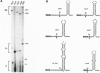Abstract
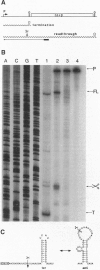
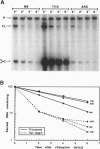
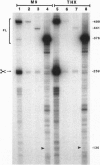
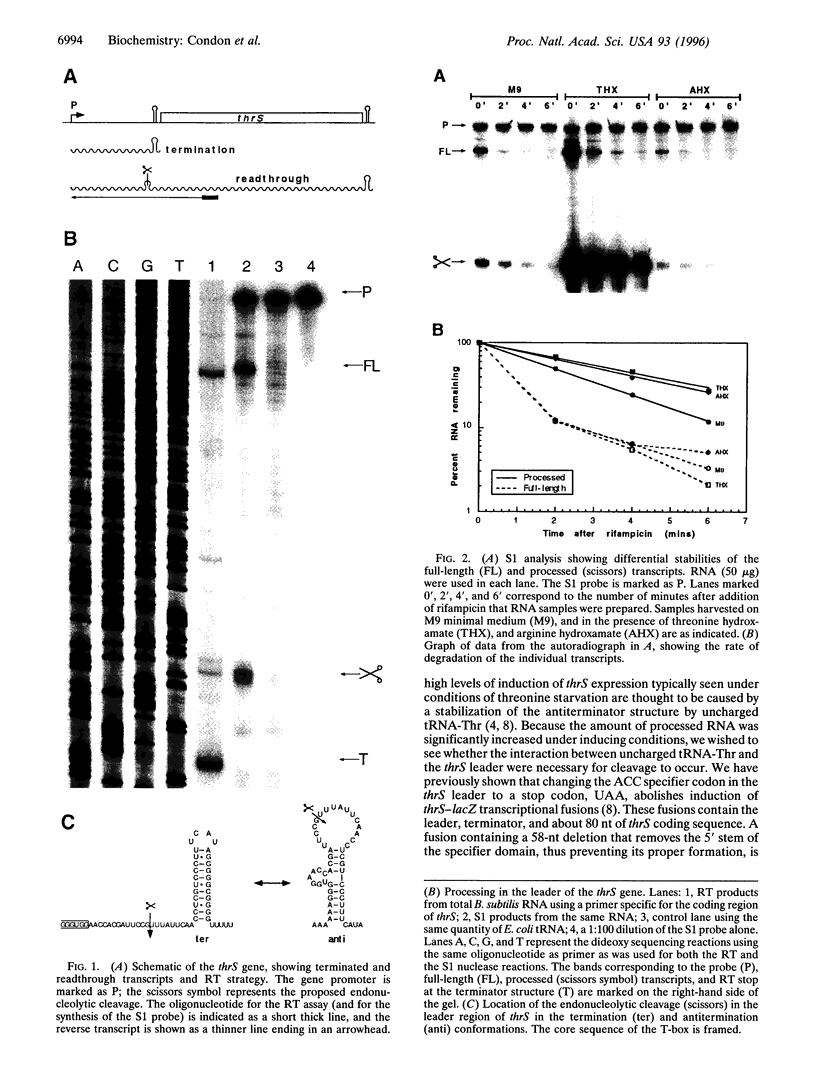
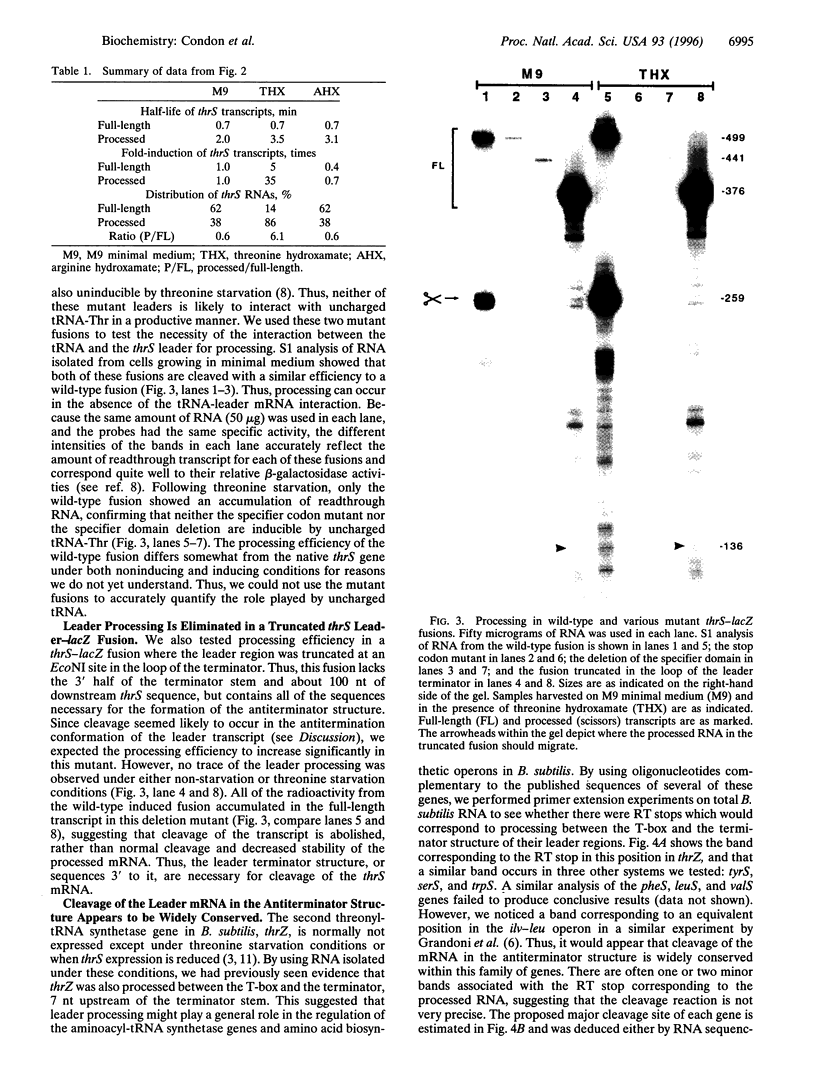
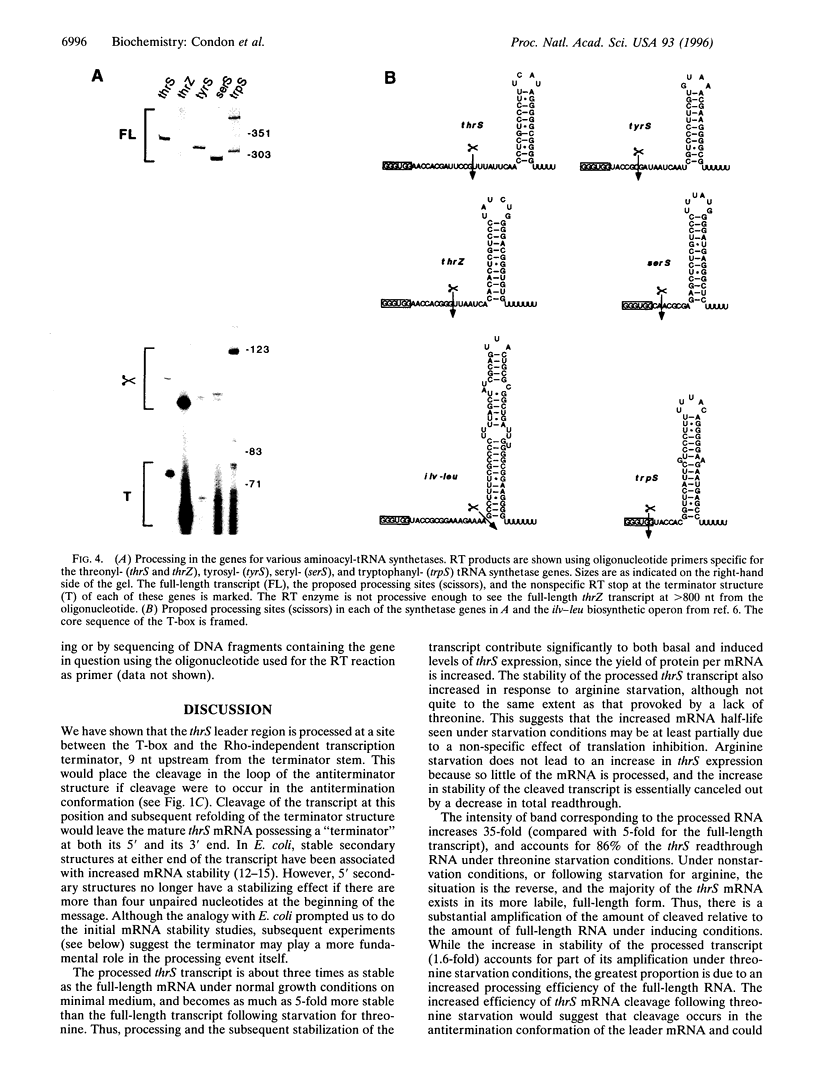
The threonyl-tRNA synthetase gene, thrS, is a member of a family of Gram-positive genes that are induced following starvation for the corresponding amino acid by a transcriptional antitermination mechanism involving the cognate uncharged tRNA. Here we show that an additional level of complexity exists in the control of the thrS gene with the mapping of an mRNA processing site just upstream of the transcription terminator in the thrS leader region. The processed RNA is significantly more stable than the full-length transcript. Under nonstarvation conditions, or following starvation for an amino acid other than threonine, the full-length thrS mRNA is more abundant than the processed transcript. However, following starvation for threonine, the thrS mRNA exists primarily in its cleaved form. This can partly be attributed to an increased processing efficiency following threonine starvation, and partly to a further, nonspecific increase in the stability of the processed transcript under starvation conditions. The increased stability of the processed RNA contributes significantly to the levels of functional RNA observed under threonine starvation conditions, previously attributed solely to antitermination. Finally, we show that processing is likely to occur upstream of the terminator in the leader regions of at least four other genes of this family, suggesting a widespread conservation of this phenomenon in their control.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Beatty J. T., Cohen S. N., Belasco J. G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988 Feb 26;52(4):609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Garrity D. B., Zahler S. A. Mutations in the gene for a tRNA that functions as a regulator of a transcriptional attenuator in Bacillus subtilis. Genetics. 1994 Jul;137(3):627–636. doi: 10.1093/genetics/137.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron N., Putzer H., Grunberg-Manago M. Expression of both Bacillus subtilis threonyl-tRNA synthetase genes is autogenously regulated. J Bacteriol. 1994 Jan;176(2):486–494. doi: 10.1128/jb.176.2.486-494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoni J. A., Zahler S. A., Calvo J. M. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J Bacteriol. 1992 May;174(10):3212–3219. doi: 10.1128/jb.174.10.3212-3219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993 Aug 13;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Henkin T. M. tRNA-directed transcription antitermination. Mol Microbiol. 1994 Aug;13(3):381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H., Brakhage A. A., Grunberg-Manago M. Independent genes for two threonyl-tRNA synthetases in Bacillus subtilis. J Bacteriol. 1990 Aug;172(8):4593–4602. doi: 10.1128/jb.172.8.4593-4602.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H., Gendron N., Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992 Aug;11(8):3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H., Laalami S., Brakhage A. A., Condon C., Grunberg-Manago M. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: induction, repression and growth-rate regulation. Mol Microbiol. 1995 May;16(4):709–718. doi: 10.1111/j.1365-2958.1995.tb02432.x. [DOI] [PubMed] [Google Scholar]