Abstract
A crucial step in the development of the vertebrate visual system is the branching of retinal ganglion cell (RGC) axons within their target, the superior colliculus/tectum. A major player in this process is the neurotrophin brain-derived neurotrophic factor (BDNF). However, the molecular basis for the signaling pathways mediating BDNF action is less well understood. As BDNF exerts some of its functions by controlling the expression of microRNAs (miRNAs), we investigated whether miRNAs are also involved in BDNF-mediated retinal axon branching. Here, we demonstrate that the expression pattern of miRNA-132 in the retina is consistent with its involvement in this process, and that BDNF induces the upregulation of miRNA-132 in retinal cultures. Furthermore, in vitro gain-of-function and loss-of-function approaches in retinal cultures reveal that miRNA-132 mediates axon branching downstream of BDNF. A known target of miRNA-132 is the Rho family GTPase-activating protein, p250GAP. We find that p250GAP is expressed in RGC axons and mediates the effects of miRNA-132 in BDNF-induced branching. BDNF treatment or overexpression of miRNA-132 leads to a reduction in p250GAP protein levels in retinal cultures, whereas the overexpression of p250GAP abolishes BDNF-induced branching. Finally, we used a loss-of-function approach to show that miRNA-132 affects the maturation of RGC termination zones in the mouse superior colliculus in vivo, while their topographic targeting remains intact. Together, our data indicate that BDNF promotes RGC axon branching during retinocollicular/tectal map formation via upregulation of miRNA-132, which in turn downregulates p250GAP.
Keywords: axon branching, axon guidance, BDNF, miRNA, p250GAP, retinotectal projection
Introduction
The visual projection represents a long-standing model system for studying the mechanisms and molecules involved in controlling axon branching and neural circuit formation. In higher vertebrates like mouse and chick, retinal ganglion cell (RGC) axons initially grow into the superior colliculus (SC) or tectum (T) in a nontopographic manner. Topographic map formation is then initiated through interstitial axon branching at and around the future termination zone (TZ). Initially, this process is rather imprecise leading to a diffuse TZ; but further extensive arborization and the removal of topographically inappropriate branches eventually leads to the formation of a mature, highly focused TZ (Feldheim and O'Leary, 2010; Suetterlin et al., 2012). Interstitial branching of RGC axons in the SC/T is promoted by brain-derived neurotrophic factor (BDNF)/tyrosine-related kinase B (TrkB) signaling, which does not, however, affect their topographic targeting (Feldheim and O'Leary, 2010; Suetterlin et al., 2012).
BDNF exerts multiple effects on neurons, in part by engaging microRNAs (miRNAs) to modulate gene expression (Numakawa et al., 2011). miRNAs are small noncoding RNAs orchestrating the expression of numerous genes by translational repression and/or degradation of mRNAs (Siegel et al., 2011; McNeill and Van Vactor, 2012; Yates et al., 2013). miRNAs are involved in virtually all processes of neural development; however, there are only a few reports identifying specific miRNAs controlling axon morphology such as axon branching in vertebrates (Pinter and Hindges, 2010; Sanuki et al., 2011; Baudet et al., 2012; Dajas-Bailador et al., 2012; Franke et al., 2012).
Here, we test the idea that BDNF promotes RGC axon branching by controlling miRNA expression. miRNA-132 is a good candidate to play such a role. It is a CREB-regulated miRNA that is induced by BDNF/TrkB signaling and neural activity (Cheng et al., 2007; Wayman et al., 2008; Remenyi et al., 2010; Wibrand et al., 2012). It modulates neurodevelopmental processes ranging from postnatal neurogenesis (Pathania et al., 2012) to neurite outgrowth (Vo et al., 2005) and spine morphogenesis (Edbauer et al., 2010; Magill et al., 2010; Tognini et al., 2011; Pathania et al., 2012; Wanet et al., 2012; for review, see Tognini and Pizzorusso, 2012). In the visual cortex, miRNA-132 is required for plasticity during the critical phase of ocular dominance columns formation (Mellios et al., 2011; Tognini and Pizzorusso, 2012).
One of the few experimentally validated target mRNAs of miRNA-132 encodes p250GAP (Vo et al., 2005; Wayman et al., 2008; Impey et al., 2010; Cambronne et al., 2012), a highly conserved GTPase activating protein that suppresses Rac function. Intriguingly, Rac has been shown to promote axon branching (Lundquist, 2003), suggesting that p250GAP might act as a negative regulator of branching.
In the present study, we show that BDNF promotes axon branching in primary neuronal cultures via an upregulation of miRNA-132 expression, which in turn downregulates p250GAP protein levels. We show that p250GAP acts as a suppressor of RGC axon branching. Importantly, in vivo knockdown of miRNA-132 demonstrates that this miRNA is also required for the formation of highly branched RGC axon terminals in the mouse SC.
Materials and Methods
Materials
Laminin was from Invitrogen, poly-d-lysine (for mouse cultures), and poly-l-lysine (for chick cultures) from Sigma. Tissue culture media and supplements were from Invitrogen. TRITC- or FITC-conjugated phalloidin was from Sigma. The anti-p250GAP antibody (i.e., anti-ArhGAP32, rabbit polyclonal antibody) and the anti-tubulin antibody were from Sigma.
miRNA and miRNA sponge vectors.
The miRNA expression and miRNA sponge vectors used are described in detail by Pathania et al. (2012). In brief, for miRNA overexpression, a mature mouse miRNA-132 sequence was inserted downstream of the U6 RNA polymerase promoter into a vector coexpressing RFP downstream of a CAG promoter based on Liu et al. (2010). A scrambled sequence was used to generate the control noncoding vector (Barkho et al., 2008).
For sponge experiments (McNeill and Van Vactor, 2012), imperfect miRNA-132 binding sites were repeated up to 20 times and cloned in the 3′-UTR of GFP. Cloning of the control sponge vector followed the same principle, with the only difference being that a scrambled binding site, not known to correspond to any known miRNA, was used (Pathania et al., 2012).
The sensor plasmid (see Fig. 3) contains only two miRNA-132 binding sites in the 3′-UTR of an RFP expression cassette (Pathania et al., 2012).
Figure 3.
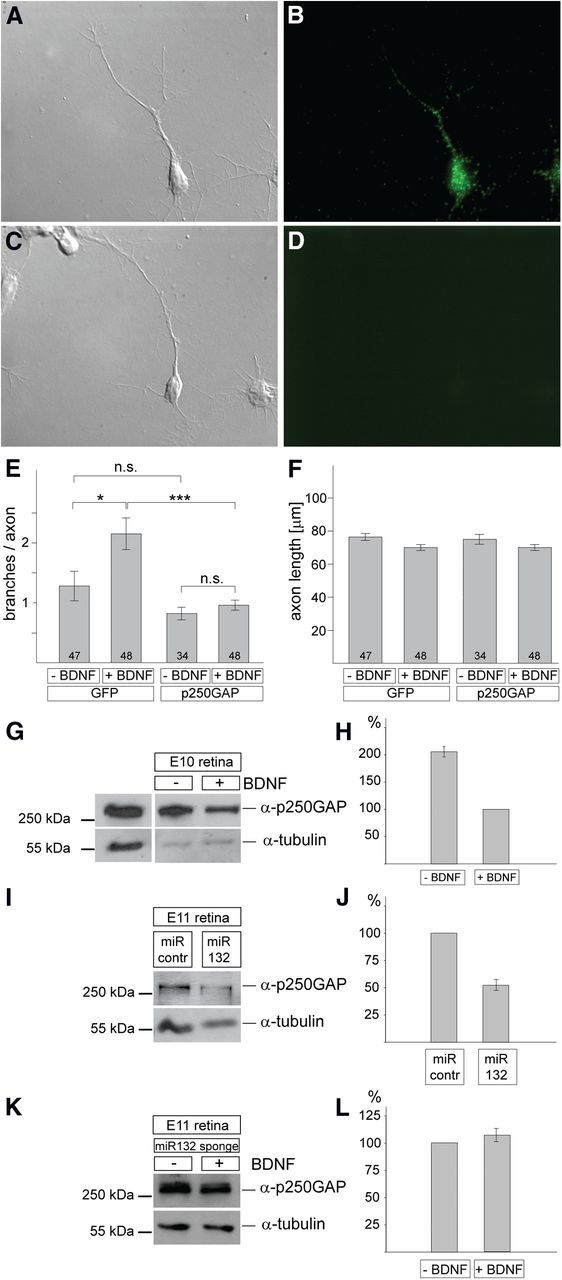
Analysis of p250GAP in chick retinal cultures. A, B, E10 chick RGC axons were stained with an anti-p250GAP antibody (B), and the corresponding differential interference contrast (DIC) picture is shown in A. C, D, A control staining omitting the anti-p250GAP antibody is shown in D with the corresponding DIC picture in C. E, Quantification of the number of branches of RGC axons after expression of p250GAP or GFP in the presence or absence of BDNF. The total number of axons analyzed for each condition is given. Overexpression of p250GAP abolishes any increase in BDNF-mediated branching. F, There is no statistically significant difference in the axon length under the various conditions. E, F, Error bars denote SEM. Significance is indicated as ***p < 0.001, *p < 0.05. n.s., Not significant. Statistical tests were performed with both Student's t test and Mann–Whitney. G, Single-cell preparations from chick E8 retina were incubated for 2 d in the presence or absence of 5 ng/ml BDNF, lysed, and analyzed for p250GAP expression by Western blot. For normalization, lysates were analyzed with an anti-α-tubulin antibody. Quantification is shown in H. The left lane shows an analysis of lysates from CHO cells transfected with the p250GAP expression plasmid to demonstrate the specificity of the antibody and the expected size of p250GAP. H, p250GAP expression is approximately twofold higher in the absence versus the presence of BDNF (average ± SEM, 2.06 ± 0.1) based on densitometric quantification of Western blots from three experiments. I, J, Single-cell preparations from E8 chick retinae were transfected with miRNA (miR)-132 or control miRNA expression plasmids and incubated for 3 d without adding BDNF. Lysates were subjected to Western blot analysis using anti-p250GAP antibodies and as a loading control anti-α-tubulin. P250GAP expression is reduced by miRNA-132 expression to 52.9 ± 9.3% relative to control miRNA expression (100%) based on densitometric quantification of Western blots from three experiments. K, L, Single-cell preparations from E8 chick retinae were transfected with miRNA-132 sponge plasmids (Fig. 2D–F) and incubated for 3 d with or without adding BDNF. Lysates were subjected to Western blot analysis using anti-p250GAP antibodies and as a loading control anti-α-tubulin. There was no statistically significant difference in p250GAP expression in the presence of BDNF (107.8 ± 7.4 relative to the expression of p250GAP in the absence of BDNF (100%) based on densitometric quantification of Western blots from three experiments. contr., Control.
p250GAP expression vector.
The principal p250GAP expression vector was a gift from Stephen Magill and R. Goodman (Oregon Health and Science University, Portland, OR) (Wayman et al., 2008). For identification of electroporated RGCs, we cloned a GFP expression cassette into the NheI site of this vector. Here, both the p250GAP and GFP expression cassettes are driven by the CMV promoter.
Methods
Primary neuronal cultures from mouse and chick retina, and their analysis.
Chick and mouse primary retinal cultures were generated as described previously (Marler et al., 2008). In brief, for mouse cultures, embryonic day 15.5 (E15.5) to E16.5 mouse embryos from timed-mated CD-1 mice were used, with the day of the plug counted as E0.5. Whole retinae from at least 20 embryos were harvested, and a single-retinal cell suspension was prepared. For chick, retinae were isolated from E8 embryos. Retinae were carefully dissociated into a single-cell suspension and finally electroporated using the Amaxa Nucleofector routinely with 5 μg DNA.
To analyze possible changes of p250GAP protein expression levels by miRNA-132, both 5 and 10 μg of DNA were used for nucleofection. Protein levels were investigated using Western blot analysis after 2 or 3 d, as described by Carvalho et al. (2006). Figure 3 shows the results from a set of experiments using 10 μg of DNA after 3 d in culture.
Retinal cells were grown in neurobasal media supplemented with B27 and l-glutamine, in the presence of penicillin/streptomycin, on coverslips coated with 10 μg/ml poly-d-lysine or poly-l-lysine; 5 μg/ml (for chick) or 10 μg/ml (for mouse) laminin; and 1 μg/ml merosin. The day of culture preparation was counted as days in vitro (DIV) 0. BDNF was added at a final concentration of 10 ng/ml on DIV 1, and the same amount was added on DIV 3 for mouse cultures; for chick cultures, 5 ng/ml BDNF was used. Cells were fixed and stained 4 h after the second BDNF addition. MicroRNA plasmids contain an RFP expression cassette, and sponge plasmids contain a GFP expression cassette (Pathania et al., 2012); thus, only RGC axons that were positive for RFP (in miRNA-132/control miRNA experiments) or GFP (in sponge/control sponge experiments) were analyzed with regard to axon length and number of branches. RGCs were identified by staining for markers expressed on RGCs such as TrkB, Brn3A, and Thy-1. Those cells with the longest axons were positive for these markers, and axons and dendrites could be clearly identified (Fig. 1B). For quantification, images were initially obtained with a 63× objective on a Zeiss M200 microscope, composite pictures were generated in Adobe Photoshop, and the length of axons and branches was determined using the NeuronJ plugin in ImageJ or MetaMorph. Only branches >5 μm were included. Data were derived from at least three independently performed experiments with n > 15 axons for each condition and were analyzed blind with regard to treatment received.
Figure 1.
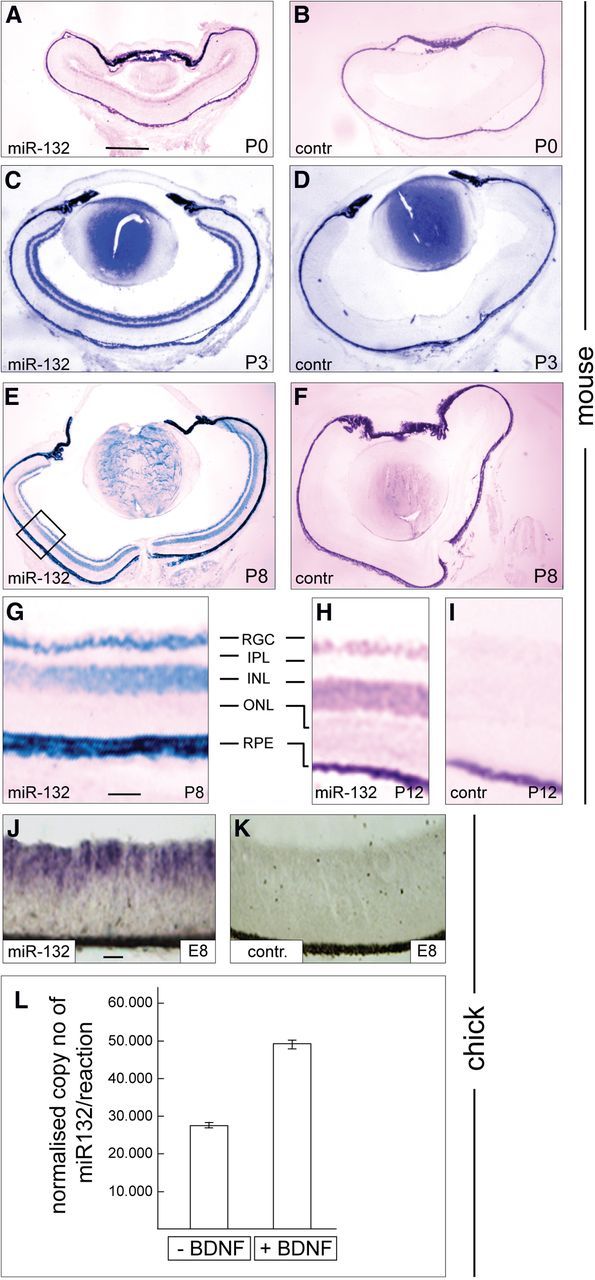
Expression of miRNA-132 in the mouse and chick retina during development of the retinotectal/collicular projection. A–I, miRNA-132 is expressed in the mouse RGC layer during development of the retinocollicular projection. Horizontal sections from mouse retinae were analyzed by RNA in situ hybridization. A, C, E, G, and H, At P0, P3, P8, and P12, miRNA-132 is expressed in the RGC layer, and from P3 onward in the INL. B, D, F, I, Using a control in situ probe (scrambled sequence of miRNA-132), at times indicated only the retinal pigment epithelium shows unspecific staining. G, Enlargement of the area marked in E to further illustrate the expression of miRNA-132 in the RGC layer. H, I, For P12 only part of the retina is shown; layers are labeled as in G. J, K, miRNA-132 expression in the E8 chick retina (horizontal sections). J, K, Expression of miRNA-132 in the inner retinal layers including the RGC layer (J), and the scrambled in situ probe does not lead to any specific signal (K). L, E8 chick retinal cultures were incubated with or without 5 ng/ml BDNF, and 24 h later RNA was isolated, reverse transcribed, and the amount of mature miRNA-132 was determined by quantitative PCR. For normalization, stable reference miRNAs were identified using geNorm software (see Materials and Methods). Data shown are the sum of five independent experiments. Error bars indicate SEM. IPL, Inner plexiform layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. Scale bars: A–F, 132 μm; G–I, 20 μm; J, K, 20 μm. contr., Control.
miRNA in situ hybridization. In situ hybridization was performed using LNA DIG-labeled miRCURY detection probes following protocols described by Karali et al. (2010) and Obernosterer et al. (2007). Sequences of the probes are CGACCATGGCTGTAGACTGTTA for miRNA-132, and GTGTAACACGTCTATACGCCCA for the scrambled negative control; both were supplied by Exiqon. In brief, cryosections were postfixed for 30 min in 4% PFA, acylated, and treated with 1 μg/ml Proteinase K for 5 min; washed extensively; and hybridized overnight at 40°C with a probe at a final concentration of 1 pm. Then slides were washed in 2× in SSC, 2× SSC/50% formamide, and PBS/T, and were incubated overnight with 1:2000 diluted anti-DIG-AP fragments. Then slides were washed five to seven times for 5 min each, and developed in NBT/BCIP; once the signal has developed, they were fixed and mounted with Dako medium.
Quantification of miRNA-132 levels after BDNF treatment.
Retinal cultures were prepared as described above from E8 chick embryos. After 16 h, 5 ng/ml BDNF was applied, and 24 h later cells were lysed. RNA was isolated including the small RNA fraction (miRNeasy, Qiagen) and analyzed using a qStandard/Exiqon protocol (www.qstandard.co.uk) with reverse transcription followed by real-time qPCR. RNA from five independent experiments was analyzed, while three stable reference miRNAs (let-7e, miRNA-124, and miRNA-223) of a pool of five candidate miRNAs were identified using geNorm software (http://genomebiology.com/2002/3/7/research/0034/) and the normalization factor for each sample was used to normalize expression values of mature miRNA-132.
Characterization of miRNA-132 sponges in primary retinal cultures.
To confirm that miRNA-132 sponges also function in retinal cells, E8 chick retinal cultures were coelectroporated with 2.5 μg of the GFP cassette containing sponge plasmids (either miRNA-132 or control) and 0.25 μg of a sensor plasmid with an RFP expression cassette containing only two miRNA-132 binding sites in the 3′-UTR of RFP. BDNF was added to the cultures on DIV1 to a final concentration of 5 ng/ml, and cultures were fixed 24 h later. Cultures were imaged using an Eclipse Ti-E inverted microscope with a 10× objective. RFP fluorescence intensity was measured using ImageJ, and results were normalized against GFP fluorescence to account for differences in cell density and transfection efficiency.
In utero electroporation.
In utero electroporation of mouse embryos essentially was performed as described by Garcia-Frigola et al. (2007). Timed-pregnant C57BL/6J females were anesthetized with a mix of oxygen-isoflurane before the abdomen was opened and the uterine horns exposed. The DNA solution (1 μg/μl plus 0.03% fast green in PBS) was injected into one eye of each embryo using a graduated pulled-glass micropipette. Five square electric pulses (35 V, 50 ms) were passed at 1 s intervals using an CUY21EDIT electroporator (NEPA GENE).
Quantification of electroporation areas and termination zones.
The developing TZs were analyzed after “scaling,” a tissue-clearing protocol that involves incubating the SC for at least 3 d in a buffer containing 4 m urea, which strikingly increases the transparency of the tissue (Hama et al., 2011). After this treatment, a full-depth confocal analysis of developing TZs (∼150 μm; 100 optical sections) could be achieved.
Three-dimensional confocal stacks of retinal and superior colliculus mounts were preprocessed by median filtering and downscaling by a factor of 2 to improve the signal-to-noise ratio. For images of the retina, all saturated voxels (predominantly cell bodies) were removed, and maximum intensity projections (MIPs) were created. For images of the superior colliculus, a mean intensity projection rather than an MIP image was calculated as it preserves axonal arbor information. Additionally, the mean superior colliculus images were denoised by estimating the noise within each mean image (fitted Gaussian distribution to the dominant distribution within the cumulative histogram of all voxels); the high-pass cutoff threshold was set to the peak center plus 1.5× the bandwidth of the Gaussian distribution. Both retinal and superior colliculus images were then binarized, and 2D cumulative histograms were created to generate percentile maps of the cumulative distributions of nonzero voxels. Images were divided evenly into predefined areas (32 × 32 μm2), and the incidence of nonzero voxels per unit area were determined. Interpolating these 2D histograms back to the image resolution (nearest neighbor), and a Gaussian smoothing filter was applied (kernel twice the linear dimension of the predefined volume). Ratios of the areas were used for subsequent statistical analysis: the area between the 90th and 50th percentiles/area of the 50th percentile. An independent-samples t test examined the differences between groups with unequal variances (Levene's test for equality of variances: retina, F = 1.4, p = 0.3; superior colliculus, F = 1.72, p = 0.2).
We verified, by analyzing sections of miRNA-132 sponge or control sponge electroporated retinae, that the percentage of electroporated RGCs in relation to the total number of electroporated cells in the retina was constant, and that the overall development of the retina was not affected by sponge expression (see Fig. 4F–J). We therefore included in our analysis the electroporated area as a whole without discriminating between RGC and non-RGC electroporated cells.
Figure 4.

Expression of a miRNA-132 sponge affects the development of termination zones in the mouse superior colliculus in vivo. A–D, Control sponge (A, B) and miRNA-132 sponge (C, D) plasmids containing a GFP expression cassette were locally electroporated into the retina at E13.5 and analyzed at P3 by confocal microscopy. A–D represent merged stacks showing parts of retina (A, C) and SC (B, D). Scale bar, 200 μm. E, Quantification of the density maps for retina and SC. We characterized the TZs by measuring the ratios between the areas of the 90th to 50th percentile and the area covering the 50th percentile in both the retina and in the SC (for details of how the density maps were calculated, see Materials and Methods and Results). The density maps for the electroporated areas in the retina do not differ statistically significantly for control sponge and miRNA-132 sponge (p = 0.468); however, there is a statistically significant difference for the TZ density distribution between control sponge and miRNA-132 sponge. Significance is indicated as * with p < 0.05. Topographic axes are indicated as follows: A, anterior; P, posterior; T, temporal; and N, nasal. The dashed line indicates the optic nerve head. F–J, Analysis of retinal morphology after electroporation of sponge-expressing plasmids. Three retinae electroporated at E13.5 with control-sponges or miRNA-132-sponges were analyzed at P3 for general anatomical structures to demonstrate that sponge expression has no influence on retina development. F, DAPI staining of a cross section of P3 retina. G, The same retina under GFP illumination highlighting areas electroporated with the miRNA-132 sponge plasmid (carrying a GFP expression cassette). H, Merged image of F and G. I, Enlargement of the area shown in H to illustrate the cellular layer organization in sponge-electroporated and nonelectroporated areas. J, A quantification of the thickness of the neuroblast layer (NBL) and ganglion cell layer (GCL) shows no significant differences between wild-type (WT) areas and areas electroporated (EP) with either the control sponge or the miRNA-132 sponge (n = 3; error bars indicate SD).
Results
Expression pattern of miRNA-132 during development of the retinocollicular projection
BDNF is expressed during retinotectal/collicular development in the SC/T and promotes axon branching of RGCs (Cohen-Cory and Lom, 2004). Since a number of reports indicate that BDNF exerts some of its functions by regulating miRNA-132, we investigated whether miRNA-132 is expressed in the appropriate spatiotemporal pattern to be involved in controlling axon branching during map formation in the mouse. Using RNA in situ hybridization experiments, we confirmed that miRNA-132 is uniformly expressed in the mouse RGC layer between postnatal day 0 (P0) and P12—the developmental phase during which RGC axons invade the SC, branch, and ultimately form TZs here (Fig. 1A–I). miRNA-132 is also expressed in the inner nuclear layer from P3 onward (Fig. 1C–I). Similarly, miRNA-132 is expressed in the chick retinal ganglion cell layer at E8 (Fig. 1J,K).
BDNF upregulates miRNA-132 expression in the retina
Next, we investigated whether BDNF affects miRNA-132 expression in the retinocollicular/tectal projection. We performed these experiments in dissociated cultures from chick E8 retinae, which were incubated with or without 5 ng/ml BDNF for 24 h, after which RNA was isolated and reverse transcribed. Changes in miRNA-132 expression level were determined by real time quantitative PCR using a series of stably expressed miRNAs for normalization (see Materials and Methods). Our data show that BDNF treatment for 24 h increases mature miRNA-132 levels approximately twofold, compared with untreated control cultures (Fig. 1L).
Overexpression of miRNA-132 results in an increase in axonal branching of mouse RGCs in vitro
Next, we investigated whether miRNA-132 is involved in controlling RGC axon branching (Fig. 2A–E). Since mouse primary retinal cultures derived from early postnatal days (during which RGC axons develop branches in vivo) grow poorly, we used cultures derived from embryonic ages when more robust axon outgrowth can be achieved.
Figure 2.

miRNA (miR)-132 controls axon branching of mouse RGCs. A, Dissociated retinal cultures from E15–E16 mouse embryos were electroporated with expression plasmids for miRNA-132 or a control (scrambled) miRNA, and were incubated for 3 d in the presence or absence of 10 ng/ml BDNF. The plasmids contain an RFP expression cassette for identification of electroporated cells (Pathania et al., 2012). Typical examples for RGC branching patterns are shown under conditions as indicated. Scale bar, 10 μm. B, Quantification of the number of branches per axon expressing miRNA-132 or an miRNA with a scrambled sequence, in the presence or absence of BDNF. The expression of miRNA-132 leads to an increase in the number of branches even in the absence of BDNF. C, Average axon length was not affected. D, Quantification of the number of branches per axon of retinal cultures expressing control sponge or miRNA-132 sponge in the presence or absence of BDNF. Axons, in which miRNA-132 function is abolished by expression of sponges, do not show an increase in branching in response to BDNF. E, The average axon length was not affected. B–E, Error bars denote SEM. Significance is indicated as ***p < 0.001, *p < 0.05. n.s., Not significant. The number of axons analyzed (from a total of three independent experiments) is shown at the bottom of the bars. Statistical analysis was done in GraphPad using one-way ANOVA and Tukey post hoc test. F, Characterization of the miRNA-132 sponge in chick retinal cultures. Single-cell cultures from E8 chick retinae coexpressing GFP-containing sponge plasmids containing multiple binding sites for either miRNA-132 or a mutated binding site (contr.), together with an RFP-containing sensor with only two binding sites for miRNA-132. Here RFP fluorescence relative to GFP fluorescence is given (see Materials and Methods). The miRNA-132 sponge, but not the control sponge, sequesters miRNA-132 away from the sensor, leading to a stronger RFP fluorescence. Error bars denote SEM. Significance is indicated as ***p < 0.001. A.U., Arbitrary units.
Plasmids expressing miRNA-132 or a control miRNA—as well as an RFP expression cassette to allow identification of successfully electroporated RGCs (Pathania et al., 2012; see Materials and Methods)—were electroporated into single-cell preparations from E15–E16 mouse retinae, which were plated on a laminin/merosin-containing substrate. The cultures were then incubated for 3 DIV in the presence or absence of 10 ng/ml BDNF, after which they were fixed and analyzed (see Materials and Methods). RGC axons and dendrites were clearly distinguishable at this time point (Figs. 2A, 3A–D), and axon length and branching were analyzed for the different conditions (Marler et al., 2008; see Materials and Methods).
RGCs cultured in the absence of BDNF usually show very little branching. However, when we overexpressed miRNA-132 in RGCs, we observed a dramatic increase in axon branching even in the absence of BDNF—an effect not seen with control miRNA (Fig. 2A,B). This increase in branching was comparable to the level of branch induction observed when control miRNA-expressing RGCs were grown in the presence of BDNF (Fig. 2B). Interestingly, transfection of retinal cells with miRNA-132 and treatment with BDNF led to a further increase in axon branching (Fig. 2B), suggesting that BDNF induces axon branching via miRNA-132-dependent but also via miRNA-132-independent pathways. The length of RGC axons was similar under all conditions (Fig. 2C).
To investigate whether miRNA-132 plays a conserved role in regulating axon branching, we then analyzed its function in chick retinal cultures. In the chick, topographically relevant interstitial axon branching in the tectum occurs between E10 and E12 (Mey and Thanos, 1992). To recapitulate this developmental time segment as well as possible, primary cultures were prepared from retinae at E8, electroporated with the relevant constructs, and analyzed for branching phenotypes after 3 DIV. Our results in chick RGCs mirror those for mouse RGCs; that is, overexpression of miRNA-132 in RGCs leads to increased axon branching (1.3 branches/axon for control miRNA vs 2.2 branches/axon for miRNA-132; p < 0.05), while axon length was not significantly affected (data pooled from three independent experiments with n > 10 axons for each condition).
Together, we conclude that the role of miRNA-132 in RGC axon branching is conserved across higher vertebrates.
Loss of miRNA-132 function results in a decrease in axonal branching of RGCs in vitro
To validate these data, we used a loss-of-function approach using “miRNA sponges” (Fig. 2D,E). Sponges are plasmids containing multiple binding/target sites for a particular miRNA with the idea that their expression sequesters these miRNAs away from their endogenous target mRNAs, and in this way relieves their translational repression (McNeill and Van Vactor, 2012). We used plasmids containing multiple copies of either the miRNA-132 binding site (called here the “miRNA-132 sponge”) or a mutated miRNA-132 binding site (called the “control sponge”; Pathania et al., 2012).
Initially, we confirmed that the miRNA-132 sponge can interfere with endogenous miRNA-132 function in primary retinal cultures, as shown previously for the Neuro2A cell line (Pathania et al., 2012). We cotransfected the GFP-containing sponges (miRNA-132 specific or control miRNA) together with an RFP-containing “miRNA-132 sensor” that harbors only two miRNA-132 binding sites instead of the >20 sites contained in the sponge constructs (Pathania et al., 2012). A quantification 2 d later showed that the sensor was found to be more highly expressed when coexpressed with the miRNA-132 sponge (as measured by RFP expression relative to GFP expression; see Materials and Methods) compared with coexpression with the control sponge. This indicates that the miRNA-132 sponge but not the control sponge sequesters endogenous miRNA-132 in RGCs, thereby alleviating the repression of miRNA-132 targets (in this case the sensor; Fig. 2F).
To investigate the effects of blocking miRNA-132 function on axon branching, these sponge plasmids were transfected into E15–E16 mouse RGCs, and cultures were analyzed in the same way as described above. We found that after transfection of the miRNA-132 sponge into RGCs, BDNF application no longer resulted in an increase in RGC axon branching, indicating that the branch-promoting effect of BDNF had been abolished (Fig. 2D). Expression of a control sponge had no effect on the increase in axon branching elicited by BDNF application (Fig. 2D). Again, there was no significant difference in axon length between the miRNA-132 sponge and control sponge expressing RGCs (Fig. 2E). These results suggest that by blocking miRNA-132 function the signaling pathway from BDNF/TrkB activation to axon branching is disrupted.
Together, our gain-of-function and loss-of-function approaches point to a role for miRNA-132 in a branch-promoting signaling axis downstream of BDNF/TrkB. Given the mode of action of miRNAs, we hypothesized that miRNA-132 exerts its branch-inducing activity in RGCs by targeting, thereby negatively regulating a repressor of axon branching.
p250GAP expression downregulates axonal branching of RGC axons
To identify mRNA targets involved in mediating the effect of miRNA-132 in axon branching, we focused our analysis on the expression pattern and function of p250GAP in RGCs (Fig. 3). p250GAP is a confirmed miRNA-132 target (Vo et al., 2005; Wayman et al., 2008; Impey et al., 2010) and a negative regulator of the GTPase Rac, which in turn promotes axon branching (Reichardt, 2006). All p250GAP mRNAs that have been identified in different species to date, including the chick and mouse, share a highly conserved binding site for miRNA-132 in their 3′-UTR; (Vo et al., 2005).
An immunocytochemical analysis showed the expression of endogenous p250GAP protein in the cell body of RGCs as well as clear axonal expression (Fig. 3A,B). To verify the specificity of the antibody, CHO cells were transfected with an expression plasmid for p250GAP and cell lysates subjected to Western blot analysis. Transfected cell lysates showed the presence of an ∼250 kDa protein when probed with the antibody against p250GAP (Fig. 3G, left).
We then overexpressed p250GAP in chick retinal cultures and analyzed its effect on RGC axon branching (Fig. 3E,F). In control experiments in which a GFP construct was transfected, BDNF treatment for 3 d led to a statistically significant increase in RGC axon branching compared with control cultures without BDNF (Fig. 3E). In contrast, RGCs transfected with the p250GAP plasmid (containing a GFP expression cassette) and treated with BDNF showed no increase in branching (Fig. 3E). Again, we did not observe any effect on axon length (Fig. 3F). For p250GAP-expressing neurons, grown both with and without BDNF, the number of branches was not significantly different from GFP-transfected neurons grown without BDNF (Fig. 3E).
Thus, p250GAP represses BDNF-mediated RGC axon branching.
Next, we investigated whether BDNF treatment affects the expression of endogenous p250GAP protein (Fig. 3G,H). Using the p250GAP-specific antibody, we analyzed lysates from chick retinal cultures grown for 2 d in the presence or absence of 5 ng/ml BDNF. Results from three experiments show that p250GAP expression is approximately twofold higher in the absence versus the presence of BDNF (Fig. 3H; factor of 2.08 ± 0.1).
Finally, we asked the question whether BDNF-induced downregulation of p250GAP is mediated by miRNA-132. To this end, E8 chick retinal cells were electroporated with miRNA-132 or a control miRNA, and incubated for 3 d in the absence of BDNF before cells were lysed and subjected to Western blot analysis. Figure 3, I and J, shows that miRNA-132 expression results in an ∼50% reduction in p250GAP protein levels (n = 3 experiments; control miRNA, 100%; miRNA-132, 52.9 ± 9.4%).
To further substantiate that BDNF and miRNA-132 act in the same signaling pathway to control p250GAP expression, we analyzed its expression levels after miRNA132 sponge transfection and incubation in the presence or absence of BDNF. The finding that we did not observe statistically significant differences in p250GAP expression levels under these conditions (Fig. 3K,L) can be taken as further evidence that BDNF and miRNA132 are indeed involved in the same pathway to control p250GAP expression.
In sum, these data strongly suggest that BDNF exerts at least some of its branch-promoting effects by upregulating miRNA-132, which in turn downregulates its target and known branching repressor p250GAP.
Effects of miRNA-132 sponge expression on the development of termination zones in vivo
After demonstrating a critical role for miRNA-132 in axon branching in vitro, we went on to investigate whether interfering with miRNA-132 function in vivo similarly affects branch formation of RGCs in the SC (Figs. 4, 5). For this, we introduced the miRNA-132 sponge or the control sponge plasmid (both containing a GFP expression cassette) into the developing E13.5 mouse retina by in utero electroporation (Garcia-Frigola et al., 2007). We then analyzed the corresponding axonal projections within the SC at P3 (Fig. 4A–E), a time point at which axon branching is at its peak, eventually leading to the formation of mature TZs at ∼P8.
Figure 5.
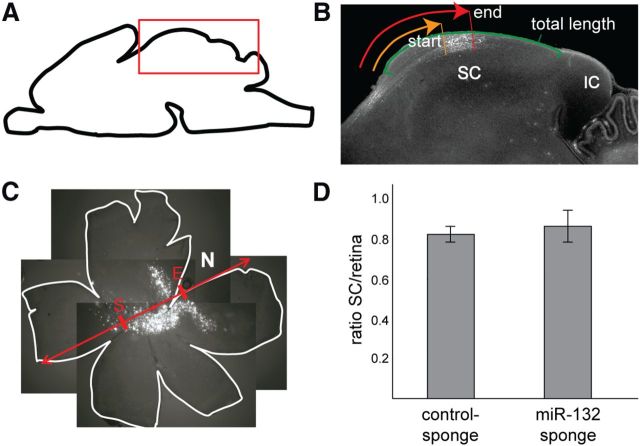
Analysis of topographic mapping of RGC axons after electroporation with miRNA-132 or control sponges. A, Gelatin-embedded brains were cut parasagittally in 100 μm sections, and GFP-positive RGC projections into the SC were analyzed along the anteroposterior axis (A–P). B, The position of the center of the TZ was calculated as the average of the start (S; orange trace) and end (E; red trace) of the TZ, as measured from the anterior border of the SC. To determine its relative topographic position, this value was in turn divided by the total length of the SC (green trace). IC, Inferior colliculus. C, Similarly, to define the electroporated area in the retina, first the retinal diameter was measured along the naso-temporal axis (N–T; red). The position of the center of the electroporated area was defined as the average of the start and end sites of the electroporated area, measured from the temporal border, and this value was in turn divided by the retinal diameter. D, To identify a potential shift in the TZ relative to the retinal position, the position of the center of the TZ in the SC was divided by the position of the center of the electroporated area in the retina. There was no significant difference in the ratios between control sponge group (mean ± SEM, 0.82 ± 0.039) and miRNA-132 sponge group (0.86 ± 0.075; n = 6 control sponge; n = 5 miRNA-132 sponge).
In utero electroporations typically led to the transfection of relatively dense clusters of RGCs (Fig. 4A,C), thus precluding the analysis of individual RGCs and their axonal arbors. However, similar to established methods using lipophilic dyes (such as DiI), we applied a population analysis and determined the characteristics of developing TZs generated by the branching of electroporated RGCs (identifiable by GFP fluorescence). In view of findings that miRNA-132 enhances RGC axon branching in vitro (Figs. 2, 3), we speculated that sponge-mediated inactivation of miRNA-132 would reduce and/or delay the formation of TZs.
Generally, immature TZs are much larger in size compared with mature TZs, and small numbers of branches are scattered at low density at and around the future TZ. Mature TZs are much more focused due to intense arborization at the correct topographic location and due to the removal of branches from topographically incorrect positions (see Introduction; Yates et al., 2001; Hindges et al., 2002). Thus, as a measure of the development of TZs, we determined the ratio between areas of low density (little branching) and areas of high density (dense branching). Accordingly, the higher this ratio, the more immature the termination zone (Fig. 4E).
For this analysis, confocal stacks of both the retina and the SC measuring GFP fluorescence were generated on a Zeiss LSM710 (typically ∼100 optical slices/150 μm depth for the SC), allowing us to determine the extent and depth of the TZs very precisely. From these data, scaled-density maps were created and related to each other (Fig. 4E; for a detailed description, see Materials and Methods). Representative data from the retinae and SCs of animals electroporated with either the miRNA-132- or the control sponge are shown in Figure 4A–D.
We found no difference in the density distribution of the electroporated area in the retina between the two experimental groups, which was expected since the DNA concentration and plasmid size were essentially the same. However, we found a statistically significant difference in the density distribution of TZs in the SC from miRNA-132 sponge-expressing RGCs vs control sponge-expressing RGCs (Fig. 4E). These data indicate that the TZs of RGCs expressing miRNA-132 sponges contain more areas of low density (little branching) than TZs from RGCs expressing the control sponge, suggesting that these TZs are less mature, which may represent either a developmental delay or a more permanent impairment (see Discussion).
Furthermore, we verified that sponge expression had no apparent effect on the development of the retina itself. Quantification of the thickness of various layers (as a measure of its development) showed no statistically significant differences between areas expressing the sponge constructs (miRNA-132 sponge or control sponge) and nonelectroporated areas in the same retinae (Fig. 4F–J).
The topographic projection of RGC axons expressing miRNA-132 sponges is not affected
Finally, we analyzed whether the TZs of miRNA-132 sponge-expressing axons formed at the correct topographic position along the anteroposterior axis (Fig. 5A–D). For this, the position of the TZs was determined on parasagittal sections from the SC (Fig. 5A,B) and correlated with the position of the electroporated area in the retina along the naso-temporal axis (Fig. 5C; see Materials and Methods). Here we used the center of the TZs, calculated as the midpoint between the start and end of the TZ (Fig. 5B; see Materials and Methods).
Our analysis showed that the expression of miRNA-132 sponges or control sponges in RGCs does not affect their topographic targeting (Fig. 5D).
Discussion
Here we show that the neurotrophin BDNF controls axon branching of RGCs (at least in part) via miRNA-132. miRNA-132 expression has a potent effect on branching since a gain-of-function approach increases RGC axon branching, while a loss-of-function approach abolishes the branch-promoting effect of BDNF, suggesting that miRNA-132 expression levels have to be finely balanced. Furthermore, BDNF upregulates miRNA-132, which in turn downregulates p250GAP, allowing branching to occur. Consistent with this, the overexpression of p250GAP in RGCs suppresses BDNF-induced axon branching. This identifies p250GAP as a target of miRNA-132, which mediates the function of BDNF. Moreover, an miRNA-132 loss-of-function approach in vivo disturbs the normal development of TZs, hinting at an axonal branching defect in the absence of miRNA-132 in vivo.
BDNF induces miRNA-132 expression
We demonstrate here that miRNA-132 is expressed in the mouse RGC layer during the developmental phase of axon branching in vivo (P0–P12), indicating that miRNA-132 is expressed in a pattern consistent with exerting a function in axon branching and formation of TZs in the mouse (Fig. 1). Equivalent expression data were also obtained for the chick, raising the possibility that the role of miRNA-132 during map development is more widely conserved in higher vertebrates (Fig. 1). This may not be surprising given the high degree of evolutionary conservation of miRNA-132 and many of its targets (Vo et al., 2005).
Furthermore, BDNF treatment for 24 h increases the concentration of mature miRNA-132 in primary retinal cultures approximately twofold (Fig. 1L), which is in line with other studies characterizing miRNA-132 regulation. Wayman et al. (2008), for example, showed that manipulation of neural activity in hippocampal neurons led to a 2.5-fold upregulation in mature miRNA-132 and a significant increase in the number of dendritic branches. This indicates that even moderate changes in miRNA-132 levels are physiologically relevant and can result in distinct biological responses (Flynt and Lai, 2008; Wayman et al., 2008).
The sustained increase in mature miRNA-132 levels in retinal cultures after 24 h of BDNF treatment (Fig. 1L) fits well with the long-term process of the formation and remodeling of RGC axon branches in the SC/T, which proceeds over a number of days in both chick and mouse (Yates et al., 2001; Hindges et al., 2002). Thus, the BDNF-induced upregulation of miRNA-132 and the corresponding downregulation of its target mRNAs/proteins (see below) appear to confer the necessary long-lasting capacity to develop axon branches.
miRNA-132 controls RGC axon branching
To study the effects of miRNA-132 on axon morphology, we used gain-of-function and loss-of-function approaches in retinal primary cultures. These experiments demonstrate that miRNA-132 expression levels are important for RGC axon branching since their upregulation leads to an increase in branching, while their inactivation abolishes BDNF-mediated branching (Fig. 2). Similar data were obtained for the chick system, where an overexpression of miRNA-132 results in an increased branching of RGC axons (see Results).
Other BDNF-induced miRNAs affecting axon morphology
Numerous miRNAs are controlled by BDNF/TrkB signaling (Numakawa et al., 2011). Of particular interest here is miRNA-9, since it controls, like miRNA-132, the morphology of developing mouse axons. miRNA-9 regulates axon extension and branching of mouse cortical neurons in a BDNF-dependent manner by targeting the microtubule regulatory protein MAP1B (Dajas-Bailador et al., 2012). Thus, the expression of both miRNA-132 and miRNA-9 is controlled by BDNF, and both miRNAs control axon morphology. However, miRNA-132 controls axon branching, whereas miRNA-9 affects both axon length and branching. The Map1B mRNA is not a predicted target of miRNA-132 (http://www.targetscan.org). Interestingly, miRNA-132 and miRNA-9 also act on the same targets, such as the FoxP2 mRNA, encoding a transcription factor associated with speech and language development (Clovis et al., 2012). Thus, miRNA-132 and miRNA-9 have shared and unique functions.
In addition to these BDNF-regulated miRNAs, there are as yet only a few other reports identifying miRNAs affecting axon morphology and guidance (Agostini et al., 2011; Baudet et al., 2012; Zhang et al., 2013). One of these describes the role of miRNA-124, which confers a time-dependent sensitivity to the repellent axon guidance molecule sema3A for Xenopus laevis RGC axons by targeting a cofactor of the neuropilin receptor CoREST (Baudet et al., 2012). Another target of miRNA-124 was identified as RhoG, which inhibits dendritic branching in hippocampal neurons by affecting cdc42, and axonal branching via Rac1 (Franke et al., 2012).
p250GAP suppresses axon branching of RGCs
Owing to the pleiotropic nature of most miRNAs, it is rarely trivial to identify which downstream targets are the key mediators in any given context. Here we demonstrate that a likely candidate to mediate BDNF function in RGCs is p250GAP, which is a well characterized target of miRNA-132 (Vo et al., 2005); crucially, functional characterizations indicate its involvement in controlling neurite morphology (Vo et al., 2005; Wayman et al., 2008; Impey et al., 2010).
Here we show that p250GAP protein is expressed during a crucial developmental period in RGCs and their axons (Fig. 3A–D), and is indeed a downstream target of miRNA-132 and BDNF. Overexpression of miRNA-132 or treatment with BDNF leads to a downregulation of p250GAP protein expression in primary retinal cultures (Fig. 3G–J). In turn, p250GAP negatively controls axon branching, since axons overexpressing p250GAP fail to show BDNF-induced axon branching (Fig. 3E,F).
Thus, our experiments identify p250GAP as a key molecule antagonizing the branch-promoting function of BDNF/miRNA-132 on RGC axons.
Local or global control of axon branching?
In situ RNA hybridization experiments did not reveal miRNA-132 expression in the axons of RGCs, even in the optic nerve head, which contains very high concentrations of RGC axon fibers (Fig. 1E). Though we cannot exclude that very low concentrations of miRNA-132 are transported into RGC axons, it indicates that miRNA-132 in the context described here exerts its function primarily in the cell body; that is globally, and not locally (e.g., in a particular axonal segment).
This would give rise to a model in which BDNF, released from cells in the SC, induces the activation and endocytosis of TrkB in RGC axons, some of which is then retrogradely transported back to the cell body (Reichardt, 2006). Here, TrkB signaling increases miRNA-132 levels and in turn downregulates p250GAP. The reduced protein levels of this branch suppressor in the cell bodies and axons would then allow the execution of the axon branching program.
However, our data also indicate that BDNF controls axon branching independent of miRNA-132 (Fig. 2B) since overexpression of miRNA-132 and treatment with BDNF induce stronger axon branching than miRNA-132 overexpression alone. Interestingly, TrkB can also signal locally, that is, induce axon branching via a local activation of signaling pathways such as the PI-3 kinase/Akt pathway (Markus et al., 2002).
Therefore TrkB may well control both global branching competence and local branch formation using miRNA-132-dependent and -independent pathways, respectively.
Knockdown of miRNA-132 in vivo delays formation of dense termination zones in the SC
A knock-out approach recently showed that inactivation of miRNA-132 dramatically decreases dendritic spine formation of hippocampal neurons, implying that this miRNA has a profound, nonredundant effect on neuronal morphology in vivo (Magill et al., 2010). Encouraged by this finding, we tested whether knockdown of miRNA-132 in vivo affects axon branching patterns of RGCs in the mouse SC. We introduced miRNA-132 sponges and control sponges (containing GFP expression cassettes) into the retina by in utero electroporation, and studied the postnatal development of RGC axon morphology in the SC.
Our analyses showed that the TZs of miRNA-132 sponge expressing axons on average contained larger areas of sparse branching than TZs from control miRNA sponge expressing axons, which appeared much more focused (Fig. 4B,D). TZ development is a multistep process. Following their initial ingrowth, RGC axons exhibit sparse and relatively widespread branching. Over the course of the first postnatal week, RGC axons form progressively more focused and highly branched “mature” TZs. Thus, our data strongly suggest that the TZs of miRNA-132 sponge expressing axons are less mature at the stage analyzed (P3). However, clarification of the underlying nature of this branching phenotype—which may also represent a more permanent impairment of branching—will require the analysis of earlier and later developmental stages of TZ development in our experimental paradigm.
Overall, our model derived from a combination of in vitro and in vivo data suggests that during initial stages of map development RGC axons express high amounts of p250GAP, which suppresses axon branching and blocks the development of TZs. Only later, when RGC axons have grown deep into the SC and are exposed to BDNF, TrkB is activated, a fraction of which is retrogradely transported to the cell body where miRNA-132 is then upregulated. This leads to the overall downregulation of p250GAP in these axons, providing them with the competence to form branches and leading eventually to the formation of mature TZs. Furthermore, our data indicate that local, miRNA-132-independent TrkB signaling triggers the local/topographically specific axon branching in concert with other guidance molecules (Marler et al., 2008). This essentially correlates with our finding that the topographic position of TZs is not affected following knockdown of miRNA-132.
Footnotes
This work was supported by Wellcome Trust program Grant 083205 and project Grant 089611, as well as by a project Grant from the Biotechnology and Biological Sciences Research Council. We thank Stephen Magill and Richard Goodman for providing an expression plasmid for p250GAP; and S. Guthrie for critical reading of this manuscript.
The authors declare no competing financial interests.
References
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkho BZ, Munoz AE, Li X, Li L, Cunningham LA, Zhao X. Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells. 2008;26:3139–3149. doi: 10.1634/stemcells.2008-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2012;15:29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Shen R, Auer PL, Goodman RH. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci U S A. 2012;109:20473–20478. doi: 10.1073/pnas.1218887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RF, Beutler M, Marler KJ, Knöll B, Becker-Barroso E, Heintzmann R, Ng T, Drescher U. Silencing of EphA3 through a cis interaction with ephrinA5. Nat Neurosci. 2006;9:322–330. doi: 10.1038/nn1655. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clovis YM, Enard W, Marinaro F, Huttner WB, De Pietri Tonelli D. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development. 2012;139:3332–3342. doi: 10.1242/dev.078063. [DOI] [PubMed] [Google Scholar]
- Cohen-Cory S, Lom B. Neurotrophic regulation of retinal ganglion cell synaptic connectivity: from axons and dendrites to synapses. Int J Dev Biol. 2004;48:947–956. doi: 10.1387/ijdb.041883sc. [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012 doi: 10.1038/nn.3082. doi: 10.1038/nn.3082. Advance online publication. Retrieved December 9, 2013. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim DA, O'Leary DD. Visual map development: bidirectional signaling, bifunctional guidance molecules, and competition. Cold Spring Harb Perspect Biol. 2010;2:a001768. doi: 10.1101/cshperspect.a001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Otto W, Johannes S, Baumgart J, Nitsch R, Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31:2908–2921. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Frigola C, Carreres MI, Vegar C, Herrera E. Gene delivery into mouse retinal ganglion cells by in utero electroporation. BMC Dev Biol. 2007;7:103. doi: 10.1186/1471-213X-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hindges R, McLaughlin T, Genoud N, Henkemeyer M, O'Leary D. EphB forward signaling controls directional branch extension and arborization required for dorsal-ventral retinotopic mapping. Neuron. 2002;35:475–487. doi: 10.1016/S0896-6273(02)00799-7. [DOI] [PubMed] [Google Scholar]
- Impey S, Davare M, Lesiak A, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, Dollé P, Banfi S. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA. Rac proteins and the control of axon development. Curr Opin Neurobiol. 2003;13:384–390. doi: 10.1016/S0959-4388(03)00071-0. [DOI] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35:65–76. doi: 10.1016/S0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- Marler KJ, Becker-Barroso E, Martínez A, Llovera M, Wentzel C, Poopalasundaram S, Hindges R, Soriano E, Comella J, Drescher U. A TrkB/EphrinA interaction controls retinal axon branching and synaptogenesis. J Neurosci. 2008;28:12700–12712. doi: 10.1523/JNEUROSCI.1915-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, Edbauer D, Sur M. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey J, Thanos S. Development of the visual system of the chick: a review. J Hirnforsch. 1992;33:673–702. [PubMed] [Google Scholar]
- Numakawa T, Richards M, Adachi N, Kishi S, Kunugi H, Hashido K. MicroRNA function and neurotrophin BDNF. Neurochem Int. 2011;59:551–558. doi: 10.1016/j.neuint.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van Vactor D, Bordey A. miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PLoS One. 2012;7:e38174. doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter R, Hindges R. Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. PLoS One. 2010;5:e10021. doi: 10.1371/journal.pone.0010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428:281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Chérasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Suetterlin P, Marler KM, Drescher U. Axonal ephrinA/EphA interactions, and the emergence of order in topographic projections. Semin Cell Dev Biol. 2012;23:1–6. doi: 10.1016/j.semcdb.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Tognini P, Pizzorusso T. MicroRNA212/132 family: molecular transducer of neuronal function and plasticity. Int J Biochem Cell Biol. 2012;44:6–10. doi: 10.1016/j.biocel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat Neurosci. 2011;14:1237–1239. doi: 10.1038/nn.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Pai B, Siripornmongcolchai T, Bittins M, Berentsen B, Ofte ML, Weigel A, Skaftnesmo KO, Bramham CR. MicroRNA regulation of the synaptic plasticity-related gene Arc. PLoS One. 2012;7:e41688. doi: 10.1371/journal.pone.0041688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Yates PA, Roskies AL, McLaughlin T, O'Leary DD. Topographic-specific axon branching controlled by ephrin-as is the critical event in retinotectal map development. J Neurosci. 2001;21:8548–8563. doi: 10.1523/JNEUROSCI.21-21-08548.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ueno Y, Liu XS, Buller B, Wang X, Chopp M, Zhang ZG. The microRNA-17–92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci. 2013;33:6885–6894. doi: 10.1523/JNEUROSCI.5180-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]