Abstract
As of mid 2013 a Medline search on “cholesterol” yielded over 200,000 hits, reflecting the prominence of this lipid in numerous aspects of animal cell biology and physiology under conditions of health and disease. Aberrations in cholesterol homeostasis underlie both a number of rare genetic disorders and contribute to common sporadic and complex disorders including heart disease, stroke, type II diabetes, and Alzheimer's disease. The corresponding author of this review and his lab stumbled only recently into the sprawling area of cholesterol research when they discovered that the amyloid precursor protein (APP) binds cholesterol, a topic covered by the Hans Neurath Award lecture at the 2013 Protein Society Meeting. Here, we first provide a brief overview of cholesterol-protein interactions and then offer our perspective on how and why binding of cholesterol to APP and its C99 domain (β-CTF) promotes the amyloidogenic pathway, which is closely related to the etiology of Alzheimer's disease.
Keywords: cholesterol, integral membrane proteins, amyloid precursor protein, C99, β-CTF, Alzheimer's disease, GPCRs, receptor, CRAC
“What has it got in its pocketses?”
The Hobbit
Distribution of Cholesterol and Other Sterols in the Tree of Life
Cholesterol is the major sterol lipid present in animal membranes. The plasma membranes of animal cells typically contain in the range 25 to 50 mol% cholesterol, whereas levels in the endoplasmic reticulum and nuclear membranes are in the range of 1 to 10 mol%, which increases to about 10 to 25 mol% in the Golgi.1–8 Most non-animal organisms, including many bacteria contain sterol-like lipids that likely play roles that are to some degree analogous to those of cholesterol in the membranes of higher organisms (see Fig. 1). Recent work has shown, for example, that the hopanoids found in many bacteria9,10 are cholesterol-like in the sense that they can promote the formation of raft-like phase-separated liquid-ordered domains in lipid vesicles.11 This is despite the amusing placement of the polar head group in hopanoids on the opposite end of the molecule from cholesterol. At the same time, very small changes in the structure of cholesterol can profoundly alter its properties. For example, reduction of the alkene moiety of cholesterol to generate coprostanol is sufficient to eliminate its ability to promote the formation of raft-like liquid-ordered phase domains.12–14 The fact that cholesterol is still 18 biochemical steps away from lanosterol (Fig. 1) belies the high degree to which is has been adapted for a variety of special roles in complex animal forms of life.15 It has been proposed that sterols represent an adaptation to the ancient advent of aerobic atmospheric conditions and may have made major contributions to the emergence of eukaryotic life on Earth.15–18 One wonders to what degree the later development of higher animal forms of life was dependent on the evolution of cholesterol.
Figure 1.
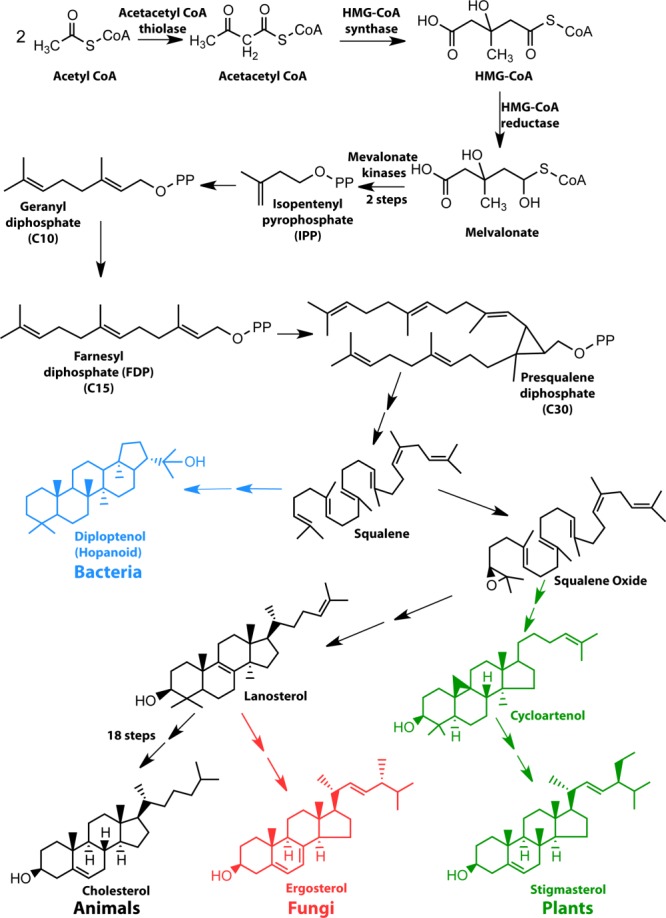
Biosynthesis of sterols in eukaryotes and sterol surrogates in prokaryotes share all steps up to the intermediate, squalene.
Cholesterol Structure
All four rings of the sterol group of cholesterol have trans ring junctions, making it a flat molecule (Fig. 2). One face of the ring system—the α face is smooth. The apposed β face is punctuated by orthogonal C18 and C19 methyl groups. At one end of the sterol ring system is the hydroxyl head group, while at the other end is an isooctyl chain, which is flexible, as illustrated by superimposing structures of cholesterol observed in several different cholesterol-protein complex crystal structures (Fig. 2). Cholesterol's topology is well suited for its integration into lipid bilayers, where it aligns itself with glycerophospholipids and sphinghophospholipids so that its isooctyl tail is near the middle of the bilayer and its 3β-OH group is at the water-membrane interface (Fig. 3). For a lipid, cholesterol's rigidity is unusual, as is the small size and modest polarity of its head group. In its bilayer configuration the cholesterol head group sits “low” in the membrane compared to the charged and more fully water-exposed head groups of phospholipids.19 Indeed, it has been shown that it is not energetically forbidden for cholesterol to spend time with its long axis in plane with the middle of the bilayer.20,21 Paradoxically, the fact that its hydroxyl head group does sit low in the bilayer interface (where the effective dielectric constant is well below that of bulk water) and not only accepts, but also can donate hydrogen bonds, enables cholesterol to participate in relatively strong attractive interactions with other cholesterol molecules and other lipids—especially sphingolipids (which also have low-sitting H-bond donor and acceptor moieties)22 (Fig. 3). Some of these interactions are probably bridged by interfacial water molecules.23
Figure 2.
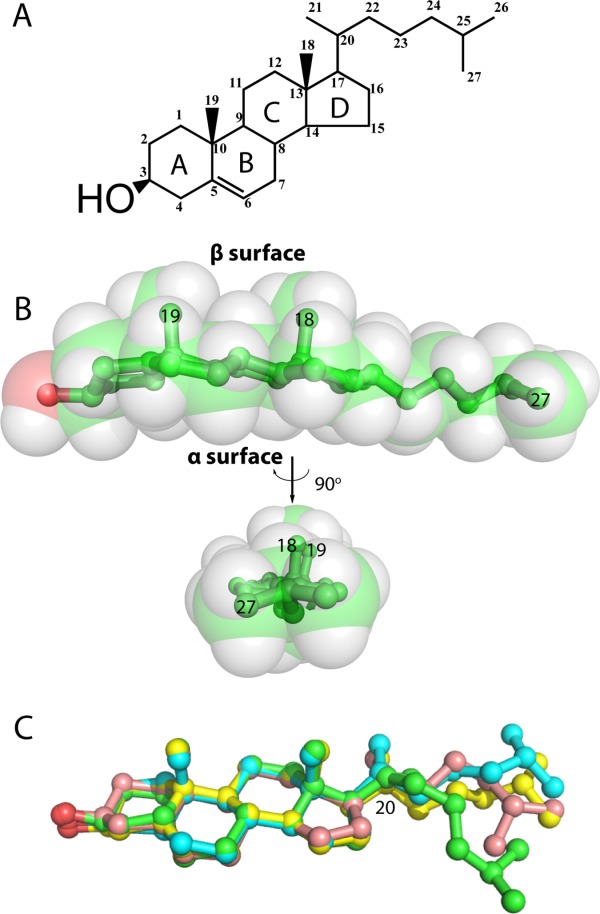
Structure of cholesterol. A) Chemical structure of cholesterol (IUPAC numbering system). B) Space-filling and stick representations of cholesterol. C) Superposition of three cholesterol molecules from different protein crystal structures demonstrates the flexibility of the tail.
Figure 3.
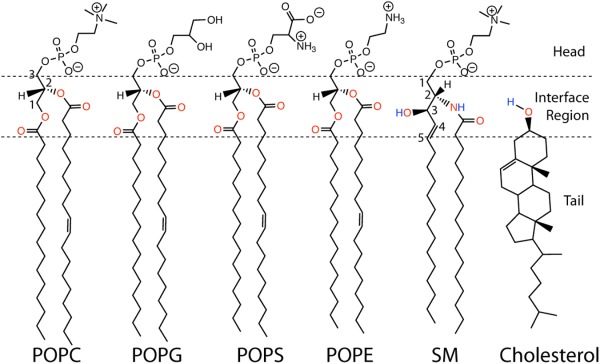
Structures of representative lipids: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphserine (POPS), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), sphingomyelin, and cholesterol. The atoms in the membrane interface region colored in red are possible hydrogen bond acceptors. Atoms in blue are possible hydrogen bond donors.
The overall rigidity of cholesterol suggests that when it interacts with more flexible molecules, such as classical glycerophospholipids, there is an entropic cost arising from cholesterol-induced dampening of motions. This is probably especially the case when the smooth and flat α face of the sterol ring system is involved. This unfavorable entropic effect may provide a driving energy potential for cholesterol to preferentially interact with other rigid molecules or to surfaces on membrane protein transmembrane domains (TMD) that are flat and smooth. It is known that cholesterol also prefers to interact with lipids with saturated acyl chains relative to unsaturated chains (with a particular abhorrence of polyunsaturation).22 This is probably because although both classes of lipid tails suffer an entropy loss upon interacting with cholesterol, the compensating Van der Waals (VDW) energy between cholesterol and extended straight chain alkanes is more favorable than for lipids having chains with one or more cis double bonds.
Cholesterol's Roles in Membrane Fluidity and Raft Formation
The structural properties of cholesterol summarized above lead to two important functions of cholesterol in membranes. First, cholesterol has a high propensity to condense with itself and other lipids, especially sphingolipids and lipids with fully-saturated acyl chains, to form domains that are colloquially referred to as “detergent-resistant membranes” (DRMs) or “lipid rafts”.24–26 These domains retain unique compositions, dynamics, and structural properties despite being surrounded by the fluid (liquid-disordered Ld) phase, although there is exchange of lipids between phases. Raft-like domains are believed to closely resemble the liquid-ordered (Lo) phase seen in bilayered lipid vesicles of certain compositions.27 In particular, while the lipid components undergo rapid lateral diffusion, the chains are extended to enable relatively tight packing between lipids. Moreover, Lo bilayers are thicker than the surrounding Ld phase.28,29 Lo domains in model membrane lipid vesicles can be both large and stable,30–32 and do not necessarily span both leaflets of the membrane.33 In living cells, raft-like domains are much more dynamic and complex, leading to substantial debate about the nature of these domains in a cellular environment.34,–39 The presence of the actin-based cytoskeleton in animal cell plasma membranes dictates that rafts in living cells are often smaller and more transient than observed in model membranes.40 One notable exception is the yeast vacuole membrane, where stable micron-sized coexisting lipid domains can be readily detected.41 Other large and abundant membrane domains are primarily raft-like in composition and physical properties, including caveolae, myelin membranes, eye lens fiber cell plasma membranes, and the apical membranes of polarized epithelial cells. Intriguingly, membrane blebs derived from the plasma membranes of cells demonstrate temperature-dependent spontaneous segregation into domains of differing order and composition, pointing to a fundamental capacity of cell membranes to demix into coexisting phases.42,43 This may reflect the proximity of biological membranes to a critical point.44,45
Raft-like assemblies are cholesterol-rich. In membranes that are phase-separated into fluid (Ld) and raft-like liquid-ordered domains, it is tempting to imagine that all of the cholesterol has partitioned into the Lo domains. However, this is not the case. Cholesterol levels in the fluid phase remain significant (albeit lower than in an adjacent Lo phase) even when there are co-existing raft-like domains.46,47 Fluid phase membranes containing abundant cholesterol retain their essential fluid phase properties: lipids undergo free lateral diffusion, acyl chains remain conformationally flexible, and lipids wobble and bob. However, the presence of cholesterol dampens these various forms of lipid dynamics to confer greater stability to the membrane, while at the same time lowering the gel-to-Ld phase transition to help maintain the bulk membrane in a fluid state at physiological temperatures.
Cholesterol as a Co-Solvent for Membrane Proteins
Membranes act as solvents for the transmembrane domains of membrane proteins. As summarized above cholesterol profoundly impacts the properties of this solvent by altering the physical properties of the bulk (Ld phase) membrane and also by both promoting and participating in formation of an alternative solvent—raft-like membrane domains—which have different physical properties and dimensions than bulk membranes.
Analysis of detergent resistant membranes and ordered phases in giant plasma membrane-derived vesicles suggests raft-like domains include both surface-anchored and, less commonly, integral membrane proteins. While identification of proteins associated with lipid rafts remains a frontier48 it appears that a majority of the proteins that associate with lipid rafts are lipid-anchored proteins.49,50 Palmitoylated proteins associate with the cytosolically-oriented leaflet of rafts while complex glycosylphosphatidylinositide (GPI) anchors associate with the extracellular leaflet. The palmitoyl chain and the lipid chains of GPI anchors are long and usually unsaturated such that, at least under some conditions, they energetically favor the Lo-like nature of rafts relative to the Ld-like properties of bulk membranes. Some non-lipidated integral membrane proteins also appear to prefer lipid rafts to the surrounding bulk membrane.51 Determination of the structural and dynamic factors that control the energetics of partitioning of membrane proteins between raft and bulk membrane domains represents a frontier area in which little is currently known.43,51–58
Modulation of the dynamics of bulk (liquid-disordered) membrane by cholesterol has been well-documented to alter the function of some membrane proteins.59–64 To cite just one example, upon absorption of a photon rhodopsin transitions to a transient equilibrium between signaling-inactive metarhodopsin I (Meta-I) and signaling-active metarhodopsin-II (Meta-II).65,66 This equilibrium is sensitive to the cholesterol concentration of the membrane, with increased cholesterol shifting the equilibrium towards Meta-I. While the mechanisms underpinning this phenomenon may be complex, a contributing factor is thought to be that cholesterol reduces the free volume available for molecular motion in the hydrophobic core of the bilayer. Since formation of Meta-II is accompanied by volume expansion of the rhodopsin TMD, a reduction of the free volume of the bilayer by cholesterol disfavors this state.
Besides impacting membrane protein function, cholesterol's modulation of the dynamics of bulk phase membranes probably also alters both the folding and stability of membrane proteins, as well as the energetics of protein oligomerization. However, so little is known about these issues that one can currently do little more than speculate, particularly since the impact of cholesterol on membrane properties is difficult to disentangle from the consequences of direct interaction of cholesterol with membrane proteins (below). It is a welcome development that methods have finally been developed to quantitate the thermodynamic stability of helical integral membrane proteins in bilayers of varying composition,67,68 which will allow systematic exploration of how varying membrane properties impact membrane protein stability.
The high concentration of cholesterol present in many membranes dictates that in addition to modulating bilayer properties, membrane proteins will be in constant solvent-like contact with cholesterol even if the protein does not form specific complexes with this lipid. This suggests that animal membrane proteins have been evolutionarily adapted to have lipid-exposed surfaces that interact in an optimal manner with cholesterol (among other selective imperatives). However, little is known about what “optimal” represents in terms of cholesterol interactions and to what degree this definition varies from protein to protein. For membrane proteins it has long been known that the “annular” layer of lipids in direct contact with protein transmembrane domains have longer lifetimes in the annular state than is expected based on transient collisions controlled merely by Brownian motion in the quasi-two-dimensional plane of the membrane (Fig. 4).59,63
Figure 4.
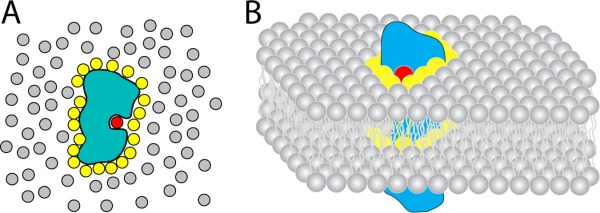
Lipids and water as solvents. A) Cartoon model of a soluble protein in a bath of water. Bulk water molecules are shown in grey; shell water molecules are shown in yellow; a bound water molecule is shown in red. B) Cartoon model of a membrane protein embedded in a membrane bilayer. Bulk lipids are shown in grey; annular lipids are shown in yellow; a bound (non-annular) lipid is shown in red.
Many membrane proteins also include sites for “non-annular interactions” that involve even longer-lived (energetically more favored) interactions of lipids, usually with a cleft in the TMD (such as at the interface between transmembrane helices) and a higher degree of specificity in terms of preferred lipid species (Fig. 4). We suggest that non-annular lipids and their interaction sites on membrane proteins should be thought of as distinct from more classical ligand or substrate binding sites. First, non-annular lipid binding sites are probably almost always occupied by a lipid, unlike the case, for example, of an enzyme that has a lipid as substrate and only approaches saturation of binding when the lipid substrate concentration is much higher than Km. Whether non-annular sites are occupied by their preferred lipid instead of less preferred lipids is determined by the local membrane lipid composition. Secondly, a number of membrane proteins appear to have multiple non-identical non-annular lipid interaction sites per subunit (e.g. Ref.69). This is unlike most cases for membrane proteins that bind a lipid as a substrate for catalysis or transport, where there is usually only one binding site per monomer (or n sites for an n-mer). Nonetheless occupation of non-annular lipid binding sites by preferred lipids may impact membrane protein stability and/or function in ways that are distinct from binding of non-preferred lipids to those same sites. Admittedly, the distinction between non-annular interactions and classical stoichiometric protein-ligand complexes (where the ligand is a lipid) is sometimes a shade of gray.
While it is evident from studies of membrane proteins such as the nicotinic acetylcholine receptor and GPCRs that cholesterol can participate in both annular and non-annular interactions, even in such well-characterized cases little is known regarding why cholesterol is favored or disfavored as a participant in such interactions relative to other lipids. This is compounded by the fact that little is known about the dynamic range of cholesterol concentrations occurring in various cellular membranes—the range of concentrations over which cholesterol varies within a given membrane during the lifetime of a cell. Furthermore, knowing the total cholesterol content of a membrane at a given time point does not provide insight into the distribution of cholesterol on inner and outer leaflets or the distribution between bulk and raft-like membrane domains.70,71 Even when the affinity of cholesterol for a protein is quantitatively known, these factors make it difficult to predict whether the impact of cholesterol on protein structure or function is likely to be constitutive under physiological conditions or whether cholesterol concentrations vary enough for the observed phenomenon to be spatiotemporally regulated by changing local cholesterol concentrations.
Specific Binding of Cholesterol to Membrane Proteins
Many proteins, both soluble and membrane-bound, bind cholesterol to form saturable stoichiometric complexes. These include enzymes that generate cholesterol as a product or employ it as a substrate, transporters that facilitate cholesterol flip-flop across the membrane and/or that deliver membrane cholesterol to soluble proteins (such as lipoproteins) for circulatory transport, and water soluble proteins that transport single cholesterol molecules from membrane to membrane.72–76 Cholesterol may bind to proteins as an allosteric modulator of protein function77–79 and possibly also to alter trafficking or sorting of membrane proteins between different organelles or membrane domains.80–82 Cholesterol also serves as a membrane receptor for certain microbial toxins as a means of recognizing target animal cells, a step that is sometimes followed by co-assembly of toxin-cholesterol complexes in the membrane.83–85 Some viral proteins recognize and exploit cholesterol during infection or budding and related membrane fusion processes.80 A recent proteomic study identified 250 candidate cholesterol binding proteins in HeLa cells.86
How does Cholesterol Bind to Proteins?
To date, 20 high-resolution structures of cholesterol-containing proteins are available in the Protein Data Bank, as summarized in Table I. All except one were determined with X-ray crystallography. These proteins are distributed between water soluble proteins and integral membrane proteins. In the former case, cholesterol is usually bound to a hydrophobic pocket in the interior of the protein [example in Fig. 5(A)]. In the case of membrane proteins, cholesterol is usually seen to be bound to a lipid-exposed hydrophobic face of the transmembrane domain [example in Fig. 5(B)]. We analyzed the occurrence of amino acids within 5 Å of cholesterol in all 19 crystal structures as summarized in Table I and Figure 6. Figure 6(A) reports the occurence of amino acids in all proteins (cyan) and in the 19 proteins of Table I (white). Given cholesterol's hydrophobicity it is no surprise that the hydrophobic residues Ile, Leu, and Phe are the three most highly occurred residues at cholesterol binding sites [Fig. 6(B)].
Table I.
Cholesterol-Containing Protein Crystal Structures in Protein Data Bank as of Mid-2013
| PDB ID | Resol (Å) | Protein | Cholesterol per subunit | Binding site | Protein contacting facea | H-bonds to 3β-OHb | π Interaction with C5=C6?c | Protein function | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1LRI | 1.45 | Cryptogein | 1 | Both | Yes | No | Sterol carrier protein | 87 | |
| 1N83 | 1.63 | Retinoic Acid-related Orphan Receptor alpha | 1 | Both | Yes | No | Nuclear hormone receptors | 88 | |
| 3GKI | 1.80 | Niemann-Pick C1 Protein | 1 | Both | Yes | Yes (F203) | Sterol carrier protein | 89 | |
| 1ZHY | 1.60 | Yeast Osh4 Protein | 1 | Both | Yes | Yes (F42, Y97) | Lipid carrier protein | 90 | |
| 3N9Y | 2.10 | Human CYP11A1 | 1 | Both | Yes | Yes (F82) | Oxidoreductase | 91 | |
| 3A3Y | 2.80 | Na+, K+-ATPase | 1 | Surface | β | Yes | No | Na+, K+-pump | 92 |
| 2ZXE | 2.40 | Na+, K+-ATPase | 1 | Surface | β | Yes | No | Na+, K+-pump | 93 |
| 4HYT | 3.40 | Na+, K+-ATPase | 2 | Surface | β | Yes | Yes (F938, W981) | Na+, K+-pump | 94 |
| 3KDP | 3.50 | Na+, K+-ATPase | 1 | Surface | β | Yes | Yes (Y39) | Na+, K+-pump | 95 |
| 3NYA | 3.16 | β2-Adrenergic Receptor | 2 | Surface | β | Yes | No | GPCR | 96 |
| 3NY9 | 2.84 | β2-Adrenergic Receptor | 2 | Surface | β | Yes | No | GPCR | 96 |
| 3NY8 | 2.84 | β2-Adrenergic Receptor | 2 | Surface | β | Yes | No | GPCR | 96 |
| 2RH1 | 2.40 | β2-Adrenergic Receptor | 3 | Surface | α | Yes | No | GPCR | 97 |
| 3PDS | 3.50 | β2-Adrenergic Receptor | 1 | Surface | α | Yes | Yes (Y70) | GPCR | 98 |
| 3D4S | 2.80 | β2-Adrenergic Receptor | 2 | Surface | β | Yes | No | GPCR | 99 |
| 4IB4 | 2.70 | 5-Hydroxytryptamine Receptor 2B | 1 | Surface | β | Yes | No | GPCR | 100 |
| 4DKL | 2.80 | μ-Type Opioid Receptor | 1 | Surface | β | Yes | Yes (Y299) | GPCR | 101 |
| 4EIY | 1.80 | Adenosine Receptor A2α | 3 | Surface | β | Yes | Yes (F255) | GPCR | 102 |
| 3AM6 | 3.20 | Rhodopsin-2 (Algal Proton Pump) | 2 | Surface | α, βd | Yes | Yes (F33Y) | GPCR-like | 103 |
Given the resolution of many of these structures, we suggest that the modeling of cholesterol into the electron density maps may sometime be ambiguous in terms of assignment of cholesterol orientation as α versus β face (see also99). Thus, the information in this column should be taken with a grain of salt.
A H-bond is assigned based on with a distance of 4 Å between the oxygen atom of 3β-OH in cholesterol and any proximal oxygen or nitrogen atoms. The orientation is not taken into consideration.
A π interaction with the C5=C6 is assumed based on distance and orientation. The aromatic ring has potential to form the π interaction with the C5=C6 double bond if it is within 5Å around and parallel to the C5=C6 double bond.
Two cholesterol molecules exist in the PDB file with one facing the protein with its α surface and the other with β surface.
Figure 5.
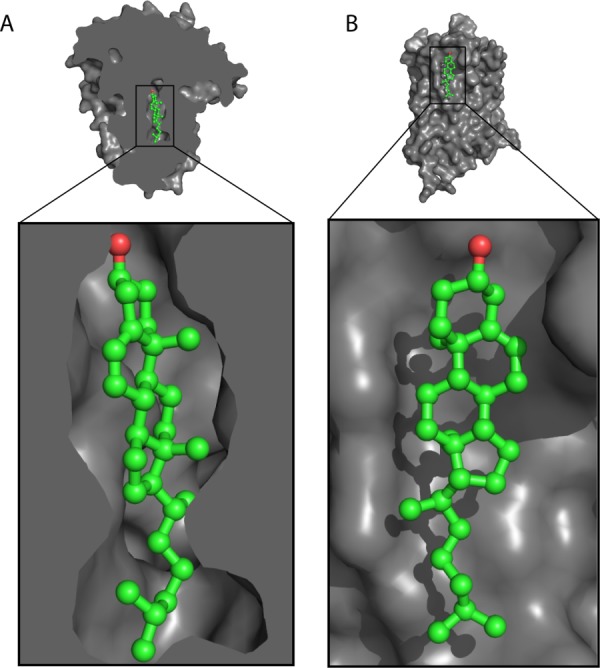
General modes of cholesterol binding to proteins. A) Cholesterol bound to a cavity in the yeast Osh4 protein (PDB ID: 1ZHY90). B) Cholesterol bound to the surface of a μ-type opioid receptor (PDB ID: 4DKL101).
Figure 6.
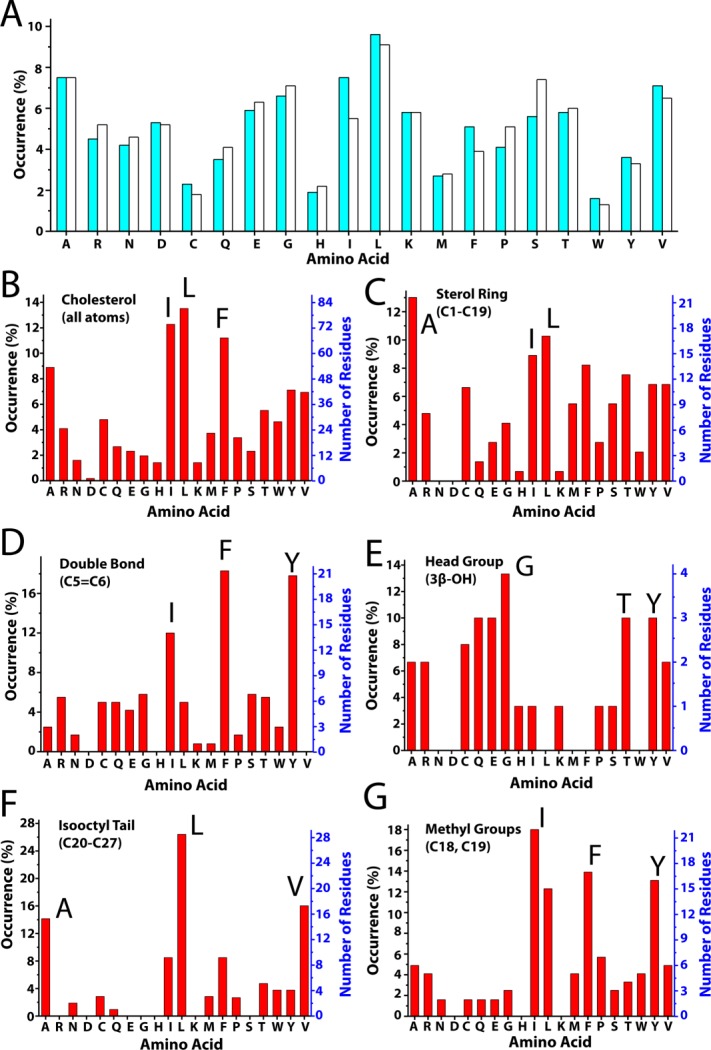
Amino acid composition of structurally-defined cholesterol binding sties. A) Occurrence of amino acids in all proteins (cyan bar) versus in the 19 cholesterol-associated proteins of Table I (white bar). B–G) Occurrence in the PDB of amino acids within 5 Å of different moieties within cholesterol. The y-axis % occurrence (left) and absolute number of observations (right) reflect the number of times that at least one atom from that amino acid (including backbone atoms) is within 5 Å of at least one atom of the indicated substituent moiety of cholesterol.
For integral membrane proteins, cholesterol may bind to a single TM helix [Fig. 7(A)], or can bind to grooves between two or more TM helices [Fig. 7(B–D)]. The most common residues that interact with the isooctyl tail of cholesterol are Ala and residues with branched side chains, Leu and Val [Fig. 6(F)]. This combination of residues can be used to form a groove in the protein surface to accomodate the tail of cholesterol. The relatively flexible isooctyl tail can adopt different conformations to fit the shape of the binding site [Fig. 7(C,D)].
Figure 7.
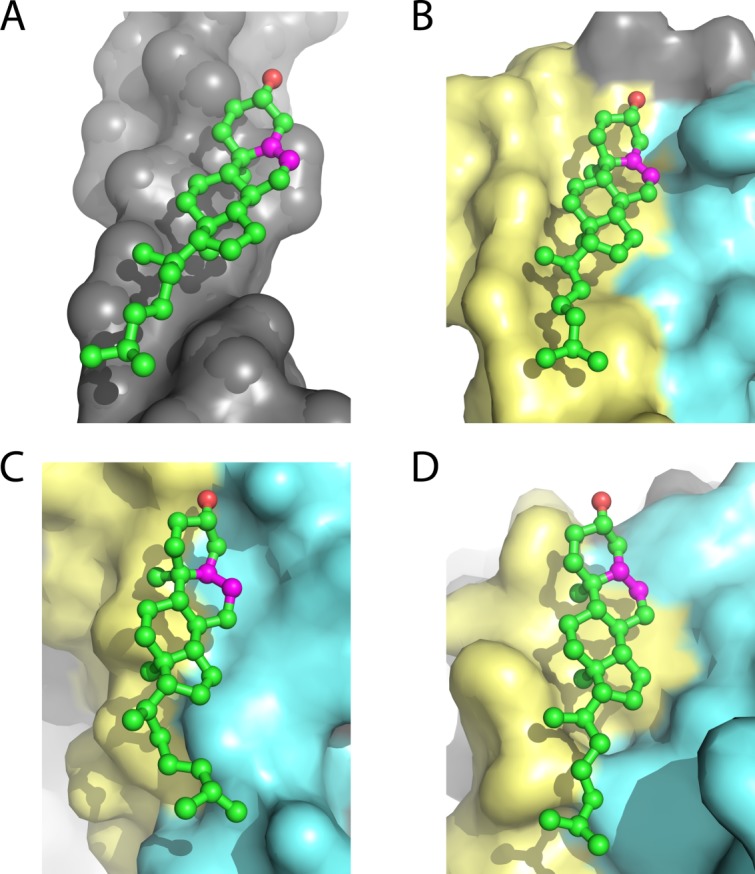
Cholesterol usually resides on the TM surface of integral membrane proteins. α) Cholesterol binds to TM helix I of the 5-hydroxytryptamine receptor 2B (PDBID: 4IB4100). B) Cholesterol binds to the groove formed by TM helices VI and VII of the μ-type opioid receptor (PDBID: 4DKL101). C) The tail of cholesterol 1 in a Na+, K+-ATPase (PDBID: 4HYT94) fits the crevice between TM helix 7 and 10. D) The tail of cholesterol 2 in a Na+, K+-ATPase (PDBID: 4HYT94) fits the gap between TM helix 8 and 10. The C5=C6 double bond in ring B of cholesterol is shown in magenta.
The sterol ring interacts almost exclusively with hydrophobic residues in both water soluble and membrane proteins [Fig. 6(C)]. For membrane proteins most of structures involve binding of the relatively rough β face of the sterol ring system to the TMD surface (Table I), with the smooth α face being free to interact with lipids (cf. Fig. 8). The β face of cholesterol has two methyl groups (C18 and C19) protruding out of the plane of the sterol ring [Fig. 2(B)]. These methyls can serve as knobs to fit in grooves or holes in the protein surface, which often involve branched residues such as Ile and Leu [Fig. 6(G)]. Given the small number of available structures it is not yet clear whether the bias in favor of a β-face/membrane protein interface will ultimately prove to be the preferred mode of interaction. An energetic rationale for why this might prove to be the case is not obvious. It should be added that direct observation of cholesterol-lipid interactions are not uncommon in membrane protein crystal structures.94,97,100,102,104
Figure 8.
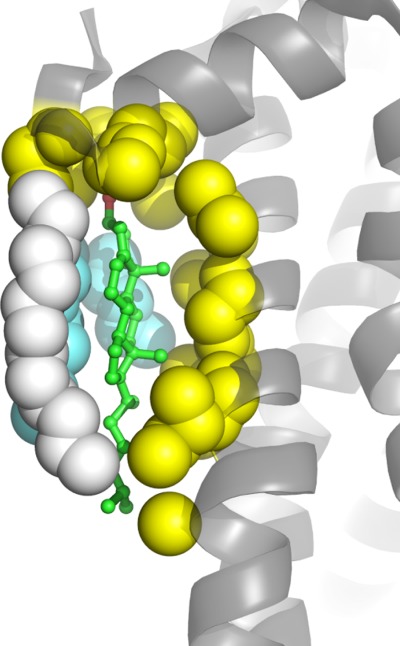
Cholesterol resides on the surface of 5-hydroxytryptamine receptor 2B (PDBID: 4IB4100) with its β-face contacting the surface of the protein (atoms shown in yellow) and its α-face contacting acyl chains of monoolein (cyan) and the C16 chain of covalently-linked palmitate (atoms shown in white).
The formation of hydrogen bonds between cholesterol and proteins is ubiquitously implied in all the available structures (Table I), in which backbone amide or polar side chains from the protein are in a suitable position to form hydrogen bonds with the 3β-OH head group of cholesterol [Fig. 9(A,B)]. In addition to simple direct hydrogen bonding to the protein, the 3β-OH sometimes also participates in hydrogen bonding networks involving water [Fig. 9(C–E)] and multiple residues from an adjacent loop or turn between two helices (Fig. 9) which, not surprisingly, often contain glycine residues [Fig. 6(E)]. For integral membrane proteins, the 3β-OH moiety of bound cholesterol has, so far, always been seen to be located near water-membrane interface.
Figure 9.
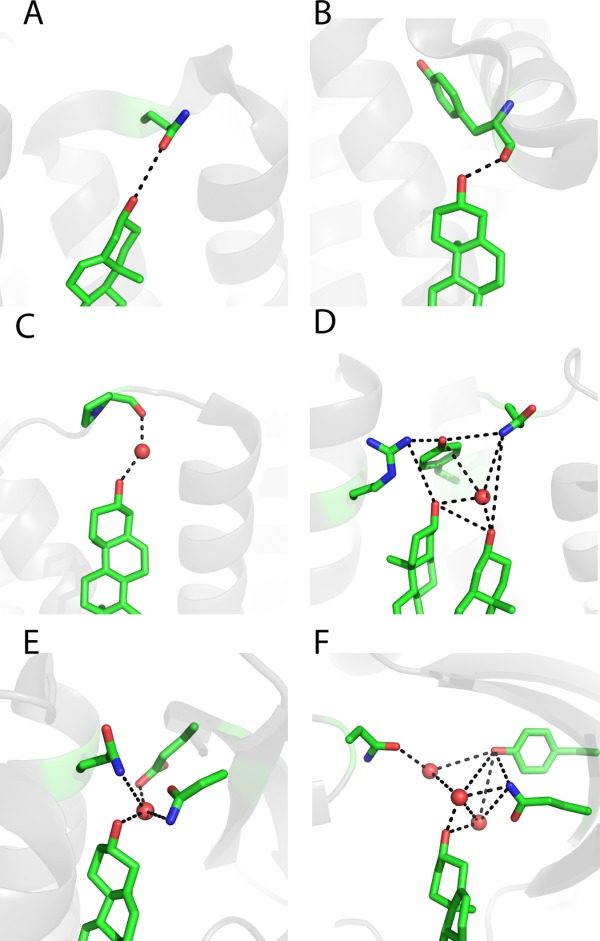
Examples of hydrogen bonds formed between the head group of bound cholesterol and proteins. A) Cholesterol forms a hydrogen bond with the side chain of Gln65 in the β2-adrenergic receptor (PDB ID: 3PDS98). B) Cholesterol forms a hydrogen bond with the backbone amide Tyr394 in the 5-hydroxytryptamine receptor 2B (PDBID: 4IB4 100). C) Cholesterol forms a hydrogen bond network with Pro309 and involves a water molecule (shown as a red ball) in the μ-type opioid receptor (PDBID: 4DKL101). D) Two cholesterols form a hydrogen bond network with Gln65, Tyr70, and Arg151 through a water molecule (shown as a red ball) in the β2 adrenergic receptor (PDBID: 3NYA96). E) Cholesterol forms a hydrogen network with Glu30, Asn41, and Gln79 in the Niemann-Pick C1 protein (PDBID: 3GKI89) mediated by a water molecule (shown as a red ball). F) Cholesterol forms a hydrogen bond network with Tyr61, Asn210, and Gln377 in human CYP11A1 (PDBID: 3N9Y91) through multiple water molecules (shown as red balls).
Aromatic residues seem to undergo three modes of interaction with the sterol ring. First, aromatic residues can stack with saturated sections of the ring system on both α- and β- faces, interactions that that may be driven by favorable Van der Waals and CH-π interactions [Fig. 10(A–D)]. Secondly, Phe and Tyr (but not Trp, see also Ref.105) often stack with the sterol at the C5=C6 double bond with the ring plan parallel with the plane of the double bond [Fig. 6(D)], strongly suggesting that aromatic residue side chains may undergo favorable π-π interactions with the C5=C6 double bond [Fig. 10(E,F)]. Third, Phe and Tyr can also interact in an orthogonal manner with the C5=C6 double bond [Fig. 10(G,H)], suggestive of favorable electric quadrupole interactions between the aromatic ring and the pi bond. For the interaction in the Niemann-Pick C1 protein illustrated in Figure 10(G), it is known that mutation of Phe203 dramatically reduces cholesterol binding affinity.89
Figure 10.
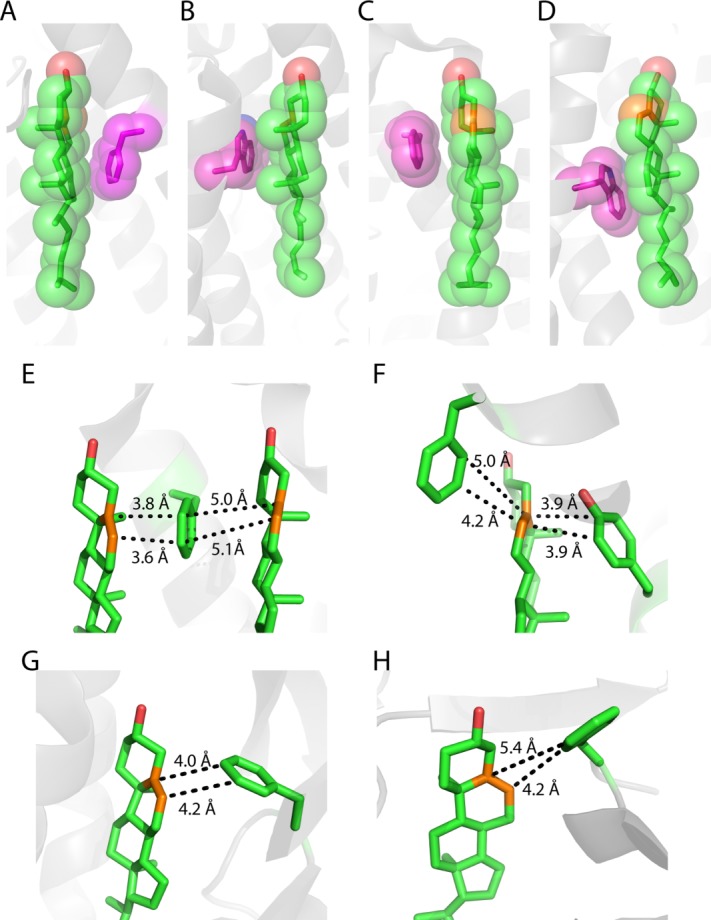
Examples of an aromatic residue (shown in magenta) interacting with the sterol ring of cholesterol. A) Phe33 interacts the α face of the sterol ring in a Na+, K+-ATPase (PDBID: 4HYT, cholesterol 1 94). B) Trp981 interacts the β-face of cholesterol in a Na+, K+-ATPase (PDBID: 4HYT, cholesterol 2 94). C) Phe255 interacts the α face of the sterol ring in the adenosine receptor A2α (PDB ID: 4EIY102). D) Trp158 interacts the α-face of cholesterol in the β2-adrenergic receptor (PDB ID: 2RH1). All carbon atoms in cholesterol and aromatic residues are displayed with spheres with a radius of 1.7 Å. E) Cholesterol sandwich packing configuration with Phe255 in the middle and cholesterols on each side in the adenosine receptor A2α (PDBID: 4EIY102). The C5=C6 double bonds (shown in orange) are parallel to the face of the aromatic ring. F) Aromatic sandwich packing configuration with a cholesterol in the middle and Tyr299 and Phe313 on each side in the μ-type opioid receptor (PDBID: 4DKL101). The C5=C6 double bond (shown in orange) is parallel to the faces of the aromatic rings. G) Interaction between the Phe203 ring of the Niemann-Pick C1 protein and the cholesterol double bond (PDBID: 3GKI89). H) Interaction of F82 of with the C5=C6 group of CYP11A1 (a cytochrome p450, PDBID: 3N9Y91).
Much effort has been devoted to searching for consensus cholesterol binding sequence motifs.106,107 This led to proposal of the “cholesterol recognition amino acid consensus (CRAC)” domain, which was first posited as a cholesterol binding motif in the C-terminal of the peripheral benzodiazepine receptor (PBR):108,109 -(L/V-X1–5-Y-X1–5-(R/K)-. Later, a reversed-CRAC motif called “CARC” was proposed to be associated with cholesterol binding: -(K/R)-X1–5-Y-X1–5-(L/V)-.110 We searched for CRAC and CARC motifs within the sequences of the 19 proteins for which crystal structures of the cholesterol-protein complex are available. The CRAC motif occurred no less than 91 times, while CARC occurred 97 times. However, only for the oxysterol binding protein Osh490 were any of these motifs located at the sterol binding site. For Osh4, two of these motifs were seen at the binding site and have very different conformations [Fig. 11(A)]. One of these two motifs is seen to adopt a beta strand in which the side chains for the signature residues of the motif are oriented away from the cholesterol binding site. The other motif is in a helical segment, with the phenol -OH group of the Tyr from the motif well-positioned to form a hydrogen bond with the cholesterol 3β-OH group. The observation that the interaction of CRAC and CARC motifs with cholesterol seems to be quite rare in the PDB complements the bioinformatic analysis of Palmer,112 who observed the CRAC motif over 5000 times in the 2100-member proteome of a cholesterol-free bacterium. The CARC and CRAC motifs appear to have little predictive value.
Figure 11.
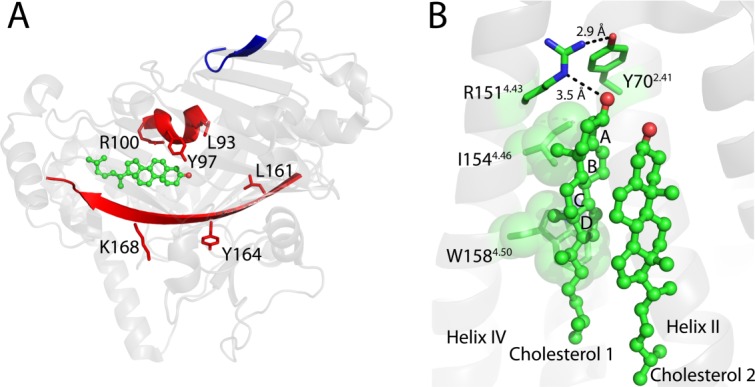
Cholesterol-proximal CRAC motifs in the Osh4 protein. A) Three CRAC motifs are highlighted in the Osh4 protein (PDB ID: 1ZHY90), with one motif (shown in blue) far from the cholesterol and the other two (shown in red) close to the cholesterol. Side chains for key residues of the closest CRAC motifs are shown. B) The cholesterol consensus motif (CCM) in the β2-adrenergic acceptor (PDB ID: 3D4S99). Cholesterol 1 binds to the CCM primarily in contact with the fourth transmembrane helix. Side chains for key residues in the CCM are shown. I1544.46 and W1584.50 are also shown in space-filling mode and interact with cholesterol 1. R1514.43 and Y702.41 may form a hydrogen bonding network, as shown with black dotted lines. The two superscript numbers for each residue reflect the Ballesteros-Weinstein numbering scheme for GPCR sites.111 The first number indicates the transmembrane helix, while the second is relative to the most conserved residue in that helix, which is designated position 50. For example, residue 4.46I is the fourth residue before the most conserved residue in helix IV (4.50W in this particular case).
The “cholesterol consensus motif” (CCM) has been proposed as a cholesterol binding site in class A GPCRs based on a crystal structure of the β2-adrenergic receptor.99 In this structure each monomer binds two cholesterols that are stacked with each other with their α-faces inward [Fig. 11(B)]. The site located on the intracellular side of the TMD in a broad and shallow cleft formed by TM helices I-IV. The head group and β-face of the first of cholesterol interacts extensively with the receptor, while the second cholesterol makes some contacts to TM helix II, but is primarily exposed for contacts with a protein-linked palmitoyl chain or with the lipid phase. The CCM motif pertains to the site for the first of these cholesterols and includes residues from transmembrane helices II and IV: a Phe or Tyr located on the second transmembrane helix (designated 2.41 using the Ballesteros-Weinstein numbering scheme) and the -(R/K)4.39-4.43-X2–6-(I/V/L)4.46-X3-(W/Y)4.50- motif on the fourth transmembrane segment. In the crystal structure of the β2-adrenergic receptor, the epsilon nitrogen of the Arg151 guanidinium group hydrogen bonds to the cholesterol 3β-OH headgroup [Fig. 11(B)]. The hydrophobic residue Ile1544.46 interacts with the first two rings (A and B) of the sterol group. The aromatic Trp1584.50 appears to contribute a very significant interaction with the cholesterol through a CH-π interaction and the edge of the fully saturated ring D [Fig. 11(B)].99 These three residues are on the same side of helix IV. The aromatic residue from the second transmembrane segment (Y702.41) is seen to undergo VDW interactions with ring A of cholesterol and also appears to hydrogen bond with Arg1514.43 [Fig. 11(B)]. In two other β2-adrenergic receptor structures cholesterol is also seen to be bound to the same cleft that is associated with CCM.97,98 In one of these structures only a single cholesterol is bound.98
In addition to the CCM site there are several other GPCR sites at which cholesterol has been observed to be bound in the currently available crystal structures. These include at least two different sites located on the extracellular side of the TMD, as observed in the adenosine A2α receptor [two sites, three cholesterols, cf. Figs. 9(D) and 10(C,E)]102 and in the mu-opioid receptor [one site, one cholesterol, Figs. 5(B), 9(C), 10(F)].101 Also seen in certain β2AR97 and 5-hydroxytrptamine receptor structures100 is a cholesterol that interacts with both the intracellular end of TM helix I and the end of surface-associated helix VIII [cf. Figs. 7(A) and 9(A,B)]. Remarkably, this cholesterol also interacts closely with one of the C16 chains that is covalently linked via a thioacyl bond to Cys residues located in helix VIII (Fig. 8). Given that raft-association of proteins often seems to be driven by protein palmitoylation,49,50 one wonder if the interactions seen in these structures in any way reflects favorable lipid-cholesterol interactions that promote raft-association in these or other proteins.
Finally, we note that a protein domain that was thought to be a cholesterol binding module, the “sterol-sensing domain” (SSD)113 is probably not an actual cholesterol binding domain based on the recent work of Motamed et al.114
Discovery of a Novel Cholesterol Binding Site in the Amyloid Precursor Protein
In 2003 the corresponding author used bioinformatic tools associated with SwissProt to search for all human membrane proteins that were known to be genetically-linked to inherited human diseases and deemed to be small enough (<350 residues) to potentially be amenable to NMR spectroscopic studies. This led to Escherichia coli expression trials for about 20 different human membrane proteins, E. coli being preferred because it is uniquely compatible with a host of NMR isotopic labeling methods. A minimalist approach was pursued in which only N- and C-terminally His6-tagged were constructs were tested. One of the proteins that expressed well was the 99 residue transmembrane C-terminal domain of the amyloid precursor protein (APP). This protein is here referred to as C99, although “β-CTF” is a common alias [Fig. 12(A)]. This protein is the product of β-secretase cleavage of APP and is the immediate precursor of the amyloid-β polypeptides (Aβ), which are released when C99 is cleaved by γ-secretase. Aβ (particularly the longer forms) is prone to form oligomers that morph into ordered cross-β amyloid fibers, ultimately leading to the formation of amyloid plaques in brain tissue. Toxicity associated with Aβ is generally thought to underlie the etiology of Alzheimer's disease.117,118
Figure 12.
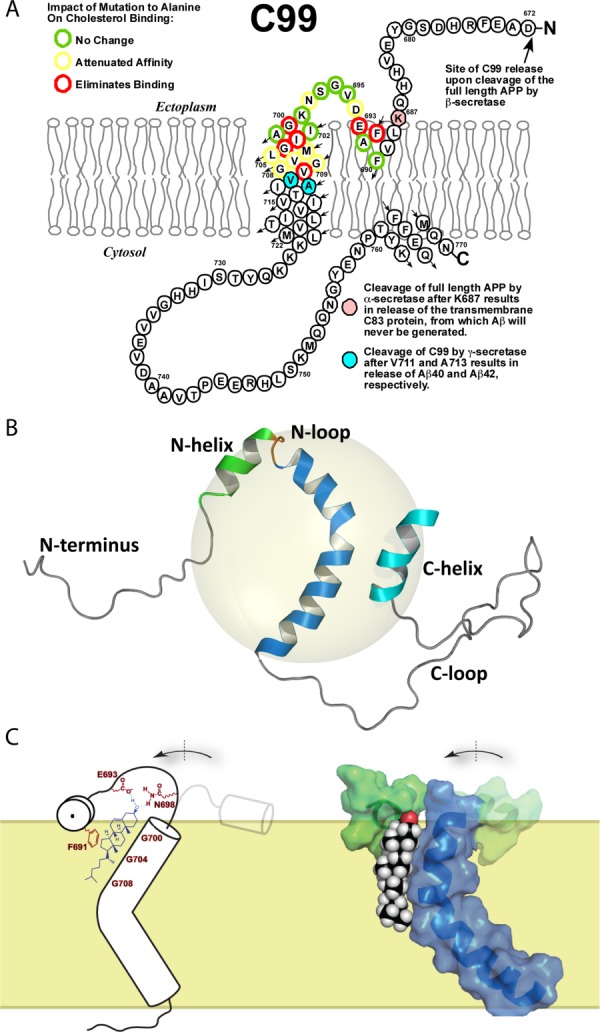
Structure of C99 and formation of a complex with cholesterol. A) The topology of C99. C99 is composed of the C-terminal 99 residues domain of the amyloid precursor protein (residues 672–770), and contains the cleavage site for α-secretase (following K687 as shown in pink) and cleavage sites for γ-secretase (following V711 and A713, as shown in cyan). Alanine-scanning of C99 residues 690 to 710 revealed which residues are critical for cholesterol binding, as indicated. This panel was adapted from Reference 115, with permission from American Chemical Society. B) Backbone structure of C99, as determined by NMR for the protein in LMPG micelles (represented with the gray sphere). This panel was adapted from Reference 116, with permission from American Association for the Advancement of Science. C) Rough model for the proposed C99-cholesterol complex and related conformational change in C99. This panel was adapted from Reference 116, with permission from American Association for the Advancement of Science (left side) and from Reference 115, with permission from American Chemical Society (right side).
While there are now literally thousands of articles on various aspects of the structures of the many forms of Aβ, there have been few structural studies of C99. We therefore decided to pursue NMR and other biophysical studies of its structure in model membranes. Early work focused on C99 in lyso-myrisotyphosphatidylglycerol (LMPG) micelles and led to the conclusion that, in addition to its TM α-helix, C99 also has a short surface-associated amphipathic “N-helix” that is located N-terminal to the TMD, as well another surface-associated amphipathic helix located at its extreme C-terminus119 [Fig. 12(A)]. We speculated that the combined N-helix, N-loop, and N-terminal half of the TMD might comprise a lipid binding site. If so, cholesterol seemed to be a likely binding partner in light of a huge literature on the cholesterol/Alzheimer's disease relationship. An undergraduate in the Sanders lab, Andrew Beel, made particularly important contributions to this work based, in part, on sifting through more than 1000 articles on this and closely related topics.
Because cholesterol exhibits only limited solubility in LMPG and most other types of micelles we initially investigated possible C99-cholesterol interactions by proxy using a water soluble derivative of cholesterol: CHOBIMALT, which is cholesterol with a tetrasaccharide attached to its head group. CHOBIMALT was observed to bind to C99 with a dissociation constant of 15 mol%.119 As an aside, we note that mole percentage or mole fraction units, rather than molarity units, are appropriate for describing thermodynamic binding equilibria involving molecules that are both associated with membranes (or micelles). A dissociation constant of 15 mol% is within the physiological concentration range of cholesterol in plasma membranes. This led us to conclude that C99 is probably a cholesterol binding protein.
That C99 binds bona fide cholesterol was demonstrated by the first author of this article, who developed conditions that allow high quality NMR spectra to be acquired for C99 following preparation in bicelles.116 Bicelles are discoidal lipid bilayer assemblies that are edge-stabilized by a detergent. Bicelles were originally developed in the lab of James Prestegard at Yale University,120 with the corresponding author of this article having the pleasure of making early contributions both while a postdoc in that lab121,122 and later as an assistant professor.123 This included the first reconstitution of membrane proteins in bicelles.124 As first shown by Minto et al., one of the virtues of bicelles is that they are capable of incorporating up to about 20 mol% cholesterol while retaining bicelle morphology.125 This enabled titration of C99 with cholesterol, using solution NMR methods to monitor binding. NMR yielded binding isotherms revealing that 1:1 saturable binding of cholesterol to C99 does indeed occur, with a Kd of 5 mol%.116 Moreover, alanine-scanning mutagenesis indentified a number of sites in C99 for which mutation eliminated or attenuated cholesterol binding [Fig. 12(A)]. A recent EPR study of C99 in lipid vesicles led to the tentative conclusion that its affinity for cholesterol binding under these even more native-liked conditions is slightly higher than in bicelles, with a Kd of about 3 mol%.115 While we have not carried out studies with full length APP (ca. 700 residues), it is very likely that this protein binds also cholesterol, with a similar affinity.
Structure of C99 and Properties of its Cholesterol Binding Site
The structure of C99 was determined in LMPG micelles using solution NMR methods, with graduate student Paul Barrett playing the central role.116 The NMR structure [Fig. 12(B)] confirmed the key features of our earlier C99 topology map, and also revealed that the transmembrane helix contains a prominent kink at the GlyGly motif located near the middle of the TMD. That this kink and the micellar topology of the protein are maintained in actual membranes was confirmed by EPR spectroscopic studies of spin-labeled C99 in lipid vesicles in collaboration with Vanderbilt colleague, Eric Hustedt. Not only were all features of the micellar structure seen to extend to lipid bilayer conditions, but EPR also revealed that the kink in C99's TM helix acts as a flexible hinge.116
The NMR-monitored titration of C99 in bicelles revealed that the N-helix, N-loop, and the N-terminal end of the TMD represent the general location of the cholesterol binding site. The identity of residues specifically involved in cholesterol binding was revealed by the alanine scanning mutagenesis experiments, with results summarized in Figure 12(A).116 The combined NMR and mutagenesis results led to two important conclusions. First, the set of C99 peaks that shifted the most in response to cholesterol binding included not only peaks for sites that are important for cholesterol binding, but also peaks for adjacent sites that were observed not to be important for binding. This strongly suggests that cholesterol binding to C99 involves a conformational change in the N-loop that connect the surface-bound N-helix and the transmembrane domain [Fig. 12(C)].
A second implication of the Ala scanning mutagenesis studies is that all three glycine residues of the G700XXXG704XXXG708 glycine zipper motif are important for cholesterol binding. The glycine zipper and related GXXXG-family motifs are well known to sometimes play a role in membrane protein oligomerization, as first recognized by Engelman and colleagues for glycophorin A.126–129 Indeed, there have been many studies of APP and C99 that have been dedicated to investigating the glycine residues of this zipper motif (see reviews in115,130). We recently carried out a study of C99 in bilayered lipid vesicles using mutagenesis, FRET, and EPR spectroscopic methods.115 This study confirmed that C99 does have a propensity to form homodimers and that the glycine zipper is central to the homodimer interface. However, the affinity of homodimerization is not avid—the observed Kd of 0.5 mol% appears to be orders of magnitude higher than the physiological concentration of C99, which means that the protein probably does not homodimerize in vivo, at least not unless other dimerization-promoting factors are present to reinforce the weak propensity of the protein for self-association. This is in contrast to the Kd observed for cholesterol binding in bicelles and vesicles (3–5 mol%), which is actually on the low end of the range of physiological cholesterol concentrations in most cellular organelles, implicating physiological relevance.
We have not yet completed a high resolution structural study of the complex between C99 and cholesterol. However, based on the binding study and mutagenesis results, some observations can be made. First, the structure of free C99 reveals that the glycines of the G700XXXG704XXXG708 zipper create an extended flat surface on the N-terminal end of the TMD. We speculate that cholesterol binding to this surface likely involves the smooth α-face of cholesterol and that this interaction is driven by a combination of Van der Waals forces and entropy. Association of two flat surfaces with each other may be entropically preferable to the dissociated state, where the rigid and flat faces of both cholesterol and C99 would interact with flexible lipid chains, thereby dampening lipid chain motions. Secondly, we propose that upon docking of the sterol ring system onto the face of the glycine zipper, that the flexible (and Gly-containing) N-loop undergoes a conformational change that swings the N-helix around to encompass the polar end of cholesterol so that hydrogen bonds between the protein and the 3β-OH can be formed [Fig. 12(C)]. The Glu693, Asp694, and Asn698 likely form hydrogen bonds with 3β-OH and/or participate in water-mediated hydrogen bond networks to the this group. Third, we speculate that Phe690 may participate in π-π interactions with the C5=C6 alkene group. That the aromatic side chain of this Phe residue could dip deep enough into the membrane to participate in such a pi interaction seems particularly plausible in light of the structure by Lau et al. of the transmembrane/cystosolic domains of the αIIb subunit of the platelet integrin.131 It was seen that a cytosolic PhePhe motif that is sequentially adjacent to the TM helix dips back into the membrane to form tertiary structural interactions with aliphatic side chains in the TM helix. It is interesting to note that C99 has a CARC motif that overlaps with the Ala-scanned segment: -K687-X3-F691-X3-V695- [Fig. 12(A)]. However, while K687 has not been mutated to determine whether it plays a role in cholesterol binding, the fact that its backbone amide resonance shifts little in response to cholesterol binding suggests it does not strongly interact with cholesterol. Also the Val695 of this motif was seen to have no impact on cholesterol binding when mutated to alanine. This suggests that the presence of this common motif in C99 is essentially unrelated to cholesterol binding.
Hypothesis for How Cholesterol Binding to C99 Promotes Amyloidogenesis and AD
Elevated cholesterol appears to promote or aggravate Alzheimer's disease through more than one mechanism.132–139 Investigating these mechanisms is complicated by, among other things, the fact that cholesterol in circulation does not cross the blood brain barrier. All the cholesterol in the central nervous system (CNS) is made in the brain.140–142
The amyloidogenic pathway of APP processing involves the cleavage of APP to produce C99 followed by the cleavage of C99 to produce Aβ. In competition with the amyloidogenic pathway is the “non-amyloidogenic pathway,” in which α-secretase cleaves APP within its Aβ domain to generate the transmembrane C83 protein. C83 is then cleaved by γ-secretase to release an apparently harmless polypeptide called P3.143–145 There is considerable evidence that elevated cholesterol levels directly activate the amyloidogenic pathway135,146,147 while actively inhibiting non-amyloidogenic cleavage of APP by α-secretase.148,149 There is also much evidence, although not without dispute, that the amyloidogenic β- and γ-secretase are preferentially localized to lipid rafts, at least under conditions where the amyloidogenic pathway is active. At the same time, it is believed that the non-amyloidogenic α-secretase resides in the bulk (non-raft) membrane (reviews in Refs.82,137, and139).
It has been hypothesized that elevated cholesterol promotes preferred partitioning of APP and C99 into raft-like membrane domains where it is more likely to encounter the amyloidogenic proteases and less likely to encounter the benign α-secretase.82,116,119 As a step towards testing this hypothesis we are examining whether binding of cholesterol to C99 leads to its preferred partitioning into raft-like domains in model membranes of well-defined compositions that contain phase-separated liquid-disordered and raft-like liquid-ordered domains.
In addition to possibly promoting amyloidogenesis by altering the phase partitioning of APP and C99, cholesterol binding to APP and C99 may also directly impact interaction of these proteins with the secretases. In the case of α-secretase, its cleavage site in APP is located immediately adjacent to the cholesterol binding site between K687 and L688 [Fig. 12(A)]. It is quite possible that access to this site by α-secretase is occluded when cholesterol is bound. Indeed, it is known that cleavage at the α-secretase site of C99 by trypsin is inhibited by the presence of cholesterol.150 There is also preliminary evidence that gamma-secretase cleavage of C99 may be directly activated by cholesterol through mechanisms that are not understood.151 That this enzyme might preferentially recognize and/or cleave the C99-cholesterol complex over free C99 is plausible (see discussion in Ref115), but has not been investigated.
Teleology of Cholesterol Binding to APP and C99
Why does cholesterol bind to the C99 and, most likely, the full length APP? An intriguing possibility is that C99 and/or APP serve as cholesterol sensors linked to signaling pathways that regulate cellular cholesterol uptake and biosynthesis. When gamma-secretase cleaves C99, the released C-terminal “APP intracellular cytosolic domain” (AICD) has been proposed, among other functions, to suppress transcription of the gene encoding the low density lipoprotein receptor (LDLR).152 Lower cell surface LDLR would reduce the amount of cholesterol imported into cells. At the same time, it has been proposed that intracellular Aβ directly or indirectly inhibits HMG-CoA reductase, the rate-limiting enzyme of the cholesterol biosynthetic pathway.153 While the evidence in support of the above proposals is not yet overwhelming, the suggestion has been made that the amyloidogenic pathway may play a role in regulating cellular cholesterol levels.154,155 If so, then APP and/or C99 may be the cholesterol sensor of these pathways.116,119 We note that if this were to prove be the case, this would not necessarily imply that the amyloidogenic pathway is the primary cholesterol regulatory system of cells. Only a modest fraction of all proteins are thought to be “essential” to their host organisms,156,157 indicating widespread functional redundancy of proteins. This helps make life robust. It is possible that the amyloidogenic pathway can serve as a backup or accessory system to help regulate cholesterol levels. This is consistent with the fact that while APP-knockout mice are smaller and less intelligent than other mice, the phenotype is not very severe (reviewed in Ref.158).
A recent and elegant study by Pierrot et al.159 has significantly advanced the notion that APP does play a role in controlling cellular cholesterol levels, although not exactly as proposed above. The primary cellular regulatory system in control of cholesterol biosynthesis and uptake has been determined by Brown, Goldstein, and co-workers79,160 to center around a membrane-anchored transcription factor called sterol regulatory element binding protein (SREBP). When freed from the membrane the soluble form of SREPB translocates to the nucleus where it binds to the sterol regulatory element in the proximal promoter of the genes encoding HMG-CoA reductase, the LDLR, and related genes of cholesterol metabolism. SREPB is normally anchored to the membrane of the endoplasmic reticulum (ER), where it is complexed with both the INSIG protein and with a cholesterol binding protein known as the SREBP cleavage activating protein (SCAP). When cholesterol levels in the ER membrane fall below 5 mol%, cholesterol dissociates from SCAP.8,114 This ligand dissociation event triggers dissociation of INSIG from the SCAP-SREPB complex, which exposes an ER-to-Golgi export site on SCAP that results in export of the SCAP-SREPB complex to the Golgi. Once in the Golgi, the transcription factor domain of SREPB is released from the membrane by successive cleavage by site-1 and site-2 proteases (S1P and S2P). Solubilized SREBP then activates transcription of the genes encoding HMG-CoA reductase, LDLR and other genes promoting increased cellular cholesterol content.
Pierrot et al.'s recent work indicates a direct role for APP in regulating the SREBP system.159 In cultured neurons it was found that increased APP or C99 but not AICD results in inhibition of SREBP cleavage in the Golgi, apparently because of formation of a complex between APP and SREBP in that compartment. This results in reduced HMG-CoA reductase and lowered cholesterol biosynthesis. At the same time, the expression of the CYP46A1 gene that encodes cholesterol 24-hydroxylase was down-regulated. This enzyme converts cholesterol to 24S-cholesterol, the main form of cholesterol that exits the brain.161 As a consequence, the cholesterol content in the cell was seen to be unchanged from APP-free conditions: the reduction of cholesterol biosynthesis was proportionately offset by decreased conversion of cholesterol to 24S-hydroxycholesterol. This suggests that APP plays a key role in maintaining the balance between cholesterol biosynthesis and degradation in neurons. Very likely it also helps control cholesterol uptake through the LDLR pathway, but this was not tested.
The ability of APP to protect SREBP from cleavage in the Golgi was eliminated when the glycines in its G700XXXG704 motif were mutated.159 This motif is, of course, central to the cholesterol binding site of APP/C99 (Fig. 12). The potential role of cholesterol binding to APP and C99 in regulating SREPB cleavage was not investigated. However, we note that if cholesterol levels in the ER are low enough (<5 mol%) for SCAP to trigger trafficking of SREPB to the Golgi, then it is conceivable that the level of cholesterol in the Golgi may vary over a range which will dictate that the fraction of C99/APP that is cholesterol-complexed may vary significantly (since its Kd for cholesterol is 3–5 mol%115,116). This is as might be expected if it this binding event play a role in the regulation of cellular cholesterol content. One wonders if it is the cholesterol-complexed form of C99/APP that protects SREBP from cleavage or whether cholesterol binding to C99 is competitive with binding of C99 to SREPB. It is interesting that APP was seen not to modulate SREBP cleavage in astrocytes, which is where most of the cholesterol in the brain is made. All told, this work suggests that APP/C99 plays an important and cell type-specific role in neuronal cholesterol homeostasis. Moreover, the cholesterol binding site of APP/C99 is central to this role. Finally, we note another recent report that intracellular Aβ can also reduce SREPB cleavage.162
Conclusions
It is clear that our understanding of the interactions of cholesterol with membrane proteins is in its infancy. Little is known about the dynamics of cholesterol levels in membranes or the dynamics of its distribution between bulk and raft-like phases. The structural and energetic principles governing partitioning of membrane proteins between raft and bulk phase membranes are not understood. Only a modest number of cholesterol-protein complex structures have been determined, with each new structure providing fresh insight. Our recognition of the numerous mechanisms that link cholesterol to a host of human diseases continues to expand; however, the more we learn the more we appreciate the complexity of these mechanisms. Moreover, even in favorable cases a long lag can be expected between basic science discoveries regarding disease-related cholesterol-protein relationships and translation into effective therapeutics. Young scientists should take heart. Numerous dragons remain to be slain.
Acknowledgments
The authors sincerely apologize to investigators who have published relevant work that was not cited in this review because of our oversight or because of space limitations.
References
- 1.Evans WH, Hardison WG. Phospholipid, cholesterol, polypeptide and glycoprotein composition of hepatic endosome subfractions. Biochem J. 1985;232:33–36. doi: 10.1042/bj2320033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambrano F, Fleischer S, Fleischer B. Lipid composition of the Golgi apparatus of rat kidney and liver in comparison with other subcellular organelles. Biochim Biophys Acta. 1975;380:357–369. doi: 10.1016/0005-2760(75)90104-6. [DOI] [PubMed] [Google Scholar]
- 3.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, Milne S, Myers D, Park H, Ryan A, Thompson BM, Wang E, Zhao Y, Brown HA, Merrill AH, Raetz CR, Russell DW, Subramaniam S, Dennis EA. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51:2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood WG, Cornwell M, Williamson LS. High performance thin-layer chromatography and densitometry of synaptic plasma membrane lipids. J Lipid Res. 1989;30:775–779. [PubMed] [Google Scholar]
- 6.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 7.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ourisson G, Albrecht P. Hopanoids. 1. Geohopanoids: the most abundant natural products on Earth? Acc Chem Res. 1992;25:398–402. [Google Scholar]
- 10.Ourisson G, Rohmer M. Hopanoids. 2. Biohopanoids: a novel class of bacterial lipids. Acc Chem Res. 1992;25:403–408. [Google Scholar]
- 11.Saenz JP, Sezgin E, Schwille P, Simons K. Functional convergence of hopanoids and sterols in membrane ordering. Proc Natl Acad Sci USA. 2012;109:14236–14240. doi: 10.1073/pnas.1212141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beattie ME, Veatch SL, Stottrup BL, Keller SL. Sterol structure determines miscibility versus melting transitions in lipid vesicles. Biophys J. 2005;89:1760–1768. doi: 10.1529/biophysj.104.049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenz JJ. Predicting the effect of steroids on membrane biophysical properties based on the molecular structure. Biochim Biophys Acta. 2012;1818:896–906. doi: 10.1016/j.bbamem.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 2000;39:843–849. doi: 10.1021/bi992543v. [DOI] [PubMed] [Google Scholar]
- 15.Mouritsen OG, Zuckermann MJ. What's so special about cholesterol? Lipids. 2004;39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 16.Galea AM, Brown AJ. Special relationship between sterols and oxygen: were sterols an adaptation to aerobic life? Free Radic Biol Med. 2009;47:880–889. doi: 10.1016/j.freeradbiomed.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Chen LL, Wang GZ, Zhang HY. Sterol biosynthesis and prokaryotes-to-eukaryotes evolution. Biochem Biophys Res Commun. 2007;363:885–888. doi: 10.1016/j.bbrc.2007.09.093. [DOI] [PubMed] [Google Scholar]
- 18.Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc London B Biol Sci. 2006;361:951–968. doi: 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannock DA, Lewis RN, McMullen TP, McElhaney RN. The effect of variations in phospholipid and sterol structure on the nature of lipid-sterol interactions in lipid bilayer model membranes. Chem Phys Lipids. 2010;163:403–448. doi: 10.1016/j.chemphyslip.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Harroun TA, Katsaras J, Wassall SR. Cholesterol is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry. 2008;47:7090–7096. doi: 10.1021/bi800123b. [DOI] [PubMed] [Google Scholar]
- 21.Kucerka N, Marquardt D, Harroun TA, Nieh MP, Wassall SR, Katsaras J. The functional significance of lipid diversity: orientation of cholesterol in bilayers is determined by lipid species. J Am Chem Soc. 2009;131:16358–16359. doi: 10.1021/ja907659u. [DOI] [PubMed] [Google Scholar]
- 22.Almeida PF. Thermodynamics of lipid interactions in complex bilayers. Biochim Biophys Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Pasenkiewicz-Gierula M, Rog T, Kitamura K, Kusumi A. Cholesterol effects on the phosphatidylcholine bilayer polar region: a molecular simulation study. Biophys J. 2000;78:1376–1389. doi: 10.1016/S0006-3495(00)76691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- 25.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 26.Elson EL, Fried E, Dolbow JE, Genin GM. Phase separation in biological membranes: integration of theory and experiment. Annu Rev Biophys. 2010;39:207–226. doi: 10.1146/annurev.biophys.093008.131238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 28.Allende D, Vidal A, McIntosh TJ. Jumping to rafts: gatekeeper role of bilayer elasticity. Trends Biochem Sci. 2004;29:325–330. doi: 10.1016/j.tibs.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Gandhavadi M, Allende D, Vidal A, Simon SA, McIntosh TJ. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent resistant rafts. Biophys J. 2002;82:1469–1482. doi: 10.1016/S0006-3495(02)75501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. Lipid rafts reconstituted in model membranes. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 32.Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiessling V, Wan C, Tamm LK. Domain coupling in asymmetric lipid bilayers. Biochim Biophys Acta. 2009;1788:64–71. doi: 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 35.Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenworthy AK. Have we become overly reliant on lipid rafts? Talking point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 2008;9:531–535. doi: 10.1038/embor.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 38.Kraft ML. Plasma membrane organization and function: moving past lipid rafts. Mol Biol Cell. 2013;24:2765–2768. doi: 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaRocca TJ, Pathak P, Chiantia S, Toledo A, Silvius JR, Benach JL, London E. Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathogens. 2013;9:e1003353. doi: 10.1371/journal.ppat.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 41.Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol. 2013;202:35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sezgin E, Kaiser HJ, Baumgart T, Schwille P, Simons K, Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 43.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veatch SL, Cicuta P, Sengupta P, Honerkamp-Smith A, Holowka D, Baird B. Critical fluctuations in plasma membrane vesicles. ACS Chem Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 45.Honerkamp-Smith AR, Veatch SL, Keller SL. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim Biophys Acta. 2009;1788:53–63. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veatch SL, Keller SL. Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 47.Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys Rev Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 48.Foster LJ. Moving closer to the lipid raft proteome using quantitative proteomics. Methods Mol Biol. 2009;528:189–199. doi: 10.1007/978-1-60327-310-7_14. [DOI] [PubMed] [Google Scholar]
- 49.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci USA. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 51.Lin Q, London E. Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J Biol Chem. 2013;288:1340–1352. doi: 10.1074/jbc.M112.415596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kahya N. Targeting membrane proteins to liquid-ordered phases: molecular self-organization explored by fluorescence correlation spectroscopy. Chem Phys Lipids. 2006;141:158–168. doi: 10.1016/j.chemphyslip.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 53.Silvius JR. Partitioning of membrane molecules between raft and non-raft domains: insights from model-membrane studies. Biochim Biophys Acta. 2005;1746:193–202. doi: 10.1016/j.bbamcr.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 54.McIntosh TJ, Simon SA. Roles of bilayer material properties in function and distribution of membrane proteins. Annu Rev Biophys Biomol Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 55.Kahya N. Protein-protein and protein-lipid interactions in domain-assembly: lessons from giant unilamellar vesicles. Biochim Biophys Acta. 2010;1798:1392–1398. doi: 10.1016/j.bbamem.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 56.Sezgin E, Levental I, Grzybek M, Schwarzmann G, Mueller V, Honigmann A, Belov VN, Eggeling C, Coskun U, Simons K, Schwille P. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim Biophys Acta. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Sengupta P, Hammond A, Holowka D, Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim Biophys Acta. 2008;1778:20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tong J, Briggs MM, Mlaver D, Vidal A, McIntosh TJ. Sorting of lens aquaporins and connexins into raft and nonraft bilayers: role of protein homo-oligomerization. Biophys J. 2009;97:2493–2502. doi: 10.1016/j.bpj.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Jafurulla M, Chattopadhyay A. Membrane lipids in the function of serotonin and adrenergic receptors. Curr Med Chem. 2013;20:47–55. [PubMed] [Google Scholar]
- 61.Barrantes FJ, Bermudez V, Borroni MV, Antollini SS, Pediconi MF, Baier JC, Bonini I, Gallegos C, Roccamo AM, Valles AS, Ayala V, Kamerbeek C. Boundary lipids in the nicotinic acetylcholine receptor microenvironment. J Mol Neurosci. 2010;40:87–90. doi: 10.1007/s12031-009-9262-z. [DOI] [PubMed] [Google Scholar]
- 62.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 65.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim Biophys Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jastrzebska B, Debinski A, Filipek S, Palczewski K. Role of membrane integrity on G protein-coupled receptors: Rhodopsin stability and function. Progr Lipid Res. 2011;50:267–277. doi: 10.1016/j.plipres.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. Proc Natl Acad Sci USA. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong H, Chang YC, Bowie JU. Measuring transmembrane helix interaction strengths in lipid bilayers using steric trapping. Methods Mol Biol. 2013;1063:37–56. doi: 10.1007/978-1-62703-583-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marius P, Zagnoni M, Sandison ME, East JM, Morgan H, Lee AG. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys J. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood WG, Igbavboa U, Muller WE, Eckert GP. Cholesterol asymmetry in synaptic plasma membranes. J Neurochem. 2011;116:684–689. doi: 10.1111/j.1471-4159.2010.07017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Meer G. Dynamic transbilayer lipid asymmetry. Cold Spring Harbor Perspect Biol. 2011;3:a004671. doi: 10.1101/cshperspect.a004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Fabiani E, Mitro N, Gilardi F, Crestani M. Sterol-protein interactions in cholesterol and bile acid synthesis. Subcell Biochem. 2010;51:109–135. doi: 10.1007/978-90-481-8622-8_4. [DOI] [PubMed] [Google Scholar]
- 73.Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013;24:342–350. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rone MB, Fan J, Papadopoulos V. Cholesterol transport in steroid biosynthesis: role of protein-protein interactions and implications in disease states. Biochim Biophys Acta. 2009;1791:646–658. doi: 10.1016/j.bbalip.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLean KJ, Hans M, Munro AW. Cholesterol, an essential molecule: diverse roles involving cytochrome P450 enzymes. Biochem Soc Trans. 2012;40:587–593. doi: 10.1042/BST20120077. [DOI] [PubMed] [Google Scholar]
- 76.Goedeke L, Fernandez-Hernando C. Regulation of cholesterol homeostasis. Cell Mol Life Sci. 2012;69:915–930. doi: 10.1007/s00018-011-0857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gimpl G, Reitz J, Brauer S, Trossen C. Oxytocin receptors: ligand binding, signalling and cholesterol dependence. Progr Brain Res. 2008;170:193–204. doi: 10.1016/S0079-6123(08)00417-2. [DOI] [PubMed] [Google Scholar]
- 78.Steck TL, Lange Y. Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 2010;20:680–687. doi: 10.1016/j.tcb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 2009;50(Suppl):S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schroeder C. Cholesterol-binding viral proteins in virus entry and morphogenesis. Subcell Biochem. 2010;51:77–108. doi: 10.1007/978-90-481-8622-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lippincott-Schwartz J, Phair RD. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu Rev Biophys. 2010;39:559–578. doi: 10.1146/annurev.biophys.093008.131357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beel AJ, Sakakura M, Barrett PJ, Sanders CR. Direct binding of cholesterol to the amyloid precursor protein: an important interaction in lipid-Alzheimer's disease relationships? Biochim Biophys Acta. 2010;1801:975–982. doi: 10.1016/j.bbalip.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hotze EM, Tweten RK. Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim Biophys Acta. 2012;1818:1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skocaj M, Bakrac B, Krizaj I, Macek P, Anderluh G, Sepcic K. The sensing of membrane microdomains based on pore-forming toxins. Curr Med Chem. 2013;20:491–501. doi: 10.2174/0929867311320040002. [DOI] [PubMed] [Google Scholar]
- 85.Gilbert RJ. Cholesterol-dependent cytolysins. Adv Exp Med Biol. 2010;677:56–66. doi: 10.1007/978-1-4419-6327-7_5. [DOI] [PubMed] [Google Scholar]
- 86.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lascombe MB, Ponchet M, Venard P, Milat ML, Blein JP, Prange T. The 1.45 A resolution structure of the cryptogein-cholesterol complex: a close-up view of a sterol carrier protein (SCP) active site. Acta Crystallogr D Biol Crystallogr. 2002;58:1442–1447. doi: 10.1107/S0907444902011745. [DOI] [PubMed] [Google Scholar]
- 88.Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 89.Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strushkevich N, MacKenzie F, Cherkesova T, Grabovec I, Usanov S, Park HW. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc Natl Acad Sci USA. 2011;108:10139–10143. doi: 10.1073/pnas.1019441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogawa H, Shinoda T, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc Natl Acad Sci USA. 2009;106:13742–13747. doi: 10.1073/pnas.0907054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 94.Laursen M, Yatime L, Nissen P, Fedosova NU. Crystal structure of the high-affinity Na+,K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc Natl Acad Sci USA. 2013;110:10958–10963. doi: 10.1073/pnas.1222308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 96.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu M, Siu FY, Liu W, Xu HE, Cherezov V, Roth BL, Stevens RC. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJ AP, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wada T, Shimono K, Kikukawa T, Hato M, Shinya N, Kim SY, Kimura-Someya T, Shirouzu M, Tamogami J, Miyauchi S, Jung KH, Kamo N, Yokoyama S. Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, Acetabularia rhodopsin II, from marine alga. J Mol Biol. 2011;411:986–998. doi: 10.1016/j.jmb.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 104.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 105.Holt A, de Almeida RF, Nyholm TK, Loura LM, Daily AE, Staffhorst RW, Rijkers DT, Koeppe RE, IInd, Prieto M, Killian JA. Is there a preferential interaction between cholesterol and tryptophan residues in membrane proteins? Biochemistry. 2008;47:2638–2649. doi: 10.1021/bi702235k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fantini J, Barrantes FJ. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Epand RM, Thomas A, Brasseur R, Epand RF. Cholesterol interaction with proteins that partition into membrane domains: an overview. Subcell Biochem. 2010;51:253–278. doi: 10.1007/978-90-481-8622-8_9. [DOI] [PubMed] [Google Scholar]
- 108.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 109.Li H, Yao Z, Degenhardt B, Teper G, Papadopoulos V. Cholesterol binding at the cholesterol recognition/interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc Natl Acad Sci USA. 2001;98:1267–1272. doi: 10.1073/pnas.031461598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baier CJ, Fantini J, Barrantes FJ. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci Rep. 2011;1:69. doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Visiers I, Ballesteros JA, Weinstein H. Three-dimensional representations of G protein-coupled receptor structures and mechanisms. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 112.Palmer M. Cholesterol and the activity of bacterial toxins. FEMS Microbiol Lett. 2004;238:281–289. doi: 10.1016/j.femsle.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 113.Kuwabara PE, Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- 114.Motamed M, Zhang Y, Wang ML, Seemann J, Kwon HJ, Goldstein JL, Brown MS. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–18012. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song Y, Hustedt EJ, Brandon S, Sanders CR. Competition between homodimerization and cholesterol binding to the c99 domain of the amyloid precursor protein. Biochemistry. 2013;52:5051–5064. doi: 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Selkoe DJ. Alzheimer's disease. Cold Spring Harbor Perspect Biol. 2011;3:a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ram P, Prestegard JH. Magnetic field induced ordering of bile salt/phospholipid micelles: new media for NMR structural investigations. Biochim Biophys Acta. 1988;940:289–294. doi: 10.1016/0005-2736(88)90203-9. [DOI] [PubMed] [Google Scholar]
- 121.Sanders CR, 2nd, Prestegard JH. Magnetically orientable phospholipid bilayers containing small amounts of a bile salt analogue, CHAPSO. Biophys J. 1990;58:447–460. doi: 10.1016/S0006-3495(90)82390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Progr Nucl Magn Reson Spectrosc. 1994;26:421–444. [Google Scholar]
- 123.Sanders CR, IInd, Schwonek JP. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry. 1992;31:8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- 124.Sanders CR, 2nd, Landis GC. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry. 1995;34:4030–4040. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- 125.Minto RE, Adhikari PR, Lorigan GA. A 2H solid-state NMR spectroscopic investigation of biomimetic bicelles containing cholesterol and polyunsaturated phosphatidylcholine. Chem Phys Lipids. 2004;132:55–64. doi: 10.1016/j.chemphyslip.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 126.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 127.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 128.Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doura AK, Kobus FJ, Dubrovsky L, Hibbard E, Fleming KG. Sequence context modulates the stability of a GxxxG-mediated transmembrane helix-helix dimer. J Mol Biol. 2004;341:991–998. doi: 10.1016/j.jmb.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 130.Wang H, Barreyro L, Provasi D, Djemil I, Torres-Arancivia C, Filizola M, Ubarretxena-Belandia I. Molecular determinants and thermodynamics of the amyloid precursor protein transmembrane domain implicated in Alzheimer's disease. J Mol Biol. 2011;408:879–895. doi: 10.1016/j.jmb.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lau TL, Dua V, Ulmer TS. Structure of the integrin alphaIIb transmembrane segment. J Biol Chem. 2008;283:16162–16168. doi: 10.1074/jbc.M801748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yanagisawa K. Cholesterol and amyloid beta fibrillogenesis. Subcell Biochem. 2005;38:179–202. doi: 10.1007/0-387-23226-5_9. [DOI] [PubMed] [Google Scholar]
- 133.Leduc V, Jasmin-Belanger S, Poirier J. APOE and cholesterol homeostasis in Alzheimer's disease. Trends Mol Med. 2010;16:469–477. doi: 10.1016/j.molmed.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 134.Chui HC, Zheng L, Reed BR, Vinters HV, Mack WJ. Vascular risk factors and Alzheimer's disease: are these risk factors for plaques and tangles or for concomitant vascular pathology that increases the likelihood of dementia? An evidence-based review. Alzheimers Res Ther. 2012;4:1. doi: 10.1186/alzrt98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fonseca AC, Resende R, Oliveira CR, Pereira CM. Cholesterol and statins in Alzheimer's disease: current controversies. Exp Neurol. 2010;223:282–293. doi: 10.1016/j.expneurol.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 136.Martins IJ, Berger T, Sharman MJ, Verdile G, Fuller SJ, Martins RN. Cholesterol metabolism and transport in the pathogenesis of Alzheimer's disease. J Neurochem. 2009;111:1275–1308. doi: 10.1111/j.1471-4159.2009.06408.x. [DOI] [PubMed] [Google Scholar]
- 137.Rushworth JV, Hooper NM. Lipid rafts: linking Alzheimer's amyloid-beta production, aggregation, and toxicity at neuronal membranes. Int J Alzheimers Dis. 2010;2011:603052. doi: 10.4061/2011/603052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Di Paolo G, Kim TW. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hicks DA, Nalivaeva NN, Turner AJ. Lipid rafts and Alzheimer's disease: protein-lipid interactions and perturbation of signaling. Front Physiol. 2012;3:189. doi: 10.3389/fphys.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Korade Z, Kenworthy AK. Lipid rafts, cholesterol, and the brain. Neuropharmacology. 2008;55:1265–1273. doi: 10.1016/j.neuropharm.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 142.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 143.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kojro E, Fahrenholz F. The non-amyloidogenic pathway: structure and function of alpha-secretases. Subcell Biochem. 2005;38:105–127. doi: 10.1007/0-387-23226-5_5. [DOI] [PubMed] [Google Scholar]
- 145.Nalivaeva NN, Turner AJ. The amyloid precursor protein: a biochemical enigma in brain development, function and disease. FEBS Lett. 2013;587:2046–2054. doi: 10.1016/j.febslet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 146.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K. Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci USA. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, Younkin LH, Younkin SG, Golde TE. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9:11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- 148.Bodovitz S, Klein WL. Cholesterol modulates alpha-secretase cleavage of amyloid precursor protein. J Biol Chem. 1996;271:4436–4440. doi: 10.1074/jbc.271.8.4436. [DOI] [PubMed] [Google Scholar]
- 149.Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci USA. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Marenchino M, Williamson PT, Murri S, Zandomeneghi G, Wunderli-Allenspach H, Meier BH, Kramer SD. Dynamics and Cleavability at the alpha-cleavage site of APP(684–726) in different lipid environments. Biophys J. 2008;95:1460–1473. doi: 10.1529/biophysj.108.129726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Osenkowski P, Ye W, Wang R, Wolfe MS, Selkoe DJ. Direct and potent regulation of gamma-secretase by its lipid microenvironment. J Biol Chem. 2008;283:22529–22540. doi: 10.1074/jbc.M801925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De Strooper B, Muller U, Shen J, Hartmann T. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- 154.Grimm MO, Grimm HS, Hartmann T. Amyloid beta as a regulator of lipid homeostasis. Trends Mol Med. 2007;13:337–344. doi: 10.1016/j.molmed.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 155.Grimm MO, Rothhaar TL, Hartmann T. The role of APP proteolytic processing in lipid metabolism. Exp Brain Res. 2012;217:365–375. doi: 10.1007/s00221-011-2975-6. [DOI] [PubMed] [Google Scholar]
- 156.Decottignies A, Sanchez-Perez I, Nurse P. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mahadevan R, Lovley DR. The degree of redundancy in metabolic genes is linked to mode of metabolism. Biophys J. 2008;94:1216–1220. doi: 10.1529/biophysj.107.118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zheng H, Koo EH. Biology and pathophysiology of the amyloid precursor protein. Mol Neurodegener. 2011;6:27. doi: 10.1186/1750-1326-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pierrot N, Tyteca D, D'Auria L, Dewachter I, Gailly P, Hendrickx A, Tasiaux B, Haylani LE, Muls N, N'Kuli F, Laquerriere A, Demoulin JB, Campion D, Brion JP, Courtoy PJ, Kienlen-Campard P, Octave JN. Amyloid precursor protein controls cholesterol turnover needed for neuronal activity. EMBO Mol Med. 2013;5:608–625. doi: 10.1002/emmm.201202215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 161.Hughes TM, Rosano C, Evans RW, Kuller LH. Brain cholesterol metabolism, oxysterols, and dementia. J Alzheimers Dis. 2013;33:891–911. doi: 10.3233/JAD-2012-121585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mohamed A, Saavedra L, Di Pardo A, Sipione S, Posse, de Chaves E. beta-amyloid inhibits protein prenylation and induces cholesterol sequestration by impairing SREBP-2 cleavage. J Neurosci. 2012;32:6490–6500. doi: 10.1523/JNEUROSCI.0630-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


