Abstract
The hypothalamus plays a key role in the regulation of feeding behavior. Several hypothalamic nuclei, including the arcuate nucleus (ARC), paraventricular nucleus, and ventromedial nucleus of the hypothalamus (VMH), are involved in energy homeostasis. Analysis of microarray data derived from ARC revealed that leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4) is highly expressed. LGR4, LGR5, and LGR6 form a subfamily of closely related receptors. Recently, R-spondin (Rspo) family proteins were identified as ligands of the LGR4 subfamily. In the present study, we investigated the distribution and function of LGR4–LGR6 and Rspos (1–4) in the brain of male rat. In situ hybridization showed that LGR4 is expressed in the ARC, VMH, and median eminence of the hypothalamus. LGR4 colocalizes with neuropeptide Y, proopiomelanocortin, and brain-derived neurotrophic factor neurons. LGR5 is not detectable with in situ hybridization; LGR6 is only expressed in the epithelial lining of the lower portion of the third ventricle and median eminence. Rspo1 is expressed in the VMH and down-regulated with fasting. Rspo3 is expressed in the paraventricular nucleus and also down-regulated with fasting. Rspos 1 and 3 colocalize with the neuronal marker HuD, indicating that they are expressed by neurons. Injection of Rspo1 or Rspo3 into the third brain ventricle inhibited food intake. Rspo1 decreased neuropeptide Y and increased proopiomelanocortin expression in the ARC. Rspo1 and Rspo3 mRNA is up-regulated by insulin. These data indicate that Rspo1 and Rspo3 and their receptor LGR4 form novel circuits in the brain to regulate energy homeostasis.
The hypothalamus is a key integrative center for peripheral and central signals controlling appetite and metabolism (1). Several distinct hypothalamic nuclei are involved in energy homeostasis, and each nucleus contains multiple neuronal types that synthesize and release different neuropeptides. Neurons in these nuclei are interconnected and also have connections to other brain areas (2). The arcuate nucleus (ARC) of the hypothalamus is a primary target of metabolic and hormonal signals from the periphery relating to energy homeostasis, because it is located in a site in the central nervous system with a permeable blood-brain barrier. Two major types of neurons located in the ARC regulate food intake. One type of neuron uses neuropeptide Y (NPY) as neurotransmitter, and intracerebroventricular (icv) injection of NPY stimulates food intake (orexigenic) (3). NPY neurons also colocalize with agouti-related protein, another potent orexigenic neuropeptide (4). Another type of ARC neuron uses α-melanocyte-stimulating hormone (α-MSH) as neurotransmitter/neuromodulator, which is derived from its precursor, proopiomelanocortin (POMC). α-MSH inhibits food intake (anorexigenic) via activation of melanocortin receptor subtype 3 (MC3R) and MC4R. POMC neurons also colocalize with another anorexigenic neuropeptide, cocaine- and amphetamine-regulated transcript (5). Agouti-related protein coproduced by NPY neurons acts as an endogenous antagonist of α-MSH on MC3R and MC4R to stimulate food intake. NPY neurons and POMC neurons from the ARC project to the paraventricular nucleus (PVN) of the hypothalamus and other brain areas. The PVN acts as an integrator of signals regulating food intake and energy expenditure. Within the PVN, NPY and POMC projections innervate neurons that express CRH and TRH, both anorexigenic (6, 7). As the first site recognized to be involved in body weight regulation, the ventromedial nucleus of the hypothalamus (VMH) plays a critical role in energy homeostasis (8). Within the VMH, neurons that express brain-derived neurotrophic factor (BDNF) are anorexigenic (9). BDNF neurons regulate energy balance downstream of MC4R (10). The lateral hypothalamus (LH) also plays a crucial role in the regulation of feeding behavior, especially in food motivation and reward. The LH receives projections from both POMC and NPY neurons of the ARC. The LH contains neurons that express the orexigenic melanin-concentrating hormone (MCH) and orexin (2, 11).
Analysis of microarray data derived from the ARC revealed that a receptor named leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4) is highly expressed in the ARC (12). This finding spurred interest, because receptors are potential drug targets for treatment of ingestive disorders. LGR4 together with LGR5 and LGR6 form a subfamily of closely related receptors sharing approximately 50% identity in amino acid sequence (13). LGR4 family receptors contain a large N-terminal extracellular domain composed of variable leucine-rich repeats, a 7 transmembrane region, and a C-terminal intracellular tail. LGR4 is composed of 951-amino acid residues and has 17 leucine-rich repeats. In the present study, we first investigated the distributions of LGR4 receptor subfamily members in the brain. Recently, R-spondin (Rspo) family proteins were identified as ligands for the LGR4 subfamily by several groups (14–17). Rspo proteins are a family of secreted proteins, which have 4 members in vertebrates (Rspo1–Rspo4). All 4 Rspo proteins are large molecules, ranging from 234 to 273 amino acids (18). The 4 Rspo proteins share the same domain structures and have a high percentage of amino acid identity. They all have the following distinct domains: an N-terminal signal peptide for secretion, 2 adjacent cysteine-rich furin-like domains, a thrombospondin type I repeat domain, and a C-terminal basic region. Rspos bind to LGR4 and LGR5 and enhance Wnt/β-catenin signaling (14–17). In this study, we also investigated the distribution of Rspos (1–4) in the rat brain and the effect of Rspos on ingestive behavior. Our data indicate that LGR4 and its ligands Rspos 1 and 3 are expressed in hypothalamic energy homeostatic areas and are involved in regulation of food intake.
Materials and Methods
Antibodies and reagents
Affinity-purified rabbit anti-Rspo1 antibody and control peptide are from Acris Antibodies. Mouse Rspo1 and Rspo3 are from R&D System. Mouse monoclonal anti-HuC/D antibody (16A11) is from Invitrogen. This antibody recognizes the embryonic lethal abnormal vision-like family members HuC and HuD, which are neuronal proteins. Labeling is visible at about the time the neurons leave the mitotic cycle (19). Further information on antibodies and control peptide is provided in Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org. The digoxigenin (Dig) RNA labeling system is from Roche.
In situ hybridization
All studies were approved by the University of Michigan Committee on Use and Care of Animals. The protocol number for animal use is PRO00001302. Male Sprague Dawley rats (280–300 g) were housed in cages with 3 rats/cage except for food intake study, in which rats were housed single caged. They were maintained at 22 ± 1°C in a room with a 12-hour light (6 am to 6 pm), 12-hour dark (6 pm to 6 am) cycle. They were fasted for 12 (overnight), 24, or 48 hours with free access to water. Fed rats had free access to food and water. Rats were anesthetized with ketamine/xylazine and perfused via the ascending aorta with 200 mL of PBS, followed by 200 mL of 4% paraformaldehyde in PBS. The brain was postfixed for 16 hours and then transferred to 20% sucrose (20% sucrose in PBS with 0.02% sodium azide) for 5 days at 4°C. The brain was embedded with 20% sucrose and Tissue-Tek optimal cutting temperature compound (2:1), and coronal sections of 14 μm were cut using a cryostat. The sections were dried overnight at room temperature and were stored at −80°C until further processing. Sections studied covered the telencephalon and diencephlon from coordinates bregma −0.80 mm to bregma −4.16 mm according to Paxinos and Watson (20). In situ hybridization was conducted using a modification of a previously described technique (21, 22).
All of the rat cDNA fragments for producing antisense and sense RNA probes (Table 1) were generated by PCR from an adult rat ARC cDNA library, except for rat Rspo2, which is from a neonatal rat brain cDNA library. All were subcloned into pBluescript SK, which has T3 and T7 RNA polymerase promoters (pBSK; Stratagene). All cDNAs were confirmed by DNA sequencing.
Table 1.
Information on Probes for In Situ Hybridization
| Genes | Access ion Number | cDNA Position | Forward and Reverse Primers Sequences | Cloning Sites in pBSK |
|---|---|---|---|---|
| LGR4 | NM_173328 | 583–1516 | F,5′GGCGCTGACCTTGGCTCTC3′ | EcoRI |
| R,5′TTCTTGGGGGCTGTTATCTTCTGT3′ | KpnI | |||
| LGR5 | NM_001106784 | 1237–1684 | F,5′ATTCACCCCAACGCTTTCTCTACG3′ | BamHI |
| R,5′TTCGGATCAGCCAGCTACCAAACA3′ | HindIII | |||
| LGR6 | NM_001062538 | 1482–1993 | F,5′GTGGCAGGCCGAGGACTTTC3′ | EcoRI |
| R,5′CGGCCGCCAGCGTGAGCAG3′ | KpnI | |||
| Rspo1 | NM_001107980 | 589–1048 | F,5′CCTGCCCGCCTGGATACTTTGAT3′ | EcoRI |
| R,5′TTCTTCCTGGTCGGGTGCCTATTG3′ | KpnI | |||
| Rspo2 | NM_001130575 | 1067–1662 | F,5′AAGGCAACCGATGGAGACGCAGTA3′ | BamHI |
| R,5′TTTCGGAAGGCAGGCAGCATA3′ | HindIII | |||
| Rspo3 | NM_001100990 | 229–676 | F,5′TGCGCGACGTGTTCAGATTAC3′ | EcoRI |
| R,5′TGGTTGGGGGACACAGGTTC3′ | KpnI | |||
| Rspo4 | NM_575261 | 392–817 | F,5′GTACCCGGGAGTGCCAGGAGGAGT3′ | EcoRI |
| R,5′GACCGGGCA-GATGGGAAAAGTAGC3′ | KpnI | |||
| HuD | NM_001077651 | 784–1244 | F,5′CAGTCCCCCAACAGGCGATAC3′ | BamHI |
| R,5′GACTTGTGGGCTTTGTTGGTTTTA3′ | HindIII | |||
| BDNF | NM_012513 | 898–1329 | F,5′CGGCCCAACGAAGAAAACCATAAG3′ | EcoRI |
| R,5′ATCCATAGTAAGGGCCCGAACATA3′ | KpnI | |||
| IR | NM_017071 | 2716–3247 | F,5′GCGCCCACAAGCCACGAAGAG3′ | BamHI |
| R,5′CGAAGATGAGGGGCCCGATGATAA3′ | HindIII |
F, forward; R, reverse.
The NPY cDNA (AI045437) in pT7T3D-PAC was purchased from Invitrogen. The rat POMC plasmid construct consisted of an 833-bp insert that included the full coding region of the POMC gene inserted into pGEM4Z (Promega). The POMC-pGEM4Z plasmid was constructed by R. C. Thompson (Neuroscience Institute, University of Michigan). The Dig- or [35S]-labeled antisense and sense RNA probes were generated using standard in vitro transcription methodology (23).
The brain sections were washed 3 times with 2× saline-sodium citrate buffer (SSC) and then digested with 0.45 μg/mL proteinase K (Invitrogen) in 100mM Tris (pH 8.0), 50mM EDTA for 15 minutes at 37°C. After brief washing with distilled water, the sections were acetylated with 0.25% acetic anhydride in 0.1M triethanolamine (pH 8.0), for 10 minutes. The sections were subsequently dehydrated through a graded series of ethanols. Antisense or sense probes were diluted in hybridization buffer (50% formamide, 3× SSC, 1× Denhart's, 200 μg/mL yeast transfer RNA, 50mM phosphate buffer [pH 7.4], 10% dextran sulfate, and 10mM dithiothreitol) to yield approximately 30 000 cpm/μL for [35S]-labeled probes, or 3 to 4 μL probes per 100 μL hybridization buffer for Dig-labeled probes. A total of 40 μL diluted probes was applied to each slide, and the sections were coverslipped. Slides were then placed in sealed plastic boxes lined with filter paper moistened with 50% formamide. The boxes were wrapped with plastic wrap and incubated at 55°C for 16 hours. For dual label in situ hybridization, the sections were hybridized with antisense Dig- and [35S]-labeled probes.
After overnight incubation, coverslips were removed by dipping in 2× SSC, and the slides were washed with 2× SSC. Slides were then incubated with 20 μg/mL ribonuclease A (Roche) in 10 mM Tris-HCl (pH 8.0, 0.5M NaCl) at 37°C for 1 hour. The slides were washed with 2×, 1×, 0.5×, and 0.1× SSC at room temperature for 5 minutes each time and then incubated in 0.1× SSC at 67°C for 1 hour. The sections were washed briefly with distilled water. For single [35S]-label in situ hybridization, the slides were dehydrated in graded alcohols and air dried. Dried slides were exposed to Kodak BioMax film. For dual label in situ hybridization, after washing with distilled water, the sections were incubated with anti-Dig antibody (Roche) conjugated with alkaline phosphatase (1:8000) overnight at room temperature. The slides were next incubated in nitroblueterazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate solution for 2–4 hours for color reactions. Then the slides were dipped in ILFORD K.5D nuclear emulsion and exposed in dark for 10–14 days at 4°C. Sections were developed with D-19 developer. Bright and dark field microscopic photos were taken and analyzed using ImageJ software. Colocalization was defined as clusters of silver grains being at least 4 times higher in the cell relative to background. Fifteen brain sections from 5 rats were counted, and percentage of colocalization was determined by dividing the colocalized neurons by total number of neurons.
Intracerebroventricular cannulation
Male Sprague Dawley rats with a body weight of 260–280 g were used. Rats were anesthetized with ketamine/xylazine and placed on a stereotaxic device with the incisor bar 3.3 mm below the interaural line according to Paxinos and Watson (20). A stainless steel 26-gauge guide cannula was implanted into the third ventricle using the following stereotaxic coordinates: 2.4 mm posterior to the bregma, 8.4 mm ventral to the surface of the skull and directly along the midline. The cannula was anchored to the skull with 4 screws and dental cement. An internal cannula was placed into the guide cannula to maintain patency. Rats were allowed to recover for 1 week. Guide cannula patency was assessed by injection of 10 ng angiotensin II in 5 μL of saline. Cannulas were considered patent if rats consumed at least 5 mL of water within 1 hour of injection. Rats with correct third ventricle cannulation were used 5 days later. All rats were handled daily to minimize stress reactions to manipulation.
Rspo1 and Rspo3 injections
After 2 hours of fasting (4–6 pm), the rats were injected with Rspo1 (1.25, 2.5, and 5 μg in 5 μL PBS) or Rspo3 (2.5, 5, and 7.5 μg in 5 μL PBS) into the third ventricle through the guide cannula. Control rats were injected with 5 μL PBS. Dark phase food intake (5–6 h from 6 pm) and 24-hour food intake were monitored. Body weight were measured before and 24 hours after injection.
In order to determine the effect of Rspo1 on hypothalamic gene expressions, rats were fasted for 12 hours (10 pm to 10 am) and then injected with either Rspo1 (5 μg in 5 μL PBS) or 5 μL PBS into the third ventricle twice (10 am and 2 pm). The rats were killed at 4 pm. Brains were embedded in powderised dry ice and then sectioned using a coronal brain matrix. Brain sections were placed on dry ice, and under ×4 magnification, the ARC was obtained using a Stoelting brain punch set. Total RNA was extracted from the brain tissues using TRIzol Reagent (Invitrogen). Around 1 μg of RNA was converted into cDNA using superscript II reverse transcriptase (Invitrogen) with an oligo dT primer. Real-time quantitative PCR (QPCR) was performed. The punches were verified to be anatomically correct, because BDNF, which is expressed in VMH, and orexin, which is expressed in LH, are undetectable with QPCR in the ARC punches (Supplemental Table 2).
Insulin injection
Rats were fasted overnight. Insulin (8 mU/rat) or saline was injected into the third brain ventricle twice at 2 hours of interval. The rats were killed 2 hours after the second injection; PVN and VMH were punched and total RNAs extracted. The punches were verified to be anatomically correct, because NPY, which is expressed in the ARC, was undetectable with QPCR in either the PVN or VMH punches (Supplemental Table 2). Rspo1 mRNA in VMH and Rspo3 mRNA in PVM were determined using QPCR.
Real-time quantitative PCR
QPCR was performed using a Rotor Gene Q real-time PCR detection system (QIAGEN). FAM-labeled fluorogenic probes and GoTaq DNA polymerase (Promega) were used. The QPCR primers for rat NPY are forward 5′-CCGCCATGATGCTAGGTAACAA-3′, probe 5′-/56FAM/AGCGAGGGTCAGTCCACACAGCC-/36-TAMTph/-3, and reverse 5′-CCAAACACACGAGCAGGGATA-3′; POMC forward 5′-CCACTGAACATCTTCGTCCTC-3′, probe 5′-/56-FAM/TGAGGCTCT/ZEN/GTCGCGGAAAGG/3IABkFQ/-3′, and reverse 5′-GAATCTCGGCATCTTCCAGG-3′; Rspo1 forward, 5′-AGAGACCCGGAAGTGTACC, probe 5′-/56-FAM/CCTGGCTCCTTGCTGTTCTTCCT/3IABkFQ/-3′, and reverse 5′-CCTTTGTGTCTCCGAGAGTTG-3′; Rspo3 forward 5′-CTCCAAACCTTTGCTGTCAGA-3′, probe 5′-/56-FAM/AAAGGGAGA/ZEN/GCGAGGGAAAAAGGG/3IABkFQ/-3′, and reverse 5′-CAGCGAGACAAGAACGTGTA-3′; BDNF forward 5′-CTGTTGGGGAGACGAGAT3′, probe 5′-/56-FAM/AGCAAAGCCACAATGTTCCACCAG/3IABkFQ/-3′, and reverse 5′-ATGCAACCGAAGTATGAAA-3′; orexin forward 5′-CGCAGAGCTAGAGCCATATC-3′, probe 5′-/56-FAM/CTGCAACCGCCACCGCTTTAG/3IABkFQ/-3′, and reverse 5′-TTAGGACTAGGACAGGGATAGAAG-3′. GAPDH was used as an internal control: forward 5′-ACCACAGTCCATGCCATCAC-3′, probe 5′-/56-FAM/CACAGCCTTGGCAGCACCAGTGGA/36-TAMTph/-3′, and reverse 5′-CAGCTCTGGGATGACCTTGC-3′. QPCR conditions were one cycle at 95°C for 5 minutes followed by 40 cycles of 95°C for 15 seconds; 56°C–58°C for 30 seconds; and 72°C for 30 seconds. Relative mRNA expression was normalized to GAPDH using the 2−ΔΔCT methodology (24).
Conditioned taste aversion test
Male Sprague Dawley rats (260–280 g) were used. The third brain ventricle was cannulated, and cannula patency was assessed with antgiotensin II injection as described above. Saccharin (0.15% wt/vol) was used as the conditioned stimulus and lithium chloride (LiCl) (0.60M, 1% body weight, injected ip) as the malaise-inducing agent (25). Conditioned taste aversion test were done according to the protocol described by Roy et al (26). Briefly, for 6 days before the onset of the experiment, rats were restricted to 60-minute water access daily until water intake stabilized. The conditioning sessions were initiated on day 7 followed by 1 test session on day 10. During the conditioning session (d 7), all rats were presented with saccharin instead of water for 30 minutes, after which they were immediately injected icv with Rspo1 (5 μg) or PBS followed by an ip injection of saline (control) or LiCl. Over the next 2 days, rats received daily access to water for 60 minutes. For the test session (d 10), all rats were presented with saccharin instead of water for 30 minutes, and water intake was compared in treatment vs control (d 10) to assess the induced aversion.
Double immunofluorescence
Dual immunofluorescence was performed to determine the expression of Rspo1 by neurons. Affinity-purified rabbit anti-Rspo1 (1:400) and mouse monoclonal anti-HuD (1:200) were used. Preabsorbed anti-Rspo1 by control peptide was use as negative control. Rat coronal brain sections of 14 μm were rinsed in PBS, 0.5% Triton X-100 3 times, 10 minutes each, and blocked in 10% serum for 1 hour, and then incubated with primary antibodies overnight at 4°C. Rhodamine (red)-conjugated goat antirabbit secondary antibody was used to visualize Rspo1. For HuD staining, biotinylated horse antimouse secondary antibody was used and then visualized with Alexa Fluor 488 (green)-conjugated avidin. Nuclei were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride.
Cell culture
Mouse hypothalamic NPY cell line (N38) was immortalized by Belsham et al (27). This cell line was purchased from Cellutions Biosystem, Inc. The cells were cultured in serum-free Neurobasal A medium supplemented with B-27 (Gibco) and human fibroblast growth factor (Biosource, Inc), 0.5 μg/mL, at 37°C supplemented with 5% CO2. RT-PCR was performed to determine the expression of NPY and LGR4 in the cell line, and the DNA sequences were confirmed by sequencing. NPY PCR primers span 3 introns: forward 5′-CCGCCCGCCACGATGCTA-3′ and reverse 5′-AACAAGGGAAATGGGGCGGAGTC-3′. LGR4 PCR primers span 1 intron: forward 5′-GAAGATAACAGCCCCCAAGACCAC-3′ and reverse 5′-CAGCGGGAAGAATATCAGCGTAAC-3′. Rspo1 (200 ng/mL) was added to the medium, and the NPY cell was incubated for 4 hours. QPCR was performed to determine NPY mRNA level as mentioned above. QPCR primers and probe for mouse NPY: forward 5′-GACAGAGATATGGCAAGAGATCC-3′, probe 5′-/56-FAM/AAGGAAAGCACAGAAAACGCCCC/3IABkFQ/-3′, and reverse 5′-TGGAAAAGTCGGGAGAA-CAAG-3′.
Statistical analysis
Values are expressed as mean ± SEM. ANOVA followed by post hoc Bonferroni test was used for statistical analysis. Significance was accepted as P < .05.
Results
LGR4 is expressed in the hypothalamus and other brain areas
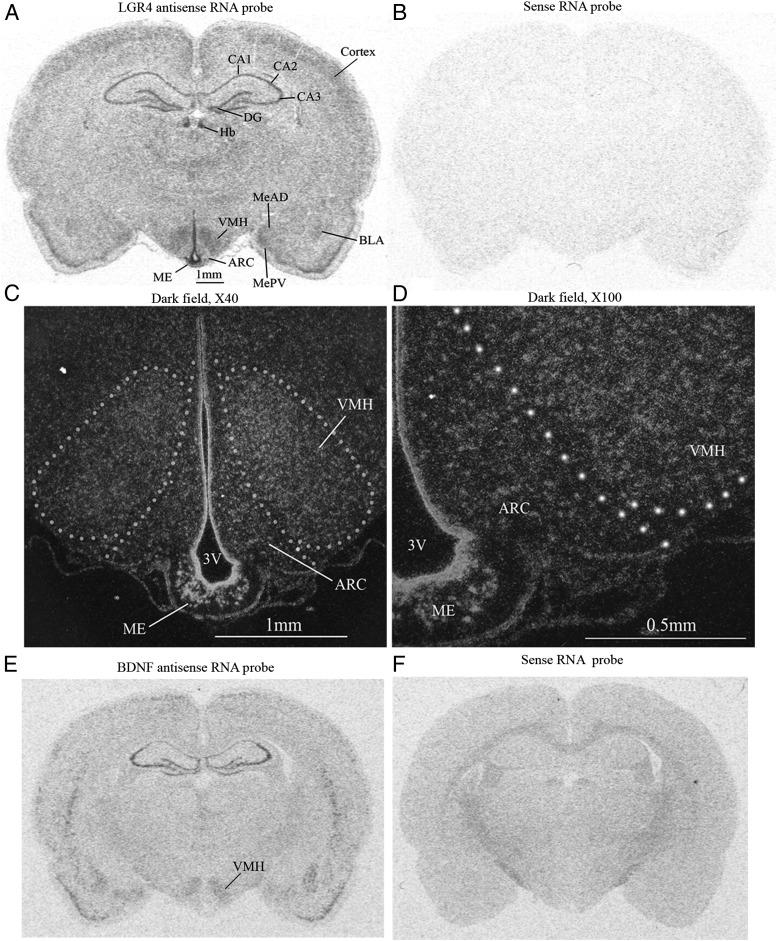
In situ hybridization revealed that LGR4 mRNA was highly expressed in the cortex, hippocampus, amygdala, and the hypothalamus (Figure 1A). In the cortex, LGR4 mRNA was expressed in layers II and III. In the hippocampus, LGR4 was expressed in CA1, CA2, CA3, and the dentate gyrus (DG).The habenular nuclei (Hbs) of the epithalamus also expressed LGR4. In the amygdala, LGR4 mRNA was expressed with high levels in the medial amygdaloid nucleus, medial amygdaloid nucleus, posterovent, and basal lateral amygdaloid nucleus.
Figure 1.
Distribution of LGR4 mRNAs in the rat brain. A, Distribution of LGR4 mRNA. CA1, CA2, and CA3, fields of hippocampus; MeAD, medial amygdaloid nucleus, anterodorsal; MePV, medial amygdaloid nucleus, posterovent; BLA, basal lateral amygdaloid nucleus, anterior. B, LGR4 sense RNA probe (negative control). C and D, Dark field photomicrographs of hypothalamus. 3V, third brain ventricle. E, Distribution of BDNF mRNA. F, BDNF sense RNA probe; n = 5.
In the hypothalamus, LGR4 is expressed in the VMH, the ARC, median eminence (ME), and the ependymocytes lining the third ventricle. Dark field microphotographs revealed that the ME and ependymocytes have the highest levels of LGR4 expression, followed by VMH and ARC (Figure 1, C and D). The expression pattern of LGR4 in the VMH overlaps that of BDNF (Figure 1E) (28).
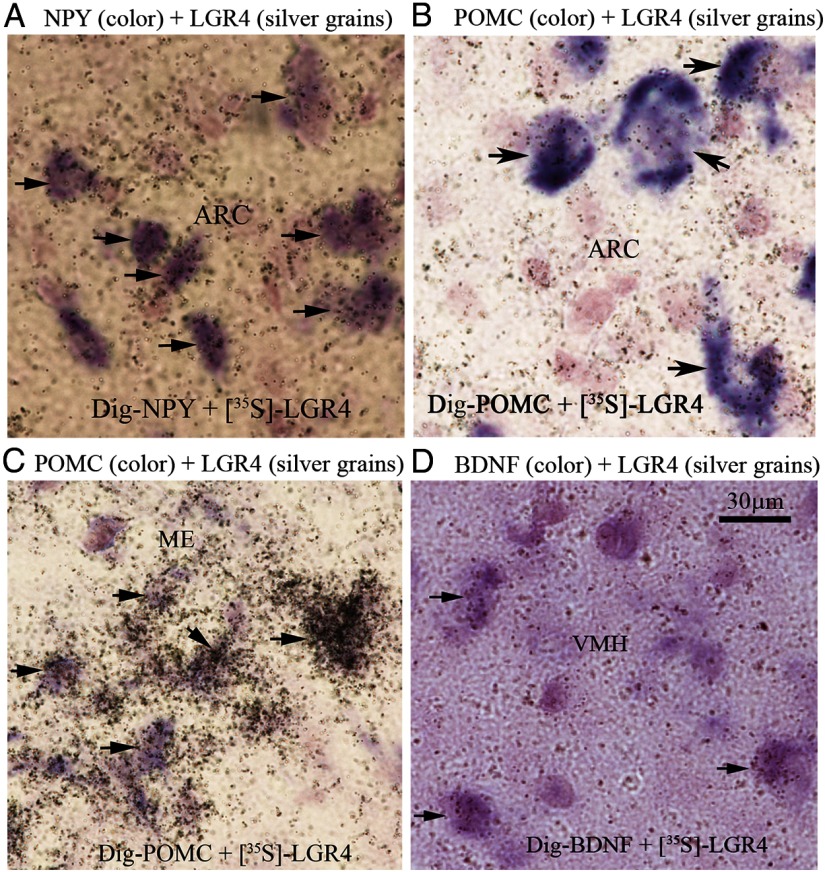
Double in situ hybridization showed that in the ARC LGR4 is expressed by most NPY neurons (96 ± 2%) and most POMC neurons (95 ± 2%) (Figure 2, A and B); NPY neurons have 40% higher levels of LGR4 expression than POMC neurons in the ARC (P < .05, n = 5). All POMC neurons in the ME express LGR4 at the highest levels (2.8-fold higher than POMC neurons in the ARC; P < .01, n = 5) (Figure 2C). In the VMH, LGR4 is expressed by the majority (77 ± 3%) of BDNF neurons (Figure 2D).
Figure 2.
Double in situ hybridization. Photomicrographs are representatives of hypothalamic areas. Silver grains indicate LGR4 mRNA in all panels. A, Purple color represents NPY mRNA. B and C, Purple color represents POMC mRNA. D, Purple color represents BDNF mRNA. Arrow heads indicate colocalization; n = 5.
There was no detectable level of LGR5 mRNA in the brain using in situ hybridization (Supplemental Figure 1A). LGR6 mRNA was found to be expressed with detectable levels only in the epithelial lining of the lower portion of the third ventricle and in the ME (Supplemental Figure 1C).
Rspo1 mRNA is expressed in VMH and other brain areas and is down-regulated with fasting in VMH
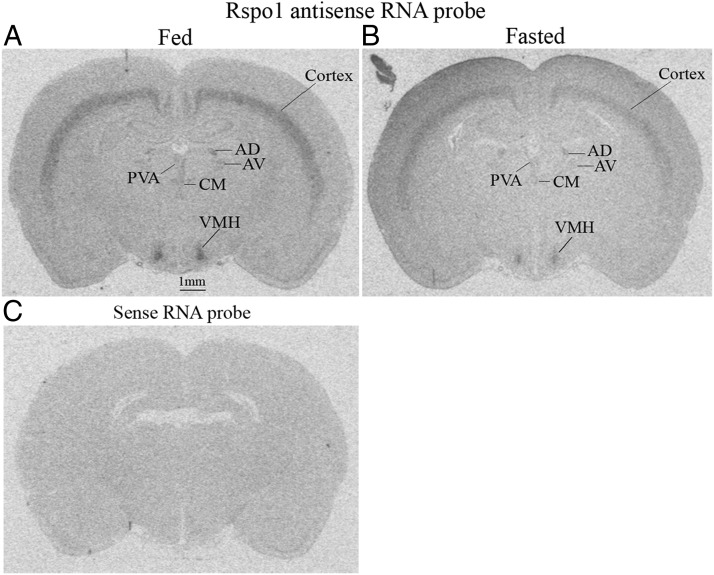
Recently, Rspo proteins were identified as ligands for the LGR4 receptor family (LGR4, LGR5, and LGR6). In order to determine whether Rspos are expressed in the brain, especially in hypothalamic areas, we performed in situ hybridization in male rat brain sections that cover the anterior through posterior hypothalamus (bregma −0.80 mm to −4.16 mm according to Paxinos and Watson). In the hypothalamus, Rspo1 was found to be highly expressed in the VMH (Figure 3, A and B). Rspo1 was also expressed in layers V and VI of the brain cortex, in the thalamus. In the thalamus, Rspo1 was expressed in the anterodorsal thalamic nucleus, anteroventral thalamic nucleus, paraventricular thalamic nucleus.
Figure 3.
Distribution of Rspo1 mRNA in the rat brain. A, Rspo1 mRNA in fed rat brain. B, Rspo1 mRNA in fasted rat brain. AD, anterodorsal thalamic nucleus; AV, anteroventral thalamic nucleus; PVA, paraventricular thalamic nucleus, anterior. C, Rspo1 sense RNA probe; n = 5.
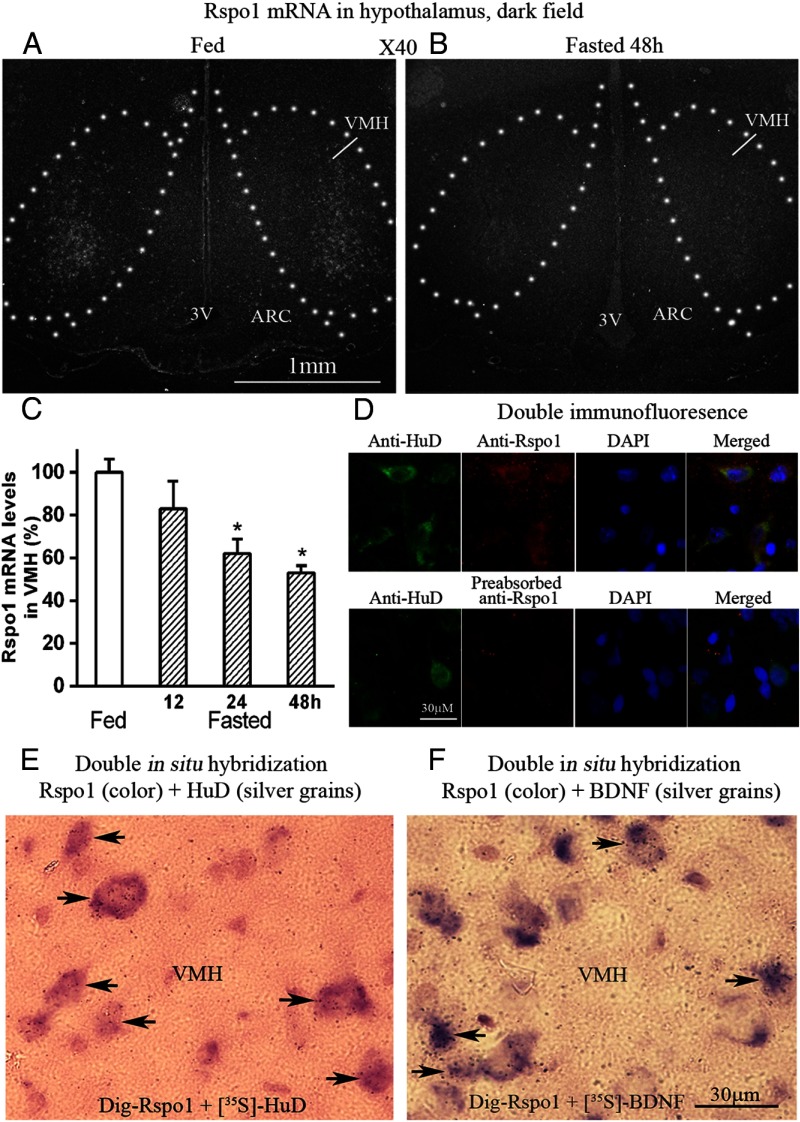
Figure 4, A and B, shows dark field microphotographs of hypothalamic areas. Clusters of silver grains, which represent Rspo1 mRNA, were concentrated in lateral portions of the VMH. Densitometry analysis of the dark field photomicrographs of rat brain sections from animals fed and fasted for different time periods revealed that Rspo1 mRNA is significantly down-regulated in the VMH by fasting in a time-dependent manner (Figure 4C).
Figure 4.
Dark field photomicrograph of hypothalamus, double in situ hybridization, and immunofluorescence. A, Fed rat. B, Fasted 48-hour rat. 3V, third brain ventricle; C, Densitomitry analysis of Rspo1 mRNA levels in VMH of fed and fasted rats. *, P < .05 compared with fed rats; n = 5. D, Photomicrograph from VMH, green represents HuD protein, red represents Rspo1 protein, blue represents nuclei. Yellow shows colocalization. E, Color represents Rspo1 mRNA, silver grains represent HuD mRNA. Arrow heads indicate colocalization. F, Purple color represents Rspo mRNA, silver grains represent BDNF mRNA n = 5. DAPI, 4',6-diamidino-2-phenylindole, dihydrochloride.
We asked whether Rspo1 is expressed by neurons in the VMH. Because HuD is a specific neuronal marker, we used Dig-labeled HuD and [35S]-labeled Rspo1 as probes to perform double in situ hybridization. For double immunohistochemistry, anti-HuD and anti-Rspo1 antibodies were used. Both double immunohistochemistry and double in situ hybridization showed that Rspo1 colocalized with HuD by 100% in the brain, indicating that Rspo1 is expressed by neurons (Figure 4, D and E).
In order to determine the nature of Rspo1-containing neurons, we performed double in situ hybridization with Dig-labeled Rspo1 and [35S]-labeled BDNF or [35S]-labeled Rspo1 and Dig-labeled NPY, POMC, MCH, and orexin. The results showed that in the VMH, about 22% (22.4 ± 2.6%) of Rspo1-positive cells also express BDNF (Figure 4F). In the ARC, only a very small portion of NPY neurons (around 3%) or POMC neurons (around 3%) express Rspo1 at low levels; most NPY and POMC neurons do not express Rspo1. Neither MCH nor orexin neurons in the LH express Rspo1 (data not shown).
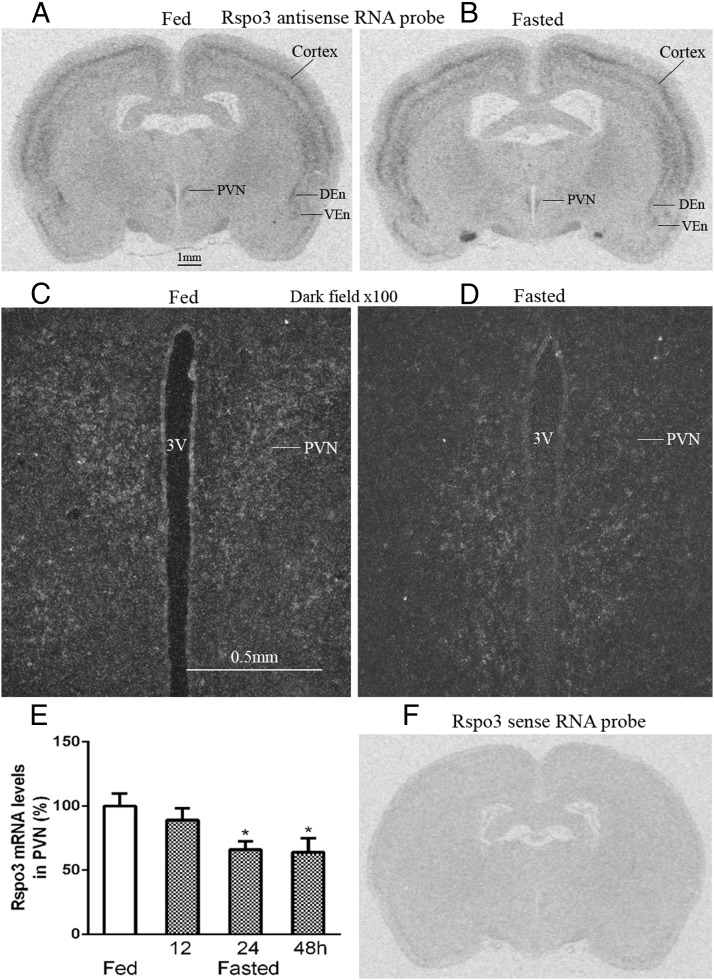
Rspo3 mRNA is expressed in PVN and other brain areas and is down-regulated in PVN with fasting
Rspo3 mRNA was found to be expressed in the PVN of the hypothalamus, brain cortex, dorsal and ventral endoform nuclei (Figure 5, A and B), DG, and polymorph layer of the DG (data not shown). Dark field photomicrographs showed that clusters of silver grains, which represent Rspo3 mRNA, were located in the PVN of the hypothalamus (Figure 5, C and D). Densitometry analysis revealed that Rspo3 mRNA levels in the PVN were down-regulated with fasting in a time-dependent manner (Figure 5E). Double in situ showed that in the PVN all CRH neurons express Rspo3 (see figure 8 below) and Rspo3 colocalized with the neuronal marker HuD (data not shown).
Figure 5.
Distribution of Rspo3 mRNAs in the rat brain. A, Fed rat brain. B, Fasted rat brain. C and D, Dark field photomicrograph of PVN areas of fed and fasted rat. 3V, third brain ventricle; DEn and VEn, dorsal and ventral endoform nuclei. E, Densitometry analysis of Rspo3 mRNA level in PVN. n = 5; *, P < .05. F, Rspo3 sense RNA probe.
Rspo2 is not detectable in the brain, and Rspo4 is only expressed in the Hb
There was no detectable level of Rspo2 mRNA in the adult rat brain (Supplemental Figure 2A). Rspo4 mRNA was only expressed in the lateral Hb that is adjacent to the dorsal third brain ventricle (Supplemental Figure 2C).
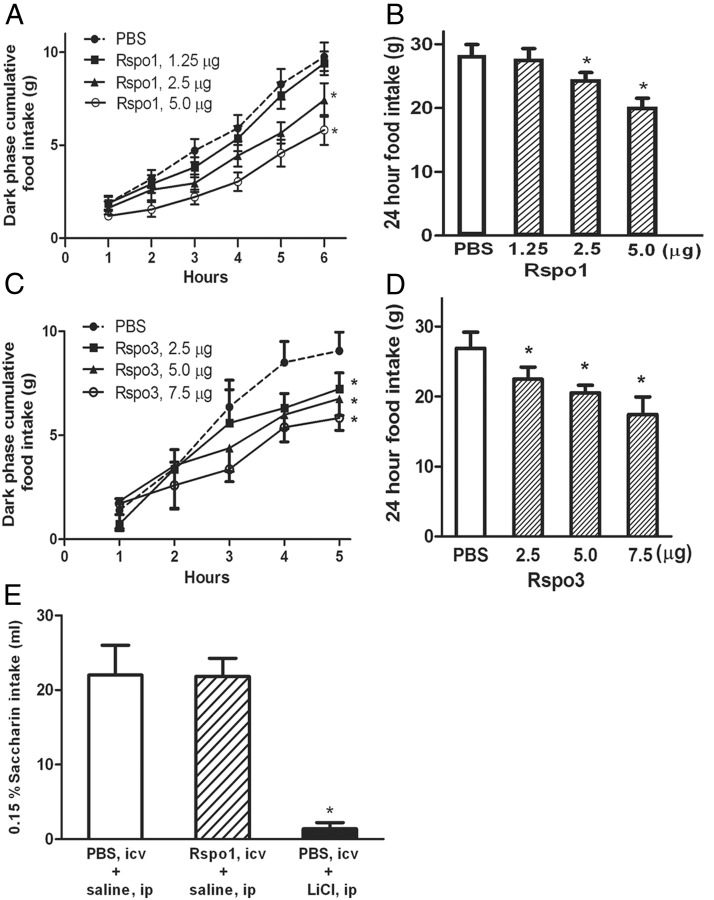
Rspo1 and Rspo3 inhibit food intake
Because LGR4 and Rspo1 and Rspo3 mRNAs were found to be highly expressed in hypothalamic energy homeostatic areas, and Rspo1 and Rspo3 mRNAs were down-regulated with fasting, we hypothesized that both Rspo1 and Rspo3 would inhibit food intake. Injection of Rspo1 into the third brain ventricle inhibited dark phase (1–6 h) and 24-hour food intake dose dependently (Figure 6, A and B). Third ventricular injection of Rspo3 had similar effects on food intake (Figure 6, C and D). Third ventricle injection of Rspo1 did not cause taste aversion in rats (Figure 6E). Compared with animals preinjection, the body weights of treated rats showed a trend of reduction, reaching statistical significance with the highest dose (5 μg) (Supplemental Figure 3) 24 hours after Rspo1 injection. There was no significant reduction in 24-hour body weight after Rspo3 injection (data not shown).
Figure 6.
Effect of Rspo1 and Rspo3 on food intake. A and B, Dark phase and 24-hour food intake after third ventricular injection of Rspo1. n = 8; *, P < .05. C and D, Dark phase and 24-hour food intake after third ventricle injection of Rspo3. n = 8; *, P < .05. E, Taste aversion test after third ventricular injection of Rspo1. n = 8; *, P < .05.
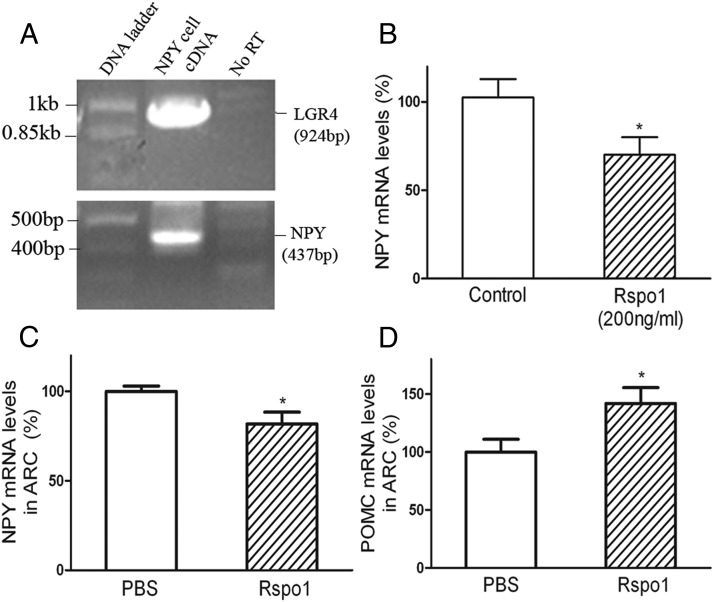
Rspo1 decreases NPY mRNA expression and increases POMC mRNA expression
In order to elucidate the mechanisms underlying the inhibition of food intake by Rspos, we studied the effect of Rspo1 on NPY gene expression both in vitro and in vivo and on POMC gene expression in vivo. Rspo1 was chosen because it was reported to be more potent than Rspo3 in receptor binding (15). Using RT-PCR, we first confirmed that a mouse hypothalamic NPY cell line did express NPY mRNA and further found that this cell line had high levels of LGR4 mRNA expression (Figure 7A, bottom and upper panels). Treatment of the NPY cell line with Rspo1 inhibited NPY gene expression (Figure 7B). Injection of Rspo1 into the third brain ventricle inhibited ARC NPY mRNA expression and increased ARC POMC mRNA expression (Figure 7, C and D).
Figure 7.
Effect of Rspo1 on NPY and POMC gene expression. A, LGR4 (upper panel) and NPY (lower panel) gene expression in hypothalamic NPY cell line. B, Rspo1 inhibited NPY gene expression in NPY cell line. n = 8; *, P < .05. C, Effect of Rspo1 on NPY expression in rat ARC. n = 6; *, P < .05. D, Effect of Rspo1 on POMC gene expression in rat ARC. n = 6; *, P < .05.
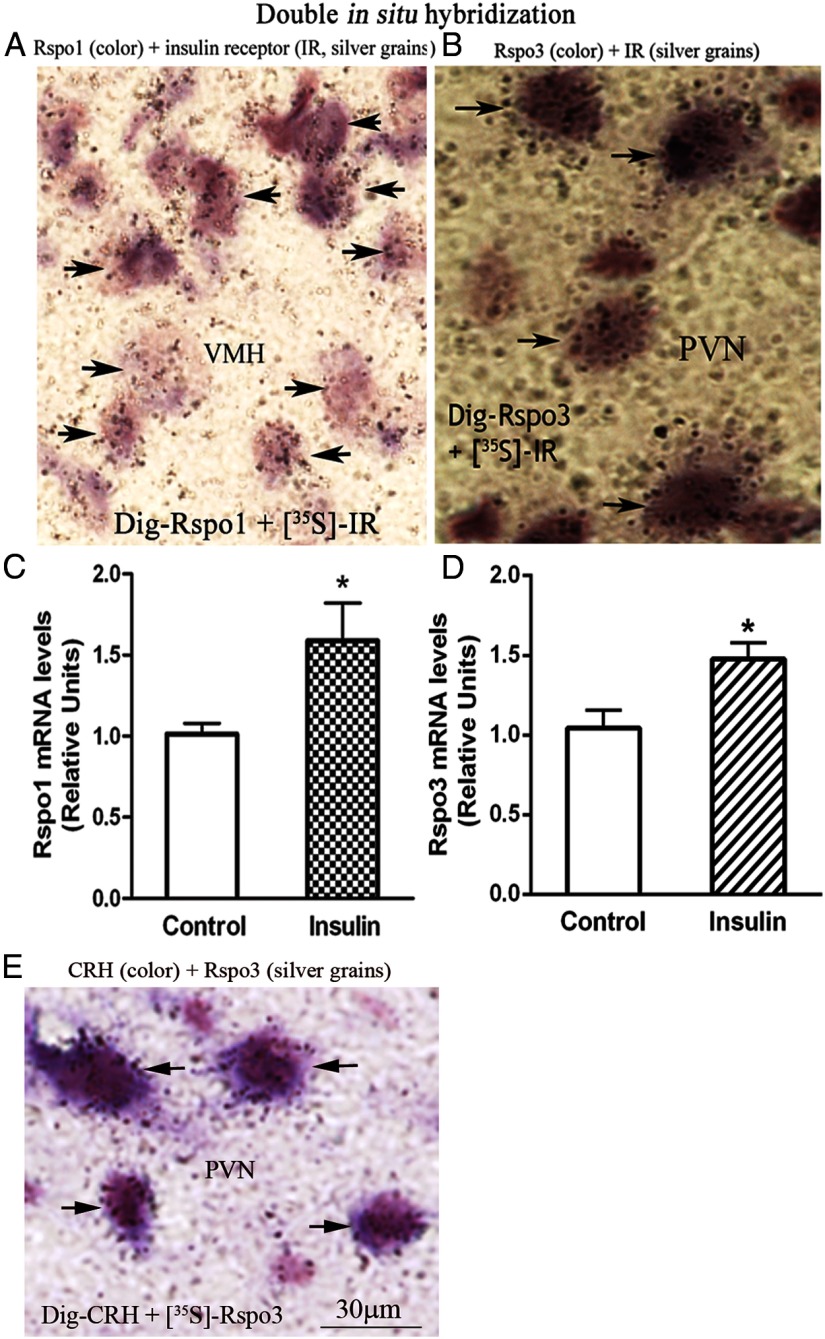
Rspo1- and Rspo3-expressing cells have insulin receptor (IR), and insulin increases Rspo1 and Rspo3 expressions
Insulin is a well-known satiety factor, which exerts its action in the hypothalamus. We asked whether Rspo1 and Rspo3 act downstream of insulin. Double in situ hybridization showed that Rspo1 and Rspo3 colocalize with IR (Figure 8, A and B). Injection of insulin into the third brain ventricle increases Rspo1 mRNA level in VMH and Rspo3 level in PVN (Figure 8, C and D).
Figure 8.
Colocalization of Rspo1 or Rspo3 with IR, Rspo3 with CRH, and effect of insulin on Rspo1 and Rspo3 expressions. A, Color represents Rspo1 mRNA, silver grains represent IR mRNA. B, Color represents Rspo3 mRNA, silver grains represent IR mRNA. Arrow heads indicate colocalization. C, Rspo1 mRNA levels in VMH. D, Rspo3 mRNA levels in PVN. n = 5; *, P < .05. E, Color represents CRH mRNA, silver grains represent Rspo3 mRNA; n = 5.
Discussion
In the present study, we systematically investigated the distribution of the LGR receptor subfamily members (LGR4–LGR6) and their recently identified ligands, Rspos (1–4), in the adult male rat brain. Both LGR4 and Rspos 1 and 3 are present in hypothalamic nuclei relevant to the control of feeding behavior, and in vivo effects of Rspo1 and Rspo3 on food intake suggest a new role for this system in regulation of food intake. The major findings that support this conclusion include: 1) LGR4 is expressed in the hypothalamus and colocalizes with NPY, POMC, and BDNF neurons; 2) Rspo1 is expressed in VMH and was down-regulated with fasting in this nucleus; 3) Rspo3 is expressed in PVN and was also down-regulated with fasting in this nucleus; 4) injection of both Rspo1 and Rspo3 into the third ventricle inhibited food intake; 5) Rspo1 decreased NPY gene expression and increased POMC expression in the ARC; and 6) insulin increased Rspo1 and Rspo3 mRNA expression.
LGR family receptors contain an N-terminal ectodomain composed of variable leucine-rich repeats, a 7 transmembrane region, and a C-terminal intracellular tail. Both LGR4 and LGR5 have 17 leucine-rich repeats, whereas LGR6 has 13. Crystal structure studies demonstrate that the leucine-rich repeat domains form horseshoe structures, and the leucine-rich repeats 3–9 modules serve as ligand binding sites (29, 30). LGR4 is expressed in a broad range of tissues, including kidney, bone/cartilage, heart, stomach, and nerve cells (31). LGR4 null mice are pre- and postnatally lethal (32, 33). It has been reported that LGR4 is involved in the development of multiple organs, including kidney (33), reproductive tract (34), and mammary gland (35). LGR5 null mice also exhibit 100% neonatal lethality associated with ankyloglossia and gastrointestinal distension (36). LGR5 has been studied as a marker of cycling stem cells in the intestine (37), whereas LGR6 has been identified as a stem cell marker in hair follicles (38).
The different levels and patterns of expression of the 3 members of LGR receptor subfamily members that we observed in the rat telencephalon (forebrain) and diencephalon (interbrain) imply that they may serve different brain functions. LGR5 was observed at very low levels of expression in the forebrain, and the lack of any specific pattern of expression implies that it plays a very limited role in central nervous system function in adult rats. The highly restricted distributions of LGR6 to ependymocytes lining the basal portion of the third brain ventricle and the adjacent ME suggest that LGR6 might have a role the regulation of neuroendocrine function. Our studies demonstrated that LGR4 is widely expressed in the brain, including the cortex, hippocampus, amygdala, and hypothalamus. Our observations are consistent with a report by Van Schoore et al (31). Using a gene trap strategy and in situ hybridization, they found that LGR4 is widely expressed in the brain and other organs of embryonic and adult mice. In our studies, the distribution of LGR4 in hypothalamic energy homeostatic areas and colocalization with key energy homeostatic neurons suggested that it might affect energy homeostasis.
Rspo1 was first identified in the NSC-19 cell line and has since been found to be transiently expressed in the dorsal portion of the neural tube during development (Rspo for roof plate-specific spondin) (39). Rspo family proteins were recently identified by several groups using different strategies, from tandem affinity purification mass spectrometry to cell-based binding assays and genome wide small interfering RNA screening (14–16). Another report used complementary genome-wide screening based on loss- and gain-of-function approaches (17). The identification of Rspos (1–4) as ligands for the orphan LGR receptor subfamily (LGR4–LGR6) was a milestone. Rspos bind to leucine-rich repeats 3–9 modules of LGR4 and LGR5 by their 2 furin-like domains, and Rspo stimulation of LGR family receptors enhances Wnt/β-catenin signaling, which plays essential roles in embryonic development and in self-renewal and maintenance of adult stem cells (14–17, 29, 30).
Rspos are large secreted proteins, ranging from 234 to 273 amino acids in length (18), similar in size to the anorexigenic factor BDNF (247 amino acids) (40). Similar to LGR4 family receptors, we found that the 4 Rspo proteins have substantially different expressions and distributions in the telencephalon and diencephalon. Rspo2 is not expressed in the telencephalon and diencephalon. Rspo4 expression is restricted to the Hbs of epithalamus. These observations indicate that Rspo2 and Rspo4 are likely to have very limited roles in the function of these brain areas. In contrast, both Rspo1 and Rspo3 are widely expressed in the telencephalon and diencephalon, although they demonstrate different patterns of expression, implying that they participate in the regulation of brain function differently. Although it cannot be ruled out that Rspo1 and Rspo3 are expressed by other cell types, the colocalization of Rspo1 and Rspo3 with the neuronal marker HuD, and the fact that a portion of BDNF neurons express Rspo1 and all CRH neurons express Rspo3 argue for the neuronal nature of Rspos-expressing cells.
These findings suggest that Rspo proteins may function as neurotransmitters or modulators. The expression of Rspo1 and Rspo3 in hypothalamic energy homeostatic areas, their down-regulation with fasting, and their up-regulation by the satiety factor insulin indicate that they may be involved in the regulation of energy homeostasis as anorexigenic factors. The inhibition of food intake observed after icv injection of Rspo1 or Rspo3 support this concept. Rspo1 was more potent than Rspo3 in inhibiting food intake. Similarly, Rspo1 has been reported to be more potent than Rspo3 in receptor binding to LGR4 (15).
Changes in Rspo1 and Rspo3 mRNA with fasting are similar to those observed for POMC mRNA (41). Using immunofluorescent staining, we observed Rspo1 protein expression in VMH and other brain areas that express Rspo1 mRNA. The function of Rspo1 and Rspo3 and their cognate receptor LGR4 expressed outside of the hypothalamus, such as in the cortex, hippocampus, and amygdala, remains unknown. One possibility is that this ligand-receptor system in these brain areas might play some role in the integration of feeding behavior, such as food rewarding (42).
In a recent study, Wang et al (43) obtained G-protein-coupled receptor 48 (LGR4) hypomutant mice that exhibited an approximately 90% knockdown efficiency of Gpr48 in the kidney and adrenal gland. Approximately half of newborns died within 24 hours of birth, with the remainder surviving. These mice exhibited polydipsia, water loss, and abnormal urine osmolality. They also demonstrated increased food intake through 24 weeks of age.
The localization of LGR4 in hypothalamic energy homeostatic areas and expression by NPY, POMC, and BDNF neurons indicates that this receptor subtype is likely to be the major mediator of Rspo1 and Rspo3 functions on food intake. The inhibition of NPY expression and induction of POMC mRNA expression in the ARC by Rspo1 may at least partially contribute to the effects on food intake by Rspo1.
In conclusion, Rspos 1 and 3 and LGR4 have functions beyond those associated with development and stem cell activity. LGR4 and its ligands also form complicated novel brain circuits in the regulation of energy homeostasis.
Acknowledgments
This work was supported by National Institutes of Health Grants 5RO1 DK054032 and 5RO1 DK043225.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ARC
- arcuate nucleus
- BDNF
- brain-derived neurotrophic factor
- DG
- dentate gyrus
- Dig
- digoxigenin
- Hb
- habenular nucleus
- icv
- intracerebroventricular
- IR
- insulin receptor
- LGR4
- leucine-rich repeat-containing G protein-coupled receptor 4
- LH
- lateral hypothalamus
- MCH
- melanin-concentrating hormone
- MC3R
- melanocortin receptor subtype 3
- ME
- median eminence
- α-MSH
- α-melanocyte-stimulating hormone
- NPY
- neuropeptide Y
- POMC
- proopiomelanocortin
- PVN
- paraventricular nucleus
- QPCR
- real-time quantitative PCR
- Rspo
- R-spondin
- SSC
- saline-sodium citrate buffer
- VMH
- ventromedial nucleus of the hypothalamus.
References
- 1. Berthoud HR, Morrison C. The brain, appetite, and obesity. Annu Rev Psychol. 2008;59:55–92 [DOI] [PubMed] [Google Scholar]
- 2. Parker JA, Bloom SR. Hypothalamic neuropeptides and the regulation of appetite. Neuropharmacol. 2012;63:18–30 [DOI] [PubMed] [Google Scholar]
- 3. Clark JT, Kalra PS, Crowley WR, Kalra SP. Neuropeptide Y and human pancreatic polypeptide stimulate feeding behavior in rats. Endocrinology. 1984;115:427–429 [DOI] [PubMed] [Google Scholar]
- 4. Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272 [DOI] [PubMed] [Google Scholar]
- 5. Vrang N, Larsen PJ, Clausen JT, Kristensen P. Neurochemical characterization of hypothalamic cocaine-amphetamine-regulated transcript neurons. J Neurosci. 1999;19:RC5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glowa JR, Gold PW. Corticotropin releasing hormone produces profound anorexigenic effects in the rhesus monkey. Neuropeptides. 1991;18:55–61 [DOI] [PubMed] [Google Scholar]
- 7. Lechan RM, Fekete C. The TRH neuron: a hypothalamic integrator of energy metabolism. Prog Brain Res. 2006;153:209–235 [DOI] [PubMed] [Google Scholar]
- 8. King BM. The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav. 2006;87:221–244 [DOI] [PubMed] [Google Scholar]
- 9. Cordeira J, Rios M. Weighing in the role of BDNF in the central control of eating behavior. Mol Neurobiol. 2011;44:441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu B, Goulding EH, Zang K, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6:736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berthoud HR, Münzberg H. The lateral hypothalamus as integrator of metabolic andenvironmental needs: from electrical self-stimulation to opto-genetics. Physiol Behav. 2011;104:29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li JY, Kuick R, Thompson RC, et al. Arcuate nucleus transcriptome profiling identifies ankyrin repeat and suppressor of cytokine signalling box-containing protein 4 as a gene regulated by fasting in central nervous system feeding circuits. J Neuroendocrinol. 2005;17:394–404 [DOI] [PubMed] [Google Scholar]
- 13. Van Loy T, Vandersmissen HP, Van Hiel MB, et al. Comparative genomics of leucine-rich repeats containing G protein-coupled receptor and their ligands. Gen Comp Endocrinol. 2008;155:14–21 [DOI] [PubMed] [Google Scholar]
- 14. de Lau W, Barker N, Low TY, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297 [DOI] [PubMed] [Google Scholar]
- 15. Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glinka A, Dolde C, Kirsch N, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruffner H, Sprunger J, Charlat O, et al. R-spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One. 2012;7:e40976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin YR, Yoon JK. R-spondin family of proteins: emerging regulator of WNT signaling. Int J Biochem Cell Biol. 2012;44:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155 [DOI] [PubMed] [Google Scholar]
- 20. Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4th ed New York, NY: Academic Press; 1998 [Google Scholar]
- 21. Schafer MKH, Herman JP, Watson SJ. In situ hybridization histochemistry. In: London ED, ed. Imaging Drug Action in the Brain. Boca Raton, FL: CRC Press; 1993:337–378 [Google Scholar]
- 22. Richardson HN, Parfitt DB, Thompson RC, Sisk CL. Redefining gonadotropin-releasing hormone (GnRH) cell groups in the male Syrian hamster: testosterone regulates GnRH mRNA in the tenia tecta. J Neuroendocrinol. 2002;14:375–383 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed New York, NY: Cold Spring Harbor Laboratory Press; 2001 [Google Scholar]
- 24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Batsell WR, Best MR. One bottle too many? Method of testing determines the detection of overshadowing and retention of taste aversions. Anim Learn Behav. 1993;21:154–158 [Google Scholar]
- 26. Roy C, Roy MC, Gauvreau D, et al. Acute injection of ASP in the third ventricle inhibits food intake and locomotor activity in rats. Am J Physiol Endocrinol Metab. 2011;301:E232–E241 [DOI] [PubMed] [Google Scholar]
- 27. Belsham DD, Cai F, Cui H, Smukler SR, Salapatek AM, Shkreta L. Generation of phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology. 2004;145(1):393–400 [DOI] [PubMed] [Google Scholar]
- 28. Larsen MH, Hay-Schmidt A, Rønn LC, Mikkelsen JD. Temporal expression of brain-derived neurotrophic factor (BDNF) mRNA in the rat hippocampus after treatment with selective and mixed monoaminergic antidepressants. Eur J Pharmacol. 2008;578:114–122 [DOI] [PubMed] [Google Scholar]
- 29. Wang D, Huang B, Zhang S, Yu X, Wu W, Wang X. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 2013;27:1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen PH, Chen X, Lin Z, Fang D, He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27:1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Schoore G, Mendive F, Pochet R, Vassart G. Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem Cell Biol. 2005;124:35–50 [DOI] [PubMed] [Google Scholar]
- 32. Mazerbourg S, Bouley DM, Sudo S, et al. Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol Endocrinol. 2004;18:2241–2254 [DOI] [PubMed] [Google Scholar]
- 33. Kato S, Matsubara M, Matsuo T, et al. Leucine-rich repeat-containing G protein-coupled receptor-4 (LGR4, Gpr48) is essential for renal development in mice. Nephron Exp Nephrol. 2006;104:e63–e75 [DOI] [PubMed] [Google Scholar]
- 34. Mendive F, Laurent P, Van Schoore G, Skarnes W, Pochet R, Vassart G. Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev Biol. 2006;290:421–434 [DOI] [PubMed] [Google Scholar]
- 35. Oyama K, Mohri Y, Sone M, Nawa A, Nishimori K. Conditional knockout of Lgr4 leads to impaired ductal elongation and branching morphogenesis in mouse mammary glands. Sex Dev. 2011;5:205–212 [DOI] [PubMed] [Google Scholar]
- 36. Morita H, Mazerbourg S, Bouley DM, et al. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2004;24:9736–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene LGR5. Nature. 2007;449:1003–1007 [DOI] [PubMed] [Google Scholar]
- 38. Snippert HJ, Haegebarth A, Kasper M, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389 [DOI] [PubMed] [Google Scholar]
- 39. Kamata T, Katsube K, Michikawa M, Yamada M, Takada S, Mizusawa H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim Biophys Acta. 2004;1676:51–62 [DOI] [PubMed] [Google Scholar]
- 40. Leibrock J, Lottspeich F, Hohn A, et al. Molecular cloning and expression of brain-derived neurotrophic factor. Nature. 1989;341:149–152 [DOI] [PubMed] [Google Scholar]
- 41. Mizuno TM, Kleopoulos SP, Bergen HT, Roberts JL, Priest CA, Mobbs CV. Hypothalamic pro-opiomelanocortinm RNA is reduced by fasting and [corrected] in ob/ob and db/db mice, but is stimulated by leptin. Diabetes. 1998;47:294–297 [DOI] [PubMed] [Google Scholar]
- 42. Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443(7109):289–295 [DOI] [PubMed] [Google Scholar]
- 43. Wang J, Li X, Ke Y, et al. GPR48 increases mineralocorticoid receptor gene expression. J Am Soc Nephrol. 2012;23:281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]