5–9). LNA probes utilize a modified ribose backbone where the 2’oxygen and 4’carbon atom are linked with a methylene bridge locking the sugar in the N-type conformation (10). This modification leads to greater thermodynamic stability and higher melting temperatures (Tm) of the probe and miRNA duplex allowing greater hybridization specificity. In addition, EDC reacts with the 5′phosphate of the miRNA, condensing it with amino groups in the protein matrix of the tissue. Without this EDC step, up to 50% of the miR-124 was released into the hybridization buffer suggesting the importance of EDC fixation especially for miRNAs expressed at low levels (11).
Here we describe an ISH assay to quantitatively measure miRNA expression in FFPE tissue microarrays (TMAs). In quantitatively measuring miRNA expression, we multiplex the ISH with cytokeratin immunofluorescence to apply the AQUA technology (15). This quantitative immunofluorescent (qIF) co-localization based approach employs DAPI and cytokeratin to establish subcellular compartments within tumor epithelia, allowing for the measurement of miRNA expression in these subcellular compartments that result in a score directly proportional to the number of molecules per unit of area. The assay permits standardization of miRNA measurement and the potential for rapid large cohort assessment, as well as testing for reproducibility in large TMA populations.
Materials and methods
miRNA in situ hybridization
FFPE tissue microarrays are first deparaffinized in xylene, rehydrated with an ethanol gradient, treated with 20 µg/mL Proteinase K (Roche Diagnostics, Indianapolis, IN, USA) for 10 min at 37°C, fixed with 4% formaldehyde (Thermo Scientific, Rockford, IL, USA) for 10 min, rinsed twice in 0.13M 1-Methylimidazole and refixed with 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, Thermo Scientific) for 1 h as described (11). Then endogenous peroxidases are blocked with 1% H2O2 for 30 min and slides are prehybridized at the hybridization temperature of 50°C for 30 min in hybridization buffer containing 50% formamide (American Bioanalytical), 5X SSC (American Bioanalytical, Natick, MA, USA), 50µg/mL Heparin (Sigma-Aldrich, St. Louis, MO, USA), 0.1% Tween 20 (Sigma), 500µg/mL yeast tRNA (Invitrogen, Carlsbad, CA, USA) adjusted to pH 6. Slides are hybridized for 1 h with 200 nM Double Digoxigenin (DIG) LNA modified probes (Exiqon, Copenhagen, Denmark) for miR-221 (Sequence: 5′-GAAACCCAGCAGACAATGTAGCT-3′), miR-21 (Sequence: 5′-TCAACATCAGTCTGATAAGCTA-3′), miR-3 4a (Sequence: 5′-ACAACCAGCTAAGACACTGCCA-3′), miR-2 05 (Sequence: 5′-CAGACTCCGGTGGAATGAAGGA-3′) and scrambled probe (Sequence: 5′-GTGTAACACGTCT- ATACGCCCA-3′) or 200 nM 5′DIG labeled miR-92a probe (Sequence: 5′-ACAGGCCGGGACAAGTGCAATA-3′) For the U6 Probe (Sequence: 5′-CACGAATTTGCGTGTCATCCTT-3′), 25 nM 5′Dig labeled probe was used. Slides are then stringently washed in 2X SSC (once at hybridization temperature then twice at room temperature for 5 min each), blocked with 2% BSA (Sigma) for 30 min and incubated with Anti-Digoxigenin-POD, Fab fragments from sheep (Roche Diagnostics) diluted 1:100 and rabbit anti-cytokeratin (Dako Corp, Carpinteria, CA, USA) diluted 1:100 in block (2% BSA in PBS) for 1 h at room temperature. Following two washes with 0.1% Tween PBS (PBS-T) and one wash in PBS for 5 min each, the miRNA signal is detected with the TSA Plus Cyanine 5 system (Perkin Elmer, Norwalk, CT, USA), the slides are washed again with PBS-T and PBS as above, and cytokeratin is detected with Alexa 546-conjugated goat anti-rabbit secondary antibody (Molecular Probes) diluted 1:100 in block for one hour, and the slides are mounted with Prolong mounting medium containing 4’,6-Diamidino-2- phenylindole (DAPI, Molecular Probes,, Eugene, OR, USA). Serial sections of small control index slides consisting of 43 breast cancer specimens were ran alongside each run to assess reproducibility and negative control scrambled probe and positive control U6 probe were also used for each run. miR-221 qISH was performed on two builds (redundant cores from different areas of same tumor specimens) of our breast cancer TMA cohort and the AQUA scores from the two cores were averaged for analysis. Any specimens with less than 0.17 mm2 tumors were excluded from analysis. miR-221 blocking oligo experiments were conducted as above. The miR-221 blocking oligo, consisting of the same sequence as endogenous mature miR-221 (sequence: 5′-AGCTACATTGTCTGCTGGGTTTC-3′) was pre-incubated at 1.5 fold excess (300nM) with the miR-221 probe for 1 h at the hybridization temperature prior to hybridization on the TMA.
Immunofluorescence
Fluorescence based immunohistochemical staining was performed as previously described (12–16). PTEN was diluted 1:100 (Cell Signaling Technologies CST138G6). c-Met antibody Met4 from George Vande Woude diluted 1:5,000 (17). ERα was diluted 1:50 (Dako 1D5). ERβ1 was diluted 1:500 (Thermo Scientific PPG5/10). Her4 (Biotechnology, Santa Cruz, CA, USA) was diluted 1:2500. Her3 (Cell Signaling Technology, Beverly, MA, USA) diluted 1:100. E2F4 (Novus SPM179, Littleton, CO, USA) was diluted 1:100. BCL2 (Dako clone 124) diluted 1:1,000. Zeb1 (Sigma Aldrich HPA027524) diluted 1:500.
Quantitative image analysis by AQUA
The AQUA method quantifies fluorescent signal within subcellular compartments as described previously (15). AQUA begins with a series of high resolution monochromatic images for each field of view using the signal from DAPI, Cy3 (cytokeratin), and Cy5 (miRNA). The AQUA algorithm first creates a binarized tumor mask (distinguishing tumor from stroma) based on the cytokeratin signal. Next subcellular compartments within the tumor mask are determined by creating the nuclear compartment (as determine by DAPI positive pixels) and subtracting the nuclear compartment from the tumor mask to create the cytoplasmic compartment. Lastly, AQUA scores for the miRNA or protein of interest are calculated by dividing the signal intensity of CY5 (scored on a scale from 0–255) by the area of the tumor mask or compartment of interest, then normalized to the illumination source intensity.
Patient cohorts
Use of human tissue in this study was approved by the Yale institutional IRB, HIC protocol 9500008219 including consent and waived consent. AQUA, Yale tissue microarrays, YTMA-49 (The Yale Breast Cancer Cohort), YTMA-79 (The Yale Lung Cancer Cohort), and microarray construction has been described previously (15–16,18). The Yale Breast Cancer Cohort consists of 619 breast cancer patients diagnosed from 1976–1982 at Yale-New Haven Hospital. The Yale Lung Cancer Cohort consists of 197 lung cancer patients who underwent surgery at Yale-New Haven Hospital between 1995–2003. Exclusion of individual histospots as a result of technical failure, attrition of sample, or missing clinical variables results in less than 100% inclusion of all tumor specimens in analyses.
Cell culture
Cell culture conditions have been previously described (15). MCF-7 and MDA-MB-231 cells were purchased from ATCC. The cells were transfected with 30 nM anti-miR-221 inhibitor or anti-miRNA negative control (Ambion, Austin, TX, USA) with Lipofectamine RNAi Max (Invitrogen) following manufacturer’s instructions and incubated for 48 h before performing ISH on coverslips or preparing RNA extracts. RNA extracts were prepared using the mirVana miRNAIsolation Kit (Ambion) following the manufacturer’s instructions. In performing ISH on coverslips, cells were washed in PBS, fixed with 1% formaldehyde for 10 min (Thermo Scientific), permeabilized with 0.2% Triton X-100 (American Bioanalytical) for 20 min on ice, refixed with 4% formaldehyde for 10 min, peroxidase blocked with 3% H2O2 for 10 min, prehybridized at 40°C for 30 min, hybridized with double DIG 100nM miR-221 or scrambled probe (Exiqon), or 50 nM U6 probe 5′DIG labeled probe (Exiqon) for 1 h, washed with 2xSSC at hybridization temperature twice then once at room temperature, then blocked with 2% BSA-PBS for 30 min, incubated for 1 h with sheep Anti-Digoxigenin-POD, Fab fragments from sheep (Roche Diagnostics) diluted 1:100 in block. Next the miRNA signal is detected with the TSA Cyanine 5 system (Perkin Elmer) and the coverslips were mounted with Prolong Gold-DAPI (Molecular Probes).
miRNA qRT-PCR
RNA concentrations were determined by the NanoDrop 2000 (Thermo Scientific), then reverse transcribed using the TaqMan microRNA Reverse Transcription Kit, and real-time PCR using the TaqMan microRNA assay kit for miR-221 (000524) or U6 (1093, Applied Biosystems). RT-PCR was performed using the CFX96 machine (BioRad, Hercules, CA). Reactions were done in triplicate along with no template control reactions, and miR-221 expression was normalized to U6 using the 2−ΔΔCT method.
Statistical analysis
All values shown are mean ± s.d. unless otherwise stated. Box plots show standard box and whiskers plots where the error bars represent the 90th and 10th percentiles. To test for differences between groups, p-values were calculated by ANOVA analysis. Survival curves were generated by Kaplan-Meier analysis and tested for significance using the Mantel-Cox log rank test. Prognostic significance for miR-221 was determined using the Cox proportional hazards model. All statistical analysis were done using Statview 5.0 (SAS Institute).
Results and discussion
A number of groups have documented the utility of miRNA ISH in formalin-fixed paraffin embedded tissue specimens using commercially available, LNA modified and DIG labeled probes (7, 9, 11). However, most studies to date have been descriptive or semiquantitative in small patient cohorts. Here we have developed a method for detecting miRNAs in situ multiplexed with 4’,6-diamidino-2-phenylindole (DAPI) and cytokeratin immunofluorescence to quantify the miRNA signal within tumor epithelia using the AQUA technology (15). By applying cytokeratin and DAPI staining to define cytoplasmic and nuclear compartments and epithelial and stromal compartments, miRNA expression can be measured in a standardized and reproducible manner without the use of feature extraction-based software or using “fold change,” the most common metric for RT-PCR based studies. Considerable effort was spent in optimizing the experimental conditions to maximize signal to noise (as evaluated by ISH of scrambled probe) without sacrificing specificity. We found the additional EDC fixation step, double DIG labeled probes, optimization of the titer of the sheep anti-DIG antibody (Roche Diagnostics), and careful increase of the stringency of post-hybridization wash were all required to provide optimal conditions. To validate our quantitative ISH (qISH) method, we assessed expression of miR-92a, miR-21, miR-34a, and miR-221, all of which have been shown to be deregulated in breast cancer (19–22). In assessment of over 400 breast cancer cases, we found a broad dynamic range in the population with AQUA scores ranging from 0–140 and variable expression patterns specific to each miRNA. Examples are shown in Figure 1. While in most cases miRNA expression was cytoplasmic, miR-221 was often localized to the nucleus, an uncommon but previously described localization for mature miRNAs (23–25). We cannot rule out the possibility of the probe recognizing the immature pre-miRNA in the nucleus (23, 24). It should be noted that the scrambled sequence probe was uniformly negative.
Figure 1. Expression of breast cancer miRNAs.
Representative examples of miR-34a, miR-92a, miR-21, miR-221, U6 positive control, and Scrambled probe negative control are shown in three patient samples on breast cancer tissue microarrays. Scale bar represents 50µm.
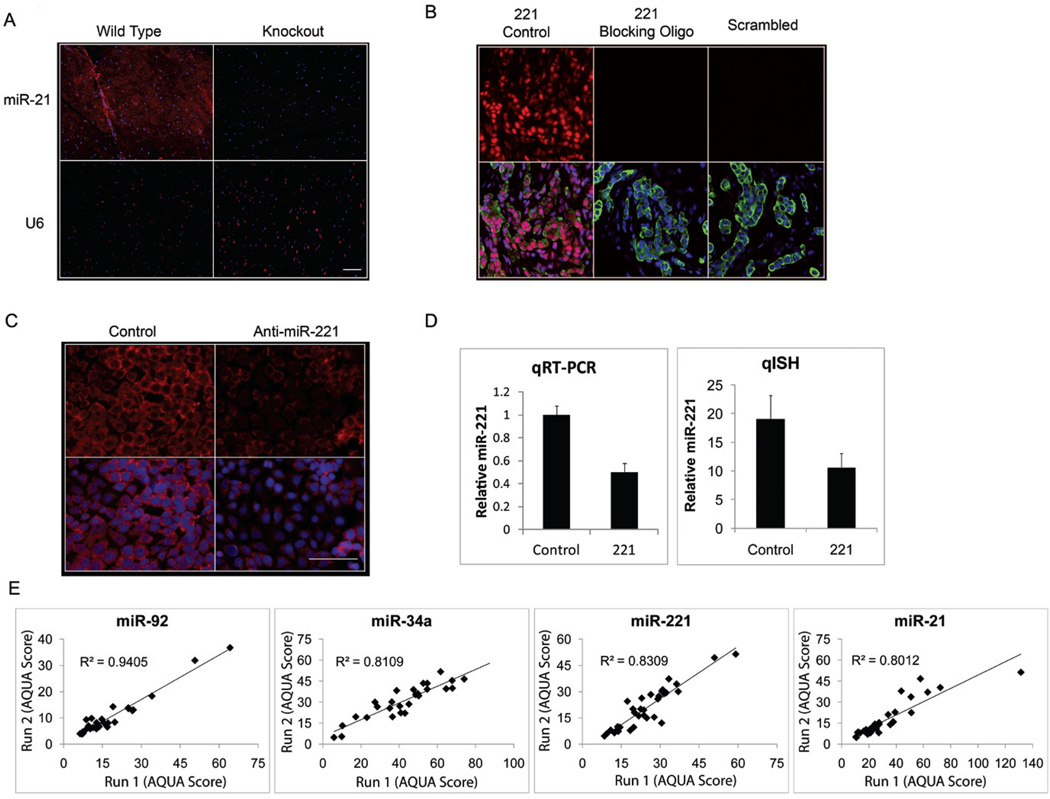
In order to prove specificity, we assessed miR-21 ISH on heart tissue from a miR-21 knockout mouse as shown in Figure 2A (26). No specific signal for miR-21 could be detected in the knockout tissue while normal wild type heart shows moderate levels of expression. We then used a miR-221 specific unlabeled blocking oligo (same sequence as the endogenous mature miR-221) to compete for miR-221 specific DIG labeled probe hybridization, and as shown in Figure 2B, no specific signal was detected in the presence of the blocking oligo. Finally, we transfected MCF-7 and MDA-MB-231, low and high miR-221 expressing cell lines respectively, with anti-miR-221 to determine quantitatively the decrease in signal measured by qISH as compared with the decrease in expression as measured by qRT-PCR. The miR-221 signal was decreased by 55.5% as measured by qISH and 49.9% as measured by qRT-PCR in the MCF-7 cells and was similarly decreased in the MDA-MB-231 cells (Figure 2C–D, and Supplementary Figure 1). To our knowledge this study represents the first time these methods have been used to validate miRNA ISH specificity. As a final validation step, we assessed reproducibility of each assay and found high correlations between near serial sections performed on different days (Figure 2E). The results from these specificity and reproducibility assays gave us the confidence to begin assays on large cohort TMAs.
Figure 2. miRNA qISH assay validation.
(A) miR-21 and U6 ISH (red) on miR-21 knockout mouse heart tissue or wild type mouse heart tissue merged with DAPI (blue). (B) Representative examples of miR-221(red) and Scrambled probe (red) ISH performed on TMAs with and without the miR-221 blocking oligo merged with DAPI (blue) and Cytokeratin (green). (C) miR-221 ISH (red) performed on MCF-7 cells transfected with 30 nM control (anti-miR negative control) or anti-miR-221 inhibitor and merged with DAPI (blue). (D) Quantification of miR-221 knockdown in MCF-7 cells by qRT-PCR normalized by U6 and qISH from 24 random fields normalized by scrambled probe. (E) Reproducibility of miR-92a, miR-34a, miR-221, and miR-21 qISH performed on different days (run 1 versus run 2) on near serial sections of the breast cancer index TMA. Scale bars represent 50µm.
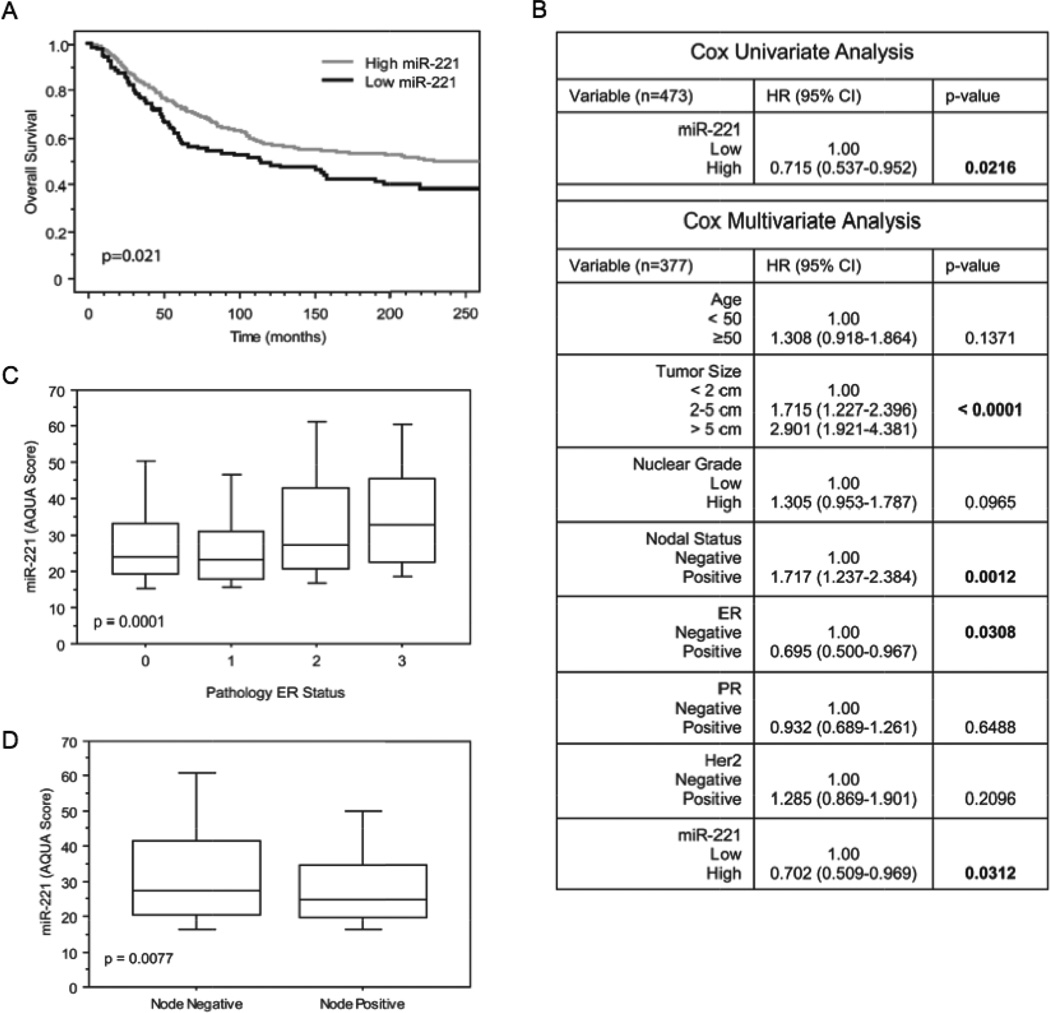
As a proof of concept for miRNAs as biomarkers, we assessed the prognostic value of each miRNA in breast cancer as evaluated by qISH. The qISH AQUA scores were classified according to quartiles to assess miRNA expression and association with survival. For miR-221, the highest three quartiles were nearly overlapping so they were collapsed and compared with the lowest quartile revealing a significantly shorter disease specific survival for the low expression of miR-221 with a p-value of 0.0210 (Figure 3A). This cutoff between high and low expression was also significant in univariate and multivariate analysis providing discovery level evidence that miR-221 expression is an independent prognostic factor in breast cancer (Figure 3B). High miR-221 expression was also associated with ER status and lymph node negativity (Figure 3 C–D). Proof of prognostic value will require more extensive analysis on larger multi-institutional cohorts along with a prospectively designed hypothesis. The other miRNAs were not significantly prognostic (data not shown).
Figure 3. Prognostic value of miR-221 in breast cancer.
(A) Kaplan-Meier survival analysis of miR-221 expression on the YTMA-49 cohort (patients with follow up and miR-221 score n=473). The comparison of breast cancer specific survival of the highest three quartiles (light line, n=354) with the lowest quartile (dark line, n=119) shows a statistically significant better prognosis for high expression of miR-221 (p value calculated by log-rank test). (B) Univariate and multivariate analysis for miR-221 shows that the prognostic value of miR-221 is independent of ERα, PR, HER2, tumor size, nuclear grade, age, and nodal status (patients with all variables and miR-221 score n=377).(C) Box plot demonstrating the distribution of miR-221 expression based on ERα expression (patients with miR-221 score and ER-α status n=514). (D) Box plot demonstrating the distribution of miR-221 expression in node positive and negative patients (patients with miR-221 score and node status n=519).
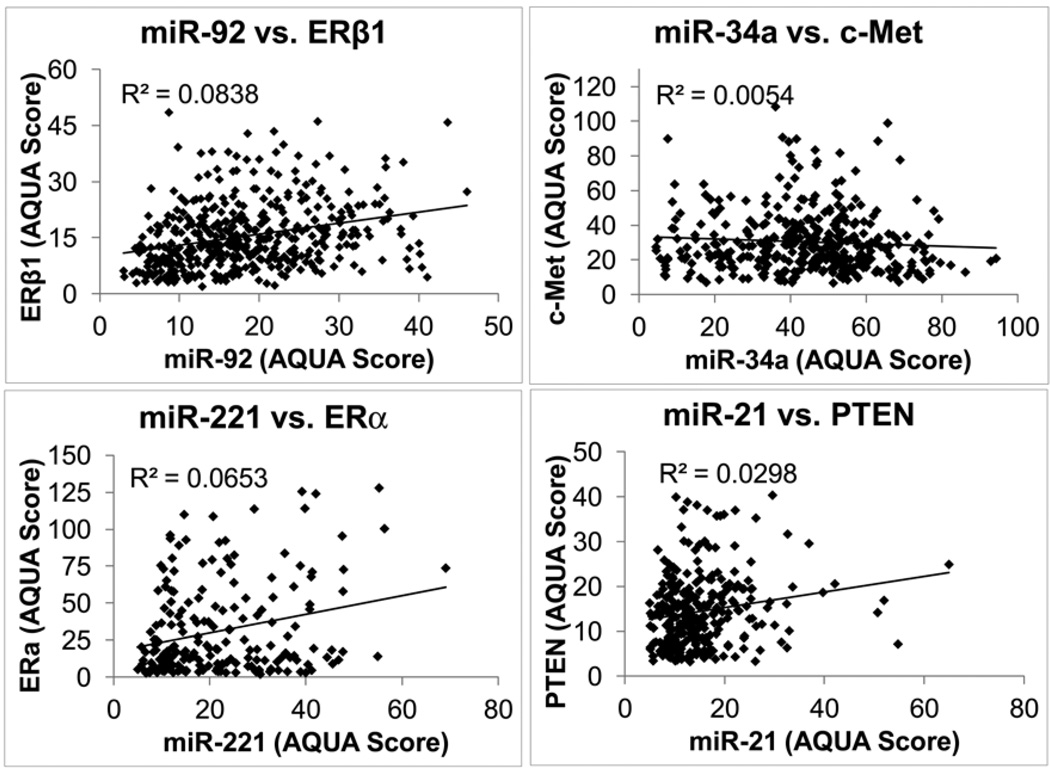
Since miRNAs are known to downregulate proteins, we selected a series of miRNA-protein pairs to evaluate this property on populations of patient specimens on TMAs. Our breast cancer cohort TMA has been extensively used by our lab and has been tested with 116 proteins previously. Thus we were able to assess this premise by comparing the AQUA scores of the protein targets to the miRNA AQUA scores. The following pairs were tested: miR-92a and ERbeta1, miR-34a and c-Met, miR-221 and Estrogen Receptor α and finally miR-21 and PTEN (Figure 4) (19, 22, 27–29). In no case was an inverse relationship observed. We also attempted to discover an inverse relationship between miR-221 and any of the 116 proteins in our database (Supplementary Table 1). While inverse relationships could be found in non-randomly selected subsets of cases, a compelling (statistically significant) inverse relationship was not discovered between miR-221 and any of the proteins in our database. To further assess this principal, we also evaluated miR-205 in non-small cell lung cancer where we had previously tested several validated targets of miR-205 including Zeb1, ERbB3, PTEN, BCL2, ERbB4, and E2F4 (Supplementary Figure 2) (30–33). Again, in 145 cases, we did not establish an inverse relationship between miR-205 and these targets, even when subclassifying tumors by histotype. Thus, in breast and lung cancer, the inverse relationships between a miRNA and its predicted targets are not readily seen in large patient cohorts.
Figure 4. Breast cancer miRNAs and association with putative protein targets.
Correlations between miR-92 and ERα1, miR-34a and c-Met, miR-221 and ERα, miR-21 and PTEN, all measured as described in the methods section on YTMA-49.
Numerous in vitro and cell line based models suggest a tight inverse relationship between a miRNA and its protein targets. Many papers are identifying new “targets” of miRNAs based on cell line luciferase assays and forced overexpression conditions. These studies convincingly describe potential targets of miRNAs in forced situations, but often lack strong evidence for biological relevance for an interaction. We sought to validate an inverse relationship between the miRNAs investigated and their targets, but were unable to find an inverse relationship in real human tumors. This lack of inverse relationships could be due to tissue heterogeneity since the protein and miRNAs assays were not done in serial sections, but often several sections away or on different cores. It is also possible that the inverse relationship is seen only by examination on an individual cell basis, which we could not discern since our method scores on the basis of a field of view. Alternatively, the regulation of protein expression may be complicated by numerous additional regulators at pre- and post- translational stages such that a single miRNA cannot be used to differentiate high versus low expression of a single protein target, except in the contrived situation of forced expression in model systems.
Given the vast regulatory network where an mRNA can be targeted by many miRNAs and miRNAs can target hundreds of mRNAs, we believe the absence of an inverse relationship in human tumors is not surprising. In each case here, we evaluated a single miRNA and compared it to the expression of one or more proteins. It is possible that in order to see decreased protein expression, multiple miRNAs are simultaneously required. A key recent report by Mukherji et al. (34) suggests a nonlinear relationship between target protein level and mRNA level where miRNAs can highly repress mRNA only when it is below a certain threshold level leading to concomitant repression of target protein level. But, if the mRNA is above the threshold level then the miRNA has almost no measurable repressive effect on the total level of mRNA, and hence no effect on the total level of protein target. They also show that this interaction has substantial cell-to-cell variability in strength of target protein repression. Thus, visualization of an inverse relationship may require simultaneous measurement of miRNA, mRNA and protein within individual cells. Even then, the Mukherji et al. work predicts an inverse relationship might only be seen in a subset of cells where the relative amounts of mRNA and microRNA are specifically coordinated. In addition, the recent evidence for “competitive endogenous RNAs” (ceRNAs) or RNAs which can act as miRNA decoys further complicates the level of regulation between a miRNA and target mRNA (35). Overall, these studies support the absence of an inverse relationship observed here.
As an example of the inverse relationship, there are several papers on miR-221 targeting and down-regulating ERα in cell line model systems (22, 36, 37). In contrast we and others have found a positive association with ERα and miR-221 in patient tissue (38). Yoshimoto et al. found that miR-221, as measured by qRT-PCR, gradually increased in expression as ER-α protein expression increased in 171 breast care patients (38). Similarly we show the same relationship by qISH in 473 patients. These observations suggest that new targets of miRNAs validated by forced-expression model systems may not be generalizable to human tumors and could benefit from more extensive validation in other systems.
Since miRNAs were discovered it has been speculated they could have value as biomarkers and numerous papers have been published where miRNAs, measured by RT-PCR show associations with outcomes (38–41). However, there are still few examples of miRNA biomarkers in widespread clinical usage. One reason for this may be the challenge of high throughput measurement by RT-PCR and the challenge of validation in high quality cohorts such that the level of evidence is sufficient to change practice. In this paper, we describe measurements of miRNAs on over 600 lung and breast cancer patients using this in situ method. We are optimistic that this approach could increase throughput sufficiently to allow the testing of miRNAs necessary for broad clinical adoption.
Although we are optimistic about the potential of this method, there are clearly limitations to the efforts to date. Most significantly, we are unable to report absolute quantification of miRNAs in ng/µg of total RNA or similar. While we are currently working toward this goal, it should be noted that the vast majority of RT-PCR based work also does not achieve absolute standardization. Although we do have a wide range of AQUA scores, the dynamic range of ISH is likely within the range of other hybridization techniques such as microarrays which cover 3–4 logs, whereas qRT-PCR has been reported to span a dynamic range of 6–7 logs (42–45). A second limitation of this method is tumor heterogeneity. While this is often not assessed in efforts using RT-PCR, we are able to compare measurements from multiple histospots from the same tumor when arrays are made at 2-fold or greater redundancy. Compared using linear regression, we find R2 values to be in the 0.4 to 0.6 range, compared with proteins in a comparable assay system where R2 values are seen to be 0.6 to 0.8. It is too early to assess the meaning of the spatial heterogeneity we observe. In the future it may be interesting to correlate this with RT-PCR and tumor geography (i.e., leading edge vs training edge, well vs poorly differentiated regions or perivascular vs hypoxic regions).
In summary, we have developed a method to quantitatively measure miRNA expression within subcellular compartments in tumor epithelia. Compared with other methods for miRNA detection and quantitative analysis, miRNA qISH has the advantages of retaining critical spatial information while increasing throughput when used in combination with tissue microarrays. In developing this method we found that the inverse relationship between miRNAs and their target proteins is difficult to identify in large patient cohorts suggesting the interaction between miRNAs and targets to be more complicated than suggested by in vitro models. Finally, as an example, we found that miR-221 may have value as a prognostic marker. While this is preliminary and exploratory with respect to its prognostic value, it provides evidence for the usage of miRNAs as tissue biomarkers.
Supplementary Material
Acknowledgments
Thanks to Eric Olson and his group for providing the miR-21 knockout mouse tissue. Thanks to George Vande Woude and his group for providing the Met4 antibody. This work was supported by NIH RO-1 grant number CA 114277 to DLR.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/113837
Competing interests
Dr. Rimm is a co-founder, consultant for and stockholder in HistoRx, the exclusive licensee of the Yale-owned AQUA technology.
References
- 1.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher FJ, Slack A. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Gupta A, Mo YY. Detection of microRNAs in cultured cells and paraffin-embedded tissue specimens by in situ hybridization. Methods Mol. Biol. 2011;676:73–83. doi: 10.1007/978-1-60761-863-8_6. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen S, Baker A, Moller S, Nielsen BS. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2010;52:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 9.Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, Conejo-Garcia JR, Kauppinen S, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin. Cancer Res. 2010;16:4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Kumar R, Wengel J. Synthesis of Novel Bicyclo[2.2.1] Ribonucleosides: 2’-Amino- and 2’-Thio-LNA Monomeric Nucleosides. J. Org. Chem. 1998;63:6078–6079. doi: 10.1021/jo9806658. [DOI] [PubMed] [Google Scholar]
- 11.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimm DL. What brown cannot do for you. Nat. Biotechnol. 2006;24:914–916. doi: 10.1038/nbt0806-914. [DOI] [PubMed] [Google Scholar]
- 13.Rothberg BE, Moeder CB, Kluger H, Halaban R, Elder DE, Murphy GF, Lazar A, Prieto V, et al. Nuclear to non-nuclear Pmel17/gp100 expression (HMB45 staining) as a discriminator between benign and malignant melanocytic lesions. Mod. Pathol. 2008;21:1121–1129. doi: 10.1038/modpathol.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welsh AW, Moeder CB, Kumar S, Gershkovich P, Alarid ET, Harigopal M, Haffty BG, Rimm DL. Standardization of estrogen receptor measurement in breast cancer suggests false-negative results are a function of threshold intensity rather than percentage of positive cells. J. Clin. Oncol. 2011;29:2978–2984. doi: 10.1200/JCO.2010.32.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 16.Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am. J. Pathol. 2010;176:2131–2138. doi: 10.2353/ajpath.2010.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anagnostou VK, Dimou AT, Botsis T, Killiam EJ, Gustavson MD, Homer RJ, Boffa D, Zolota V, et al. Molecular classification of nonsmall-cell lung cancer using a 4-protein quantitative assay. Cancer. 2011;118(6):1607–1618. doi: 10.1002/cncr.26450. [DOI] [PubMed] [Google Scholar]
- 18.Knudsen BS, Zhao P, Resau J, Cottingham S, Gherardi E, Xu E, Berghuis B, Daugherty J, et al. A novel multipurpose monoclonal antibody for evaluating human c-Met expression in preclinical and clinical settings. Appl. Immunohistochem. Mol. Morphol. 2009;17:57–67. doi: 10.1097/PAI.0b013e3181816ae2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Nakhle H, Burns PA, Cummings M, Hanby AM, Hughes TA, Satheesha S, Shaaban AM, Smith L, Speirs V. Estrogen receptor {beta}1 expression is regulated by miR-92 in breast cancer. Cancer Res. 2010;70:4778–4784. doi: 10.1158/0008-5472.CAN-09-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 21.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 22.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 24.Jeffries CD, Fried HM, Perkins DO. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011;17:675–686. doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 26.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 30.Barh D, Parida S, Parida BP, Viswanathan G. Let-7, miR-125, miR-205, and miR-296 are prospective therapeutic agents in breast cancer molecular medicine. Gene Ther. Mol. Biol. 2008;12B:189–206. [Google Scholar]
- 31.Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7:300–304. doi: 10.4161/rna.7.3.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, Denicola G, Webster KA, et al. In Vivo Identification of Tumor-Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H. Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res Treat. 2011;130(1):331–339. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
- 39.Duncavage E, Goodgame B, Sezhiyan A, Govindan R, Pfeifer J. Use of microRNA expression levels to predict outcomes in resected stage I non-small cell lung cancer. J. Thorac. Oncol. 2010;5:1755–1763. doi: 10.1097/JTO.0b013e3181f3909d. [DOI] [PubMed] [Google Scholar]
- 40.Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, Strobel P, Riedmiller H, Kneitz B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int. J. Cancer. 2010;127:394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 41.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang JS, Simon VA, Feddersen RM, Rakhshan F, Schultz DA, Zschunke MA, Lingle WL, Kolbert CP, Jen J. Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays. BMC Genomics. 2011;12:144. doi: 10.1186/1471-2164-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shippy R, Sendera TJ, Lockner R, Palaniappan C, Kaysser-Kranich T, Watts G, Alsobrook J. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics. 2004;5:61. doi: 10.1186/1471-2164-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang F, Hajkova P, Barton SC, O’Carroll D, Lee C, Lao K, Surani MA. 220-plex microRNA expression profile of a single cell. Nat. Protocols. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.