Optical imaging techniques have evolved from basic preclinical to enabling clinical technology platform for the diagnosis and treatment of human diseases. A major bottleneck for the clinical translation of light based imaging and therapeutic techniques is the limited penetrability of light in deep tissue. This drawback has led to widespread attention and renewed interest in Cerenkov emission, especially in the area of molecular imaging, as a tissue depth-independent spontaneous light source.[1] Cerenkov radiation (CR) is a photophysical phenomenon where continuous wave (CW) light in the 250–600 nm range is emitted by charged particles such as positrons and electrons when travelling faster than light in a dielectric medium with refractive index >1.[2] Generally, appreciable CR is produced as a ‘byproduct’ of highly energetic radionuclide or external beam radiation that are used in various clinical interventions. However, due to significant tissue autofluorescence, absorption and scattering of UV and visible light, Cerenkov luminescence imaging (CLI) faces steep challenges such as rapid signal attenuation and low fluence rates. Thus, coupling CR with fluorophores that emit in the optical window at wavelengths between 650 nm and 950 nm will minimize tissue autofluorescence and enhance detection of light from deep tissue.
Recently, positron emission tomography (PET) isotopes such as Copper-64 (64Cu), Fluorine-18 (18F) and Gallium-68 (68Ga), as well as external beam irradiators such as clinical linear accelerators, have been used to excite fluorophores both in vitro and in vivo.[3] Unfortunately, the CW emission of CR increases the probability of photobleaching organic dyes, which generally have low photostability and narrow absorption spectrum for efficient energy transfer from Cerenkov emission. In this regard, quantum dots (QDs) are viable alternatives to organic dyes because of their excellent photostability, high quantum yield, large Stokes shifts and broad excitation spectra that can capture both the UV and visible light from CR.[4] Recent reports have shown that CR can excite QDs both in vitro and in vivo through Cerenkov radiation energy transfer, where CR serves as the energy donor and the QDs as the energy acceptor.[3b,5] These studies were based on the random interactions between CR and fluorophores.
In this study, we explored the potential application of CLI in the development of activatable probes by programming and controlling the distance between fluorophore and radionuclide. To develop activatable probes and also a hybrid nanoruler based on Cerenkov radiation-associated distance dependent luminescence, we designed new molecular beacons using QDs as CR acceptor, DNA as spacer and 64Cu as source of CR. Both QDs and DNA offer significant advantages to developing molecular rulers.[6] Three prerequisites are essential for the success of this design: (i) specificity (ii) programmability and (iii) rigidity. DNA uniquely possesses these attributes. The programmable nature of DNA self-assembly serves as an excellent platform for developing molecular probes and beacons. Unlike conventional FRET, CR energy transfer can occur over a long range, requiring the consideration of DNA’s persistence length needed to maintain the validity of the distance-based model. Persistence length of DNA is the length of DNA that behaves as a rigid rod. It is ~5 nm for single stranded (ss) DNA and 10 times higher for double stranded (ds) DNA (~50 nm or 100 bp) at a given salt concentration.[7] Therefore, we used DNA hybridization systems that maintained linearity over the model range to minimize coiling or flexibility that resulted in the inaccuracies observed in previous ssDNA models.[8]
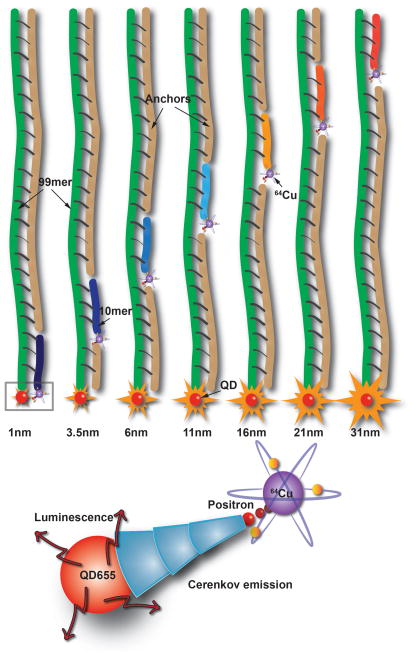
The probe design consists of streptavidin coated QD655 that was conjugated to biotin modified 99 base long ssDNA (99mer). Streptavidin coated QDs were used to primarily provide rigidity to the design because other modifications such as carboxyl and amine functions are typically conjugated to the QD surface using flexible PEG linkers, opening the possibility for misalignment of DNA. Seven 10 base long ssDNA (10mer) sequences complementary to the 99mer were designed to anneal distinctly at locations 1–10, 7–17, 15–25, 30–40, 44–54, 59–69 and 88–98 bases on the 99mer, where 1 is closest to the QD. Assuming that the distance between two adjacent base pairs is 0.34 nm, the estimated distance of the 10mers’ respective anneal locations corresponds to 1.15 nm, 3.65 nm, 6.15 nm, 11.15 nm, 16.15 nm, 21.15 nm and 31.15 nm. In calculating the metric distances, the length of the linkers used for the bioconjugation was also considered. Non-cross hybridizing sequences were selected to prevent any errors in the probe assembly process.[9] A chelator, DOTA (1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid) with a functional azide group, was attached to the 10mers that controls the distance of the chelated radionuclide from the QD surface (Figure 1). Due to its optimal half-life (12.7 h) and significant Cerenkov emission, 64Cu was used to implement the design. Copper free [2+3] cycloaddition strategy (‘click’ chemistry) was used for bioconjugation. This strategy was primarily chosen for its simplicity, high selectivity in aqueous media, prevention of non-specific Cu adduct, modularity and stability of the reactants and final products.[10] Amino group modified 10mer ssDNA (0.8 μM) were activated with DBCO-PEG4-NHS ester and conjugated to azide-DOTA after chelation with 64Cu (~50 μCi, 156.25 nM). The seven unique 10mer ssDNA sequences were similarly prepared for annealing with the QD-99mer adducts. Complementary ssDNA ‘anchors’ were designed to hybridize to the 99mer to ensure maintenance of a persistent dsDNA through the entire length of the 99mer (Figure 1). In addition, the electrostatic repulsion between the phosphate groups of the multiple ssDNA copies on each QD favours long persistence rigidity.
Figure 1.
Schematic showing the design of CRaDLe using QD conjugated to 99mer ssDNA and seven 10mer ssDNA with 64Cu, annealing at distinct distances from the QD surface. Positrons and electrons emit Cerenkov radiation that excites QD655 for luminescence imaging (not drawn to scale).
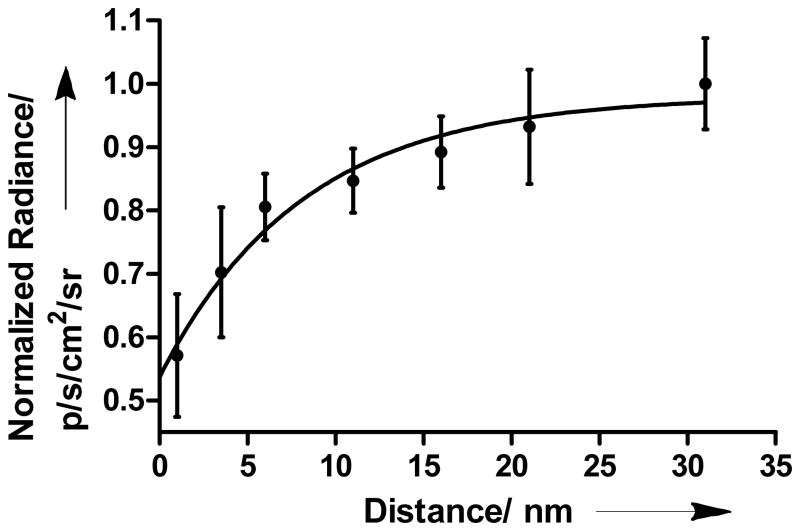
Distance measurements of 64Cu-DOTA-ssDNA (10mer) at specified locations on the 99mer-QD adduct were determined through luminescence using IVIS Lumina XR multimodal image capturing system (Figure S3 in Supporting Information). All experiments were performed in buffered aqueous solutions to maintain the persistence length of DNA. Positive control consisting of an admixture of QDs and 64Cu-DOTA without DNA linkers generated maximum radiance in the Cy5.5 channel (690 nm – 770 nm). No significant radiance was observed from the wells containing 64Cu-DOTA alone in this channel. At 1 nm distance, QD emission was 33±5% of the initial (control) value, representing the lowest radiance value observed in our model. Increasing the distance between radionuclide and QD surface from 1 nm to 31 nm resulted in a significant increase in the emission (Figure 2). In comparison to the control, the corresponding percentage values were 41±6, 47±3, 49±3, 52±3, 55±5, and 58±4 for 3.5 nm, 6 nm, 11 nm, 16 nm, 21 nm and 31 nm, respectively.
Figure 2.
Plot depicting the exponential increase in QD655 radiance as a function of 64Cu distance from the QD surface. Non-linear regression fitting analysis was applied after background correction. Radiance was normalized to the maximum value.
The above trend suggests the existence of distance-dependence in CR induced luminescence, which is attributable to a variety of factors. Since Cu(II) is known to quench the fluorescence of organic dyes, QDs and fluorescent proteins,[11] we investigated the effect of Cu(II) on the QD signal attenuation. There was no noticeable difference in the QD655 radiance between the DOTA chelated or unchelated (free) 64Cu (data not shown). However, fluorometric titrations of unchelated “cold” Cu(II) with QD655 revealed a 25% reduction in the emission at a molar ratio of 1:1 (Figure S4 in Supporting Information). In contrast, only 13% reduction in luminescence was determined for unlinked chelated Cu(II). This can be attributed to enhanced contact quenching as a result of random collisions between Cu(II) ions and QD surface. However, the probability of contact quenching occurring in the QD-DNA-Cu constructs is low because of the presence of multiple rigid dsDNAs per particle. This prompted further investigation into the possible distance dependent quenching effect of ionic copper. Distance studies using DNA linked “cold” DOTA-Cu(II) showed the QD emission reduced to 56±5% of the initial value at 1 nm. The corresponding percentage values were 64±5, 75±3, 77±7, 79±5, 84±8, and 93±6 for 3.5 nm, 6 nm, 11 nm, 16 nm, 21 nm and 31 nm, respectively (Figure 3a). Based on these findings, the quenching phenomenon is likely due to photoinduced electron transfer process or energy transfer or a combination of these factors.
Figure 3.
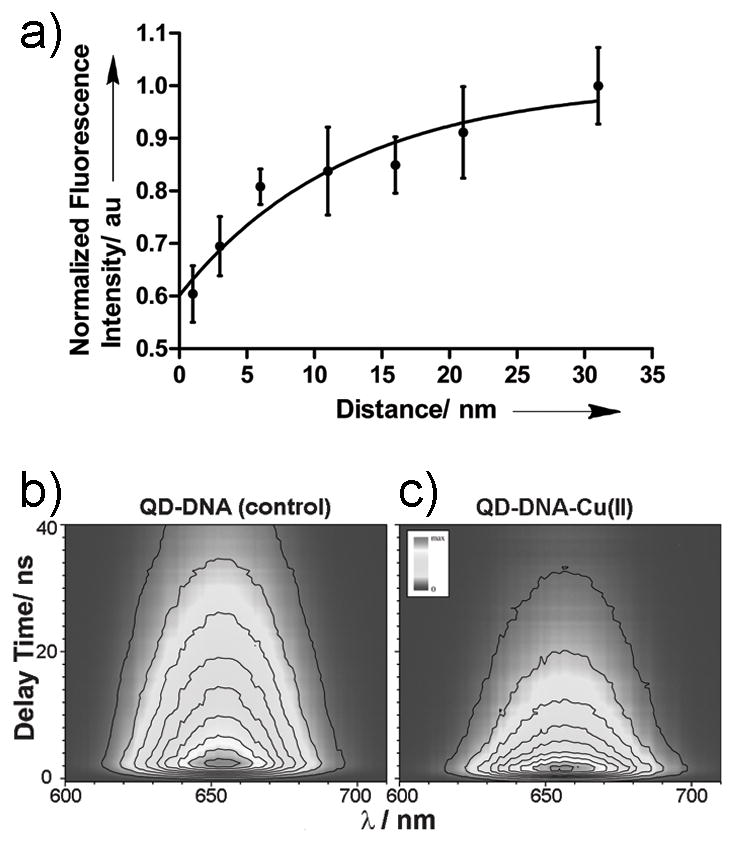
a) Plot depicting the validation of the distance based model with “cold” Cu(II) through steady-state fluorescence intensity measurements. Non-linear regression fitting analysis was applied. b) TRF of the QD-DNA without Cu(II). c) TRF of the QD-DNA attached to Cu(II) at a distance of 1 nm from QD.
To better understand the underlying phenomenon, picosecond time-resolved fluorescence (TRF) spectroscopy studies were conducted. Figures 3b and 3c show, respectively, the fluorescence decay contours of QD-DNA (denoted as control) and QD-DNA-Cu(II) with Cu cation attached to DNA strand at a distance of 1 nm from the QD nanoparticle. Difference in fluorescence decay of both samples is evident. Both samples were measured under identical conditions. Thus, the presence of Cu(II) in the QD vicinity influences the emissive recombination of the photogenerated electron-hole pairs. Results of data fitting shown in Figure S6 (Supporting Information) demonstrate that density of the electron-hole pairs which recombine with the emission of photons is relatively low.
In addition, the fluorescence decay has primarily exponential character with a decay rate constant k = 2.7×10−2 (ns)−1, corresponding to the fluorescence lifetime of τ = 38.4 ns (for details see the Supporting Information). In contrast, the proportion of this component was reversed for QD-DNA-Cu(II), with the exponentially decaying component (Figure S6 in Supporting Information) accounting for only 25% of initially emitted (at t = 0) photons. Domination of the second order decay character in the fluorescence dynamics of QD-DNA-Cu(II) suggests that initial concentration of the deeply trapped electrons responsible for the emission of photons during recombination process is significantly higher than in the QD-DNA control sample. This result is surprising because such effect can be achieved by increasing excitation intensity. However, the excitation intensity remained exactly the same during all TRF measurements. Alternatively, similar effect could be expected if mobility of the photogenerated carriers is increased through the so-called trap-filling effect.[12] Probably, the observed phenomenon can be achieved by introducing an additional non-radiative recombination path for photogenerated electrons that are still in conduction band and do not contribute to fluorescence. This should decrease the carrier density and increase mobility of non-trapped electrons and holes, accelerate the trapping process, and trigger non-exponential decay of fluorescence. The putative path suggests an ultrafast electron transfer to Cu(II) via DNA, facilitating charge separation, which is followed by rapid non-radiative recombination with non-trapped hole. This sequence of events leads to charge recombination from the valence band in the QD.
Detailed characteristics of the kinetic components obtained from the fluorescence decay fitting of the samples with Cu(II) attached at different distances from the QD are provided in Table S1 (Supporting Information). No clear relationship of the decay rates from a distance between QD and Cu(II) was found. However, the decay rate of the exponential component is clearly larger for all Cu(II) attached samples compared to the control. Lifetime of the exponentially decaying component of the fluorescence decay shortens from 38.4 ns to the average value of 28.6 ns. It suggests that also some sort of resonant energy transfer between the QD nanoparticle and Cu(II) occurs and provides additional non-radiative decay path for excitation and additional fluorescence quenching. Cu(II) absorption spectum taken in buffered solution exhibits a broad featureless band spanning both visible and NIR regions, substantially overlaping with the QD655 emission spectrum (Figure S5 in Supporting Information). The overlap provides energy funnelling direction, making energy transfer between QD and Cu(II) energetically feasible. However, due to the low molar extinction coefficient of Cu(II) (ε750 = 1.20 × 104 dm3 mol−1cm−1), the contribution of energy transfer towards quenching QD is most likely minimal.
Some of the events in the early delay times after excitation cannot be resolved by TRF measurements. To obtain this additional information, a femtosecond time-resolved absorption spectroscopy (TRA) was applied (for details, see the Supporting Information). However, no significant differences in the transient absorption spectra and dynamics between the samples was observed (Figure S7 in Supporting Information). These results are less surprising. Intensity of the amplified laser beam used for TRA is ~3000 times higher than that used for TRF measurements. Unfortunately, under such conditions subtle differences in excited state dynamics expected to be observed are masked by the overwhelming trap filling effect induced by high intensity excitation.
We therefore suggest that QD-DNA-64Cu construct undergoes static quenching with electron transfer as the dominant phenomenon. Duplex DNA is known to assist ultrafast short and long range electron transfer process through its π-framework, which is facilitated by base stacking.[13] However, a DNA strand could also mediate the observed quenching. Distance-dependent quenching has been reported for ethidium/rhodium intercalators as donor/acceptor pair in duplex DNA and hairpins.[14] To assess the possible contribution of DNA to QD quenching, we labelled streptavidin coated QDs with biotin-DOTA-64Cu directly, without using DNA assembly. QD-streptavidin admixed with azide-DOTA-64Cu was used as control. Streptavidin-biotin interfacial platforms have been used as “bridges” for electron transfer.[15] The observed radiance of QD-avidin-biotin-DOTA-64Cu complex was ~41% of the control (Figure S8 in Supporting Information). This quenching efficiency is similar to the DNA assembled at 1 nm distance from the QD, suggesting that the DNA does not contribute significantly to the observed quenching phenomenon. In addition, we did not observe any change in radiance when the QD-99mer construct, without the complementary 10mer strand, was admixed with azide-DOTA-64Cu. Additionally, a full length 99mer complementary ssDNA with a DBCO internal modification at 88th base, corresponding to 31 nm distance, was used to correlate the luminescence and fluorescence measurements with the 10mer and anchor based design at the same distance. Essentially, no significant difference between the 99mer and 10mer based design was observed in both, luminescence (64Cu) and fluorescence (Cu(II)), measurements (Figure S9 in Supporting information). By conforming to the distance based model, these findings suggest that the quenching efficiency is influenced by the length of the linkers used for connecting the electron donor to the electron acceptor, as long as they maintain the electron transfer process.
Therefore, the observed distance dependence changes in QD luminescence using 64Cu is primarily driven by the quenching efficiency of Cu(II) through ultrafast electron transfer. This places 64Cu in a unique position among PET isotopes because of its potential to serve both as a light source through CR and a broad range luminescence quencher through energy and photoinduced electron transfer mechanisms. This dual role could maximize the utility and performance of optical-nuclear activatable probes powered by CR.
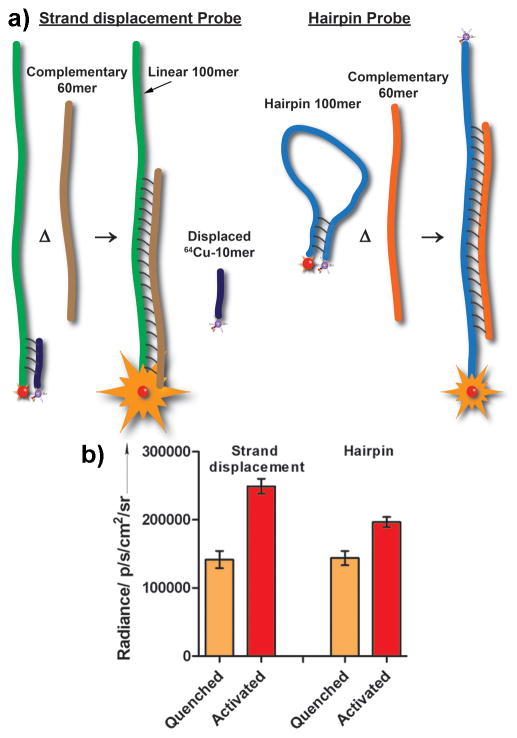
To validate this model, we used both the strand-displacement hybridization and hairpin probe approaches. The strand displacement hybridization, also known as Yin Yang probes, fundamentally operate like hairpin probes, where the fluorophore (QDs) on 3′ end of ssDNA is in close proximity to a quencher on the 5′ end of its complementary sequence.[16] In hairpin, both of these sequences form the ‘stem’, connected by a ‘loop’ ssDNA sequence that is usually designed to complement the target ssDNA (Figure 4a). In the strand displacement model, we observed a 45±7.5% increase in radiance after annealing the target 60mer to the 99mer, displacing the 10mer-DOTA-64Cu complex (Figure 4b).
Figure 4.
a) Schematic of the strand displacement and hairpin probes. b) Comparison of luminescence readings for strand displacement probe and hairpin probe after target DNA hybridization activation event.
When an activatable probe is envisaged, obtaining the highest SNR is highly desired. In the case of strand displacement probes, the design aims to release the isotope completely into the surrounding milieu. This implies that the sphere of influence will play a deterministic role in obtaining a high SNR. Sphere of influence can be loosely defined as the volume of liquid in the isotope’s immediate vicinity in which photon flux of CR is optimal for exciting fluorophores. Currently, the sphere of influence for individual PET isotopes is uncertain. Factors such as the diffusion rate, half-life, and internal dynamics of cellular events could contribute to the redistribution of an isotope and the diffusion of fluorophores away from the sphere of influence. In such cases, real-time tracking of the probe to record the activation event will be required. This can be especially challenging for in vivo-based detection systems. Using a hairpin probe could obviate this potential problem.
In the hairpin probe design, a 100mer was used to simulate the 1.15 nm distance construct in its native state and the 31 nm distance construct after annealing with its target and linearization. The 5′ end was biotinylated for binding with QD-streptavidin and the 3′ end was modified with an amine group for conjugating a DBCO based linker for ‘clicking’ with azide-DOTA. Upon annealing with its 60mer complement, luminescence increase of 22±6.5% relative to the initial value was observed (Figure 4b). This agrees with the ~25% increase observed in our model nanoruler, where there was ~33% and ~58% quenching at 1.15 nm and 31 nm distance respectively, relative to the control. Although a complete quenching of the luminescence would be desirable prior to the probe activation, the background luminescence could facilitate tracking of the probes using in vitro and in vivo systems. However, complete luminescence quenching and higher activation luminescence can be obtained by incorporating a dark quencher labelled with 64Cu and using DNA probes exceeding 30 nm, respectively.
In summary, we have demonstrated the feasibility of utilizing 64Cu to design and construct optical nanorulers and activatable probes. The design was based on using DNA as a linker to control the distance of 64Cu from the QD655 surface. Short DNA 10mers were used for annealing on a 90mer at seven distinct locations, ranging from 1 nm to 31 nm. The increase in radiance at 1 nm and 31 nm was ~33% and ~58%, respectively, relative to the initial value. The results suggest that luminescence quenching was mediated by ultrafast CR induced electron transfer from the QD to Cu(II). This model was validated by using strand displacement model, where ~45% increase in luminescence upon activation was observed compared to the hairpin model, which had a modest ~22% increase relative to the initial value. The ensemble of the results holds promise for the potential use of the distance model to report molecular processes. The usable working life of 64Cu based probes will be determined by the initial activity achieved after probe assembly and the sensitivity of the imaging system. The relatively short half-life of 64Cu and the activity (< 150 μCi) used to develop the probes described in this study can be classified as relatively non-hazardous and slightly “radiotoxic”. However, safety considerations are required for handling radioisotopes under all circumstances, and all the experiments with radioactive materials were conducted with the approval of Washington University Radiation Safety Committee.
Supplementary Material
Footnotes
This study was supported in part by grants from the US National Institutes of Health (NIBIB R01 EB008111, NCI R01 CA171651 and SIG 1S10RR031625-01) and the National Science Foundation (CCF 0963742). Time-resolved fluorescence and absorption experiments were performed in Ultrafast Laser Facility of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy (DE-SC 0001035). We thank Dr. Mikhail Berezin, Dr. Monica Shokeen, Dr. Yuan-Chuan Tai, and Sergey Komarov for helpful discussions.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Nalinikanth Kotagiri, Department of Radiology, Washington University School of Medicine, 4525 Scott Avenue, St. Louis, MO 63110 (USA), Fax: (+1) 314-362-8599
Dr. Dariusz M. Niedzwiedzki, Photosynthetic Antenna Research Center, Washington University in St. Louis, One Brookings Drive, St. Louis, MO 63130, USA
Mr. Kohtaro Ohara, Department of Radiology, Washington University School of Medicine, 4525 Scott Avenue, St. Louis, MO 63110 (USA), Fax: (+1) 314-362-8599
Prof. Dr. Samuel Achilefu, Email: achilefus@mir.wustl.edu, Department of Radiology, Washington University School of Medicine, 4525 Scott Avenue, St. Louis, MO 63110 (USA), Fax: (+1) 314-362-8599, Homepage: http://www.orl.wustl.edu
References
- 1.a) Robertson R, Germanos MS, Li C, Mitchell GS, Cherry SR, Silva MD. Phys Med Biol. 2009;54:N355–65. doi: 10.1088/0031-9155/54/16/N01. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu H, Ren G, Miao Z, Zhang X, Tang X, Han P, Gambhir SS, Cheng Z. PLoS One. 2010;5:e9470. doi: 10.1371/journal.pone.0009470. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Spinelli AE, Kuo C, Rice BW, Calandrino R, Marzola P, Sbarbati A, Boschi F. Opt Express. 2011;19:12605–18. doi: 10.1364/OE.19.012605. [DOI] [PubMed] [Google Scholar]; d) Hu Z, Ma X, Qu X, Yang W, Liang J, Wang J, Tian J. PLoS One. 2012;7:e37623. doi: 10.1371/journal.pone.0037623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jelley JV. Br J Appl Phys. 1955;6:227–232. [Google Scholar]
- 3.a) Axelsson J, Davis SC, Gladstone DJ, Pogue BW. Med Phys. 2011;38:4127–4132. doi: 10.1118/1.3592646. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dothager RS, Goiffon RJ, Jackson E, Harpstrite S, Piwnica-Worms D. PLoS One. 2010;5:e13300. doi: 10.1371/journal.pone.0013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sunderesan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Zhang X, Xing B, Han P, Gambhir SS, Cheng Z. Small. 2010;6:1087–91. doi: 10.1002/smll.200902408. [DOI] [PubMed] [Google Scholar]
- 6.a) Morgner F, Geibler D, Stufler S, Butlin NG, Lohmannsroben HG, Hilebrandt N. Angew Chem Int Ed. 2010;49:7570–7574. doi: 10.1002/anie.201002943. [DOI] [PubMed] [Google Scholar]; b) Chhabra R, Sharma J, Wang H, Zou S, Lin S, Yan H, Lindsay S, Liu Y. Nanotechnology. 2009;20:485201. doi: 10.1088/0957-4484/20/48/485201. [DOI] [PubMed] [Google Scholar]
- 7.Hays JB, Magar ME, Zimm BH. Biopolymers. 1969;8:531–536. [Google Scholar]
- 8.Parak WJ, Pellegrino T, Micheel CM, Gerion D, Williams SC, Alivisatos AP. Nano Lett. 2003;3:33–36. [Google Scholar]
- 9.Deaton R, Kim JW, Chen J. Appl Phys Lett. 2003;82:1305–07. [Google Scholar]
- 10.Best MD. Biochemistry. 2009;48:6571–84. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 11.a) Brunner J, Kraemer R. J Am Chem Soc. 2004;126:13626–7. doi: 10.1021/ja047252a. [DOI] [PubMed] [Google Scholar]; b) Chen Y, Rosenzweig Z. Anal Chem. 2002;74:5132–8. doi: 10.1021/ac0258251. [DOI] [PubMed] [Google Scholar]; c) Rahimi Y, Goulding A, Shrestha S, Mirpuri S, Deo SK. Biochem Biophys Res Commun. 2008;370:57–61. doi: 10.1016/j.bbrc.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshihara T, Katoh R, Furube A, Tamaki Y, Murai M, Hara K, Murata S, Arakawa H, Tachiya M. J Phys Chem B. 2004;108:3817–3823. [Google Scholar]
- 13.a) Barton JK, Kumar CV, Turro NJ. J Am Chem Soc. 1986;108:6391–6393. [Google Scholar]; b) Wan C, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc Natl Acad Sci U S A. 1999;96:6014–6019. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Kelley SO, Holmlin E, Stemp EDA, Barton JK. J Am Chem Soc. 1997;119:9861–9870. [Google Scholar]; b) Lewis FD, Wu T, Zhang Y, Letsinger RL, Greenfield SR, Wasielewski MR. Science. 1997;277:673–676. doi: 10.1126/science.277.5326.673. [DOI] [PubMed] [Google Scholar]
- 15.Azzaroni O, Alvarez M, Mir M, Yameen B, Knoll W. J Phys Chem C. 2008;112:15850–15859. [Google Scholar]
- 16.Li Q, Luan G, Guo Q, Liang J. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.