Abstract
Patients with chronic granulomatous disease (CGD) suffer from recurrent, life-threatening bacterial and fungal infections of the skin, the airways, the lymph nodes, liver, brain and bones. Frequently found pathogens are Staphylococcus aureus, Aspergillus species, Klebsiella species, Burkholderia cepacia and Salmonella species. CGD is a rare (∼1:250 000 births) disease caused by mutations in any one of the five components of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in phagocytes. This enzyme generates superoxide and is essential for intracellular killing of pathogens by phagocytes. Molecular diagnosis of CGD involves measuring NADPH oxidase activity in phagocytes, measuring protein expression of NADPH oxidase components and mutation analysis of genes encoding these components. Residual oxidase activity is important to know for estimation of the clinical course and the chance of survival of the patient. Mutation analysis is mandatory for genetic counselling and prenatal diagnosis. This review summarizes the different assays available for the diagnosis of CGD, the precautions to be taken for correct measurements, the flow diagram to be followed, the assays for confirmation of the diagnosis and the determinations for carrier detection and prenatal diagnosis.
Keywords: CGD, DHR test, mutation analysis, NADPH oxidase, NBT test
Introduction
Patients with chronic granulomatous disease (CGD) suffer from a variety of recurrent bacterial and fungal infections (for a review see [1]). These infections occur most commonly in organs in contact with the outside world – the lungs, gastrointestinal tract and skin, as well as in the lymph nodes that drain these structures. Because of both contiguous and haematogenous spread of infection, a wide range of other organs can be affected, most notably the liver, bones, kidneys and brain. In approximately two-thirds of patients, the first symptoms of CGD appear during the first year of life in the form of infections, dermatitis (sometimes seen at birth), gastrointestinal complications (obstruction or intermittent bloody diarrhoea due to colitis) and a failure to thrive. The clinical picture can be quite variable, with some infants suffering from several of these complications, whereas others appear to be far less ill. In some cases, the presenting symptoms of CGD can be mistaken for pyloric stenosis, food or milk allergy or iron-deficiency anaemia.
Pneumonia is the most common type of infection encountered in CGD in all age groups and is caused typically by Staphylococcus aureus, Aspergillus species, Burkholderia cepacia and enteric Gram-negative bacteria. Aspergillus and other fungal infections of the lung also pose difficult challenges because they typically require prolonged treatment (3–6 months). Cutaneous abscesses and lymphadenitis represent the next most common types of infection in CGD and are caused typically by S. aureus, followed by various Gram-negative organisms, including B. cepacia complex and Serratia marcescens. Recurrent impetigo, frequently in the perinasal area and caused by S. aureus, usually requires prolonged courses of oral and topical antibiotics to clear. Hepatic (and perihepatic) abscesses are also quite common in CGD and are caused typically by S. aureus. Patients usually present with fever, malaise and weight loss. Osteomyelitis is another important infection in CGD and can arise from haematogenous spread of organisms (S. aureus, Salmonella spp., S. marcescens) or contiguous invasion of bone, seen typically with non-A. fumigatus pneumonia, such as A. nidulans spreading to the ribs or vertebral bodies. Perirectal abscesses are also common in CGD patients, and once formed can persist for years despite aggressive anti-microbial therapy and fastidious local care. Other frequently encountered catalase-positive microbial agents are Escherichia coli species, Listeria species, Klebsiella species, Nocardia and Candida species.
CGD patients usually manifest their symptoms at an early age, in the first 2 years of life. However, due to the diverse genetic causes of the disease (see below), some patients may also present later in life. Most CGD patients (about 80%) are male, because the main cause of the disease is a mutation in an X-chromosome-linked gene. However, defects in autosomal genes may also underlie the disease and cause CGD in both males and females.
CGD is caused by the failure of the patients' phagocytic leucocytes to kill a wide variety of pathogens. This is due to a defect in these phagocytes in producing reactive oxygen species (ROS), which are needed for the killing process. In normal phagocytes, these ROS are generated by an enzyme called nicotinamide adenine dinucleotide phosphate (NADPH) oxidase. This enzyme is composed of five subunits, two of which are in resting cells localized in the plasma membrane and three in the cytosol. The two membrane-bound subunits are a transmembrane glycoprotein (gp) with a molecular mass of 91 kD, called gp91phox (phox for phagocyte oxidase) and another transmembrane protein with a molecular mass of 22 kD, called p22phox. These two proteins form a heterodimer and are dependent upon each other's presence for maturation and stable expression. This heterodimer is called cytochrome b558 because gp91phox contains two haem groups with an absorbance peak at 558 nm. The three cytosolic subunits (p40phox, p47phox and p67phox) form a heterotrimer that translocates to cytochrome b558 upon cell activation (e.g. by binding of micro-organisms or chemotactic factors to membrane receptors). As a result, the conformation of gp91phox is slightly changed, which enables NADPH in the cytosol to bind and donate electrons to this protein. These electrons are then transported within gp91phox to molecular oxygen on the apical side of the membrane. In this way, superoxide – the one-electron reduction product of oxygen – is generated within the phagosome that engulfs the ingested micro-organism, or on the outside of the phagocyte. From superoxide, other ROS, such as hydrogen peroxide, can be generated.
The exact mechanism of pathogen killing within the phagosome is not known. From the killing defect seen in CGD phagocytes, it is clear that ROS play an important role, but whether this is a direct role through formation of hypochlorous acid from hydrogen peroxide and chloride, catalysed by myeloperoxidase, or an indirect role through facilitating the release of proteolytic enzymes from the granules in the phagocytes [2], or a combination of these mechanisms, remains to be established. Most CGD pathogens share the property of producing catalase; as such, they degrade the hydrogen peroxide that they themselves generate. It has therefore been suggested that catalase-negative organisms, by supplying the CGD phagocytes with microbial hydrogen peroxide, might complement the hydrogen peroxide deficit in CGD phagocytes, thus inducing killing of the microbes themselves. Catalase production was thus thought to be an important microbial pathogenicity factor in CGD. However, this hypothesis must be viewed in the context that the majority of all pathogens contain catalase (with the important exception of streptococci). This view has been challenged further by the retained virulence of Aspergillus and staphylococci rendered genetically deficient for catalase production [3,4]. In addition, individuals with the quite common deficiency of myeloperoxidase do not suffer from CGD-like symptoms.
The genes encoding the five NADPH oxidase components are CYBB (located on the X chromosome) for gp91phox, and the autosomal genes CYBA for p22phox, NCF1 for p47phox, NCF2 for p67phox and NCF4 for p40phox (Table 1). About 70% of the CGD patients have a mutation in CYBB (most of them hemizygous males, but a few heterozygous females with skewed expression of their mutation are also known). The remainder of the patients have a mutation in NCF1 (about 20%), in CYBA (about 5%) or in NCF2 (about 5%). Only one patient is known with a mutation in NCF4. A mutation in any of these five genes can cause CGD. If the mutation leaves some residual NADPH oxidase activity intact, the clinical expression of the disease is less serious [5] and the chance of survival of the patient is larger [6] than in the case of total oxidase deficiency. This depends upon the gene mutated, the type of mutation and the position of the mutation within the gene. In general, mutations in NCF1 lead to a milder form of CGD (later presentation, milder clinical expression, better chance of survival) than mutations in any of the other genes. For genetic counselling and prenatal diagnosis, mutation analysis of the CGD genes is mandatory.
Table 1.
Properties of the phagocyte respiratory burst oxidase (phox) components
| gp91phox | p22phox | p47phox | p67phox | p40phox | |
|---|---|---|---|---|---|
| Synonyms | β-chain | α-chain | NCF1 | NCF2 | NCF4 |
| Heavy chain | Light chain | ||||
| Nox-2 | |||||
| Amino acids | 570 | 195 | 390 | 526 | 339 |
| Molecular weight (kDa) | |||||
| Predicted | 65·0 | 20·9 | 44·6 | 60·9 | 39·0 |
| By PAGE | 70–91 | 22 | 47 | 67 | 40 |
| Glycosylation | Yes (N-linked) | No | No | No | No |
| Phosphorylation | Low level | Yes | Yes | Low level | Low level |
| PI | 9·7 | 10·0 | 9·5 | 5·8 | 6·4 |
| mRNA(kb) | 4·7 | 0·8 | 1·4 | 2·4 | 1·4 |
| Gene locus | CYBB | CYBA | NCF1 | NCF2 | NCF4 |
| Xp21.1 | 16q24 | 7q11.23 | 1q25 | 22q13.1 | |
| Exons/span | 13/30 kb | 6/8·5 kb | 16/40 kb | 11/15 kb | 10/18 kb |
| Cellular location in resting neutrophils | Specific granule membrane; plasma membrane | Specific granule membrane; plasma membrane | Cytosol; cytoskeleton | Cytosol; cytoskeleton | Cytosol; cytoskeleton |
| GenBank Accession no. | X04011† | M21186, J03774 | M25665, M26193 | M32011 | U50720-U50729 |
CYBA: cytochrome-b alpha; CYBB: cytochrome-b beta; NCF: neutrophil cytosol factor; PAGE: polyacrylamide gel electrophoresis.
This GenBank Accession number refers to the sequence as originally published. The complete corrected sequence (encoding an additional 64 amino acids) has not been deposited in GenBank.
Treatment should be started immediately after CGD has been definitely diagnosed, or even before. This involves determination of the exact complicating infections and selection of the most appropriate antibiotic or anti-fungal therapy. Surgical drainage (and sometimes excision) of infected lymph nodes and abscesses involving the liver, skin, rectum, kidney and brain is often necessary for healing, particularly for the visceral abscesses. Daily prophylaxis with Bactrim and/or Itraconazole is recommended during infection-free periods. For more detailed treatment options, the interested reader is referred to Roos et al. [1]. One of the main reasons to make a rapid diagnosis of the severe forms of CGD is that such patients may be treated successfully with a bone marrow transplant [7–9]. A few reports suggest that gene therapy may eventually be successful both in X-linked and autosomal CGD [10,11]. Thus, there are many reasons to identify precisely the genetic defect in patients with CGD.
Patient diagnosis
NADPH oxidase activity
Patients suspected of suffering from CGD (Table 2) must be diagnosed by the inability of their blood phagocytes to generate reactive oxygen species. This can be performed in various ways.
Table 2.
Clinical indications for chronic granulomatous disease (CGD) patients and carriers
| (a) When should a patient's neutrophil nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity be tested? |
| The patient has frequent suppurative cervical lymphadenitis, skin infections, anal infections, osteomyelitis, liver abscesses, central nervous system (CNS) or brain infections, or obstructions of airways, urinary–genital tract or digestive tract. These symptoms usually manifest during the first 10 years of life, but may also start later. Infectious organisms are Stapylococcus aureus, Escherichia coli, Aspergillus ssp., Salmonella ssp., Klebsiella ssp., Burkholderia ssp., Nocardia ssp. and/or Serratia marcescens, some of which are opportunistic and in themselves already form a reason to consider CGD. Some of the infections may provoke sterilizing hyperinflammatory responses, e.g. in fulminant aspergillosis reactions. Other infections in tissue organs may cause more smouldering granulomatous reactions. The latter may relate to the fact that the patients may also have inflammatory bowel disease symptoms or rheumatoid arthritis symptoms. Lymphocyte abnormalities and complement abnormalities are not present |
| (b) When should a female be tested for CGD carriership or for being a CGD patient? |
| 1. The woman is a relative (mother, sister, daughter, maternal aunt, maternal grandmother) of a CGD patient |
| 2. The woman has symptoms of CGD (see Table 2a) |
| 3. The woman is a relative of a CGD patient and has discoid lupus symptoms |
| Carriership or occurrence of CGD should be tested functionally [NADPH oxidase activity in the neutrophils with a per-cell assay, e.g. nitro-blue tetrazolium (NBT) slide test or dihydrorhodamine-1,2,3 (DHR) assay] and genetically |
Usually, purified blood neutrophils [12] are used for these tests, but total leucocytes or even diluted full blood can also be used. Blood can be sent by courier to the testing laboratory, but several precautions must be taken.
Ethylenediamine tetraacetic acid (EDTA) or heparin blood can be used for NADPH oxidase activity testing and for preparation of neutrophil lysate for NADPH oxidase component expression by Western blot. In the case of EDTA blood, the neutrophil fraction purified from it must be recalcified and left for 30 min at room temperature before NADPH oxidase activity can be measured. For DNA preparation, EDTA blood is superior.
The blood transport must take place in polypropylene tubes (completely filled) and at room temperature. This means, for instance, no transport in plane cargo compartments.
The blood must arrive at the place of investigation within 48 h after vena puncture, preferably within 24 h.
A control blood sample must be shipped together with the sample from the presumed patient and/or relative(s). All assays must be performed in parallel with the control cell preparation.
The NADPH oxidase enzyme that is affected in CGD reduces molecular oxygen to the one-electron radical superoxide (O2−), which is subsequently reduced further to hydrogen peroxide (H2O2). The reducing equivalents for this reaction are derived from NADPH, which is converted into NADP+ and H+. Therefore, the complete reaction runs:
The activity of the NADPH oxidase enzyme can be measured by its consumption of oxygen, its generation of superoxide or the subsequent production of hydrogen peroxide. In the different assays discussed below, the phagocytes must be incubated with a certain stimulus to activate the NADPH oxidase in these cells, because in resting phagocytes this enzyme is inactive. Frequently used stimuli are phorbol myristate acetate (PMA, a soluble, receptor-independent stimulus of protein kinase C), serum-treated zymosan particles (a particulate stimulus that binds to Fc-gamma receptors and complement receptor-3 on the cell surface) and the bacterial peptide formyl-methionyl-leucyl-phenylalanine (fMLP), binding to fMLP receptors on the cell surface and activating the NADPH oxidase when the cells have been ‘primed’ with platelet-activating factor (PAF).
Oxygen consumption
Oxygen consumption can be measured with an oxygen electrode [13], but this is a time-consuming and relatively insensitive method that is no longer used for CGD diagnostics. It is the most quantitative method of oxidase measurements, but for CGD diagnosis a simple yes (activity) or no (no activity) usually suffices. Assays for superoxide or hydrogen peroxide are generally employed instead.
Superoxide production
Superoxide generation can be measured by its ability to reduce ferricytochrome c, nitroblue tetrazolium, isoluminol or lucigenin.
The ferricytochrome c reduction is followed spectrophotometrically at 550 nm, because the difference in extinction coefficients of ferricytochrome c (0·89 × 104 M/cm) and its reduction product ferrocytochrome c (2·99 × 104 M/cm) is the largest at that wavelength. The contribution of superoxide to the reduction process must be quantified by adding superoxide dismutase (SOD). This enzyme catalyzes the second reaction shown above, and thus prevents superoxide from reacting with ferricytochrome c. Any reduction of ferricytochrome c in the presence of SOD is superoxide-independent and must therefore be subtracted from the total reduction to obtain the superoxide-dependent contribution. The assay relies upon the excretion of superoxide by activated phagocytes because it takes place extracellularly, in the medium surrounding the cells. A detailed protocol for this reaction, with isolated neutrophils activated with PMA in a microtitre plate, can be found in [14].
Nitroblue tetrazolium (NBT) is a pale yellow dye that can be reduced by superoxide to the black, insoluble formazan. This reaction takes place inside activated phagocytes, thus leaving cells with an active NADPH oxidase stained by formazan deposits that cannot leave the cells. This property has made NBT an ideal agent to judge the oxidase activity of individual cells, which is especially useful for carrier detection of X-linked CGD (see section Oxidase activity or protein expression in single cells). CGD patients usually show no or very little formazan deposition in any cell [15]. The test can be performed as a microscopic slide assay or can be made more quantitative by lysing the cells in KOH afterwards and dissolving the formazan in dimethyl sulphoxide (DMSO), which can then be measured spectrophotometrically at 620 nm. Water-soluble derivatives of NBT also exist and can be used to measure superoxide production online, as with the ferricytochrome c assay. Detailed protocols for these assays can be found in [14]. Care should be taken with neutrophils derived from shipped blood, in which superoxide derived from damaged mitochondria may lead to a false-positive NBT result [16].
A number of reagents is known to react with superoxide, to be excited by this process and then to release energy in the form of light (chemiluminescence). Among these are lucigenin (bis-N-methyl-acridinium nitrite) and isoluminol (6-amino-2,3-dihydro-1,4,-phtalazinedione). Isoluminol does not pass membranes and therefore detects exclusively extracellular superoxide. For this reaction, addition of a peroxidase to the reaction mixture is required. Chemiluminescence assays are highly sensitive and can therefore be carried out with very few cells. Protocols, also for microtitre plate assays, can be found in [14,17].
Hydrogen peroxide generation
Hydrogen peroxide (H2O2) has oxidizing properties; such reactions are catalyzed by peroxidases (although these enzymes can also use superoxide as a substrate). Well-known H2O2-detecting agents are dihydrorhodamine-1,2,3 (DHR), 10-acetyl-3,7-dihydroxyphenoxazine (resorufine, Amplex Red) and 5-amino-2,3-dihydro-1,4-phtalazinedione (luminol). DHR enters the cells freely and is oxidized intracellularly to rhodamine-1,2,3, which emits a bright fluorescent signal at 585 nm when excited by light with a wavelength of 488 nm [18–20]. This oxidation reaction is peroxidase-dependent and thus relies upon the activity of myeloperoxidase or eosinophil peroxidase in the phagocytes. In case of myeloperoxidase (MPO) deficiency, a not uncommon condition, the DHR assay with neutrophils will give a negative result, which may be misinterpreted as an NADPH oxidase deficiency, i.e. as CGD [21]. The assay is carried out in a flow cytometer and thus measures the fluorescent signal from each separate cell, which can again be used for detection of carriers of X-CGD (see section Oxidase activity or protein expression in single cells). Care should be taken to select neutrophils by their scatter characteristics and gate out apoptotic cells to avoid a false bimodal fluorescence pattern that might be mistaken for a mosaic of oxidase-positive and -negative neutrophils. It is a highly sensitive and reliable assay that can be performed with as little as 0·2 ml of blood. For a detailed protocol, see [14].
Amplex Red does not enter cells and therefore detects only H2O2 excreted by the phagocytes. For this reason, a peroxidase is added to the assay mixture. Amplex Red is oxidized to the brightly fluorescent resorufin, which can be detected at 580 nm after excitation at 530 nm. The assay can be carried out in a microtitre plate on a plate reader with a fluorescence detector. This assay is also highly sensitive, reliable and easy to perform. It offers the advantage of testing cells online for their response to a number of stimuli (PMA, zymosan, serum-treated zymosan, PAF/fMLP) over a prolonged time-period. This is a distinct advantage when testing cells from CGD patients with hypomorphic mutations, such as X91− CGD patients, which show less NADPH oxidase activity than normal cells but distinctly more than cells from ‘classical’ CGD patients. For details, see Protocol 1. It should be kept in mind that the Amplex Red assay is not really a quantitative assay, as it overestimates low NADPH oxidase activities. This may be due to catalase in the neutrophils more efficiently removing high than low levels of intracellularly formed H2O2 before it can be detected in the extracellular medium. An alternative assay for such patients is the ferricytochrome c assay (see section Superoxide production), which can also be used with various NADPH oxidase stimuli.
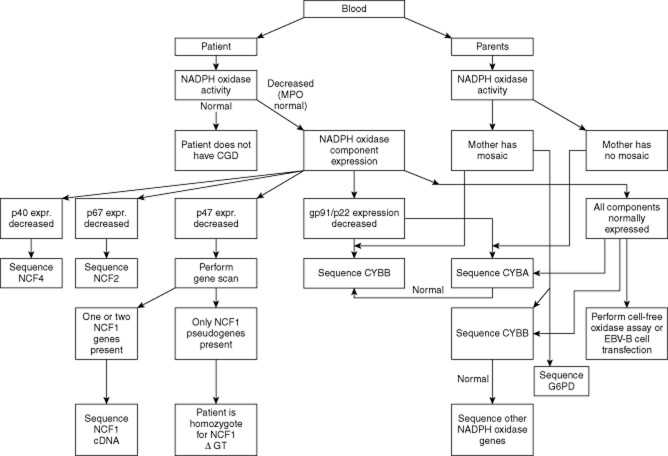
Fig. 1.
Flow diagram for complete molecular diagnosis of chronic granulomatous disease (CGD).
Protocol 1. Amplex Red assay [nicotinamide adenine dinucleotide phosphate (NADPH)-oxidase activity measurement]
NB: Control cells should also be tested!
Materials:
Microplate reader: Genios Plus, Tecan
96-well microtitre plates, flat-bottomed, white polystyrene: Costar
Amplex Red: Molecular Probes, cat no. A-12212, 5 mg
Horseradish peroxidase (HRP): Sigma, cat no. P-8250, 5000 U
Zymosan: MP Biomedicals
Serum-treated zymosan (STZ): see Goldstein et al., J Clin Invest 1975; 56:1155–63
Phorbol myristate acetate (PMA): Sigma
Formyl-methionyl-leucyl-phenylalanine (fMLP): Sigma
Platelet-activating factor (PAF): Sigma
Prepare 20 mM Amplex Red in dimethylsulphoxide (DMSO), aliquots of 12·5 μl in −20°C
Prepare 200 U/ml HRP in phosphate-buffered saline (PBS), aliquots of 25 μl in −20°C
Solutions:
-
Prepare ×2 reaction mix:
Add 1 ml of HEPES to the Amplex Red aliquot and 1 ml of HEPES to the HRP aliquot and transfer to 3 ml of HEPES medium to make 5 ml of ×2 reaction mix
-
Prepare working solutions of the different stimuli:
unstimulated: HEPES medium
Zymosan: 4 mg/ml, ready for use
STZ: add 450 μl of HEPES medium to 300 μl of 10 mg/ml STZ
PMA: add 4 μl of PMA (100 μg/ml) to 996 μl of HEPES medium
fMLP: add 4 μl of fMLP (1 mM) to 996 μl of HEPES medium
PAF/fMLP: add 4 μl of PAF (1 mM) and 4 μl of fMLP (1 mM) to one Eppendorf vial of 992 μl of HEPES medium
Dilute cell suspension to 1 × 106/ml in HEPES medium
Method:
Open ‘Amplex Red’ mode on plate reader (Ex 535 nm, Em 590 nm, interval 30 s, 61 cycles, 2 s of shaking before and in between cycles, 37°C)
-
Pipette (no air bubbles!!) in white 96-well plate (do not use outer wells)
100 μl of ×2 reaction mix
50 μl of cell suspension
Place 96-well plate in plate reader, and click ‘plate in’ (preincubation at 37°C).
Click after 5 min ‘plate out’
Pipette 50 μl of stimulus (no air bubbles!!)
Click ‘Start’ directly (NB: reaction to fMLP is very quick and transient)
Luminol is a ROS probe with chemiluminescent properties. It enters cells and therefore detects both intra- and extracellular H2O2. By adding SOD and catalase, to remove extracellular O2− and H2O2, the reaction can be made specific for intracellular ROS. The luminol assay relies, again, on the availability of intracellular peroxidase and thus again carries the danger of misdiagnosing MPO deficiency for CGD. Detailed protocols for this assay can be found in [14,17].
NADPH oxidase component expression
Because the NADPH oxidase is composed of multiple subunits, five of which may be affected by mutations leading to CGD (Table 1), further differentiation of the diagnosis is needed before mutation analysis can be performed. CGD subgroup investigation begins with FACS analysis of intact neutrophils with monoclonal antibody 7D5 against gp91phox, because the epitope for this antibody is on an extracellular region of gp91phox [22]. If that is normal, immunoblot analysis of neutrophil fractions or intracellular staining in permeabilized neutrophils with antibodies against the five NADPH oxidase components is indicated. In cases where one of the cytosolic components (p47phox or p67phox) is missing, one can be confident that the gene carrying the mutation has been identified.1 However, if one of the membrane-bound components (gp91phox or p22phox) is missing, the other is always undetectable as well, because these subunits stabilize each other for full maturation and expression. In that case, distinction can often be made by searching for a mosaic pattern of NADPH oxidase activity or gp91phox expression in the neutrophils of female relatives of the patient, because the gene for gp91phox is located on the X chromosome (see section Carrier detection).
FACS analysis
For a protocol, see [23].
Western blotting
See Protocol 2.
Protocol 2. Western blotting of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase components
NB: A lysate from the control cells should also be made!
Materials:
Diisopropyl fluorophosphate (DFP)
1 N NaOH
Protease inhibiter mix (PIM, 25×; Roche)
×2 sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS/PAGE) sample buffer (2 × SB)
β-mercaptoethanol (if not already in 2 × SB)
0·5 M ethylenediamine tetraacetic acid (EDTA)
-
Antibodies: monoclonal antibody (mAb) 48 anti-gp91phox from Sanquin Reagents, Amsterdam
mAb 449 anti-p22phox from Sanquin Reagents, Amsterdam
mAb clone 10 anti-p47phox from Santa Cruz, cat. no. SC-17845
pAb rabbit anti-human p67phox from Upstate, cat. no. 07-002
Preparations:
Place heating block in fumehood at 95°C
If not already present, add 2% v/v β-mercaptoethanol to 2 × SB
-
Prepare PIM/EDTA: 40 μl of 25 × PIM
20 μl of 0·5 M EDTA
940 μl of PBS
Place 1 ml vial of 2 × SB and 1 ml vial of PIM/EDTA in heating-block
NB: DFP waste (pipette tips, supernatants) overnight in 1 N NaOH to inactivate DFP!
Method (in flowhood):
Resuspend PMNs at 5-10 × 106 in 1 ml of PBS
Add 1 μl of DFP, leave on ice for 10 min
Centrifuge, 10 s Eppendorf centrifuge, 21 000 g
Resuspend pellet in 50–100 μl PIM/EDTA 95°C, and place vial directly in heating-block (NB: supernatant should be added to 1 N NaOH!)
When all pellets are resuspended, add 50–100 μl 2 × SB (95°C) and leave vials (closed with screwing cap) on 95°C for 30 min, and store thereafter at −20°C
Western blot details:
Standard SDS/PAGE, 10% gel, and transfer to nitrocellulose
Blocking with 5% milk powder (1 h at room temperature); primary antibodies (2–4 μg/ml) against gp91, p22, p47 and/or p67 (overnight, 4°C); conjugates horseradish peroxidase (HRP)-labelled (Amersham) followed by visualization by enhanced chemiluminescence (ECL), or conjugate fluorescent-labelled (Licor), followed by scanning by Odyssey (dual fluorescent scanner, Licor)
NADPH oxidase component activity
If all five subunits of the NADPH oxidase are detectable, even though the activity of this enzyme is absent or strongly decreased, the possibility of so-called (+) variants must be considered. These are caused by mutations in any of the five subunits that leave the protein expression intact but destroy the enzymatic activity of the assembled oxidase complex. In that case, direct sequencing of all five genes can be considered. Alternatively, a cell-free oxidase assay may be used to distinguish a defect in a cytosolic component (p40phox, p47phox or p67phox) from a defect in a membrane-bound component (gp91phox or p22phox). For this assay, neutrophil membranes from the patient are mixed with neutrophil cytosol from a healthy donor (or vice versa), incubated with NADPH and γS-GTP, and activated with an amphiphilic agent [low concentrations of sodium dodecyl sulphate (SDS) or arachidonic acid] [27]. The resulting oxidase activity can be measured by superoxide formation or oxygen consumption and is used to localize the defect to either the cytosol or the membrane fraction. Identification of the mutated gene that causes the defect in NADPH oxidase activity can also be made if transfection of the patient's Epstein–Barr virus (EBV)-transformed B lymphocytes with retroviral vectors that contain the wild-type cDNA restores this activity [28].
Cell-free assay
For a detailed protocol, see [27].
Transfection of EBV-B cell lines
For a protocol, see [28].
Mutation analysis
The disease-causing mutation should be determined in every CGD patient. This is necessary for undisputable proof of which gene is affected, and as such the basis for genetic counselling. Carriers of the disease without clinical symptoms can only be diagnosed reliably by mutation analysis. Also, in case prenatal diagnosis or gene therapy is an option in the family, this information must be available. When patients are transplanted with stem cells from a family member, it is important to know that this donor is not carrying the mutation. Finally, this information helps investigators to link medical expression of CGD to the genetic cause.
Genomic DNA and RNA can be extracted from the mononuclear leucocyte fraction [peripheral blood mononuclear cells (PBMC)] obtained as a side product during neutrophil purification [12].
Gene sequencing
The CYBB, CYBA, NCF2 and NCF4 genes (for properties see Table 1) can be analysed from genomic DNA by polymerase chain reaction (PCR) amplification and sequencing. NCF1 is more difficult, because it is accompanied on each side by one pseudo-NCF1 gene. These pseudo-NCF1 genes are >99% homologous to NCF1 but lack a GT sequence at the start of exon 2, which induces a frame-shift and a premature termination of protein synthesis. Therefore, NCF1-specific PCR is difficult, because the primers have to contain NCF1-specific sequences at the segregating points between NCF1 and its pseudogenes. It is recommended, therefore, to first perform a gene scan [29] to determine whether only GT-deletion-containing pseudogenes are present or whether one or two NCF1 genes are present in the patient's DNA. In the latter case, sequencing of NCF1 cDNA usually reveals another mutation than the GT deletion in NCF1, which must then be confirmed with genomic DNA after NCF1-specific amplification of the relevant part of NCF1 [30]. If an entire exon is deleted without the presence of a mutation in the bordering exons, a splice-site mutation may be present in the bordering introns in the genomic DNA. This, too, must be analysed in NCF1-specific PCR amplicons. For protocols see [29,30].
Screening methods
Some investigators apply screening for a mutation in a PCR product to select the fragment to be sequenced. For this purpose, single-strand conformation polymorphism analysis [31], denaturing high-pressure liquid chromatography [32] or high-resolution melting analysis [33] can be used. Single-strand conformation polymorphism (SSCP) is based on the difference in electrophoresis profile between denatured patients' PCR products and wild-type PCR products in a polyacrylamide gel. PCR products with an aberrant migration pattern are then sequenced. Denaturing high-pressure liquid chromatography (DHPLC) is based on heteroduplex formation between a PCR product from a patient with a wild-type PCR product. In case the two PCR products differ, the elution profile of the heteroduplex over a column will differ from the profile seen with a wild-type homoduplex. Such PCR products are then sequenced. High-resolution melting analysis is based on the difference in melting curves between hetero- and homoduplexes. However, as a lack of aberrant signal does not guarantee a wild-type sequence in the patient's PCR product in any of these methods, such screening assays are not generally applied.
Confirmation assays
Splice-site mutations found in genomic DNA should be confirmed for their effect on mRNA splicing by analysing the lack of one or more exons in the cDNA of the patient. Also, the presence of large deletions, usually based on the lack of PCR product formation, should be confirmed by an independent assay, such as multiplex ligase-dependent probe amplification [34] or array comparative genomic hybridization [35]. Restriction fragment length polymorphism (RFLP) analysis is also possible, but this technique is tedious, requires a great deal of freshly purified genomic DNA and does not always lead to unequivocal results. Multiplex ligase-dependent probe amplification (MLPA), with a set of probes annealing at different positions, analyses which parts of a gene or gene-surrounding sequences are still present. In array comparative genomic hybridization (ACGH), DNA from a test sample and from a normal reference sample are labelled differently with fluorescent dyes and are then hybridized to a set of probes on a glass slide. The ratio of the fluorescence intensity of the test DNA to that of the reference DNA is then calculated, to measure the copy number changes for a particular gene or gene fragment.
Carrier detection
Carrier detection for autosomal forms of CGD can best be performed at the DNA level, by searching for the family-specific mutation, because NADPH oxidase protein expression and oxidase activity are close to normal in these carriers. Only in the X-linked form of CGD can the (female) carriers usually, but not always, be detected by a mosaic pattern of gp91phox-positive and -negative phagocytes, correlating with NADPH oxidase-positive and -negative cells (Table 2b). This is caused by the process of X-chromosome inactivation at an early stage of fetal development in all cells from female individuals. The X chromosome inactivated in a certain cell will also be inactive in all daughter cells derived from that cell. The inactivation process may hit either the wild-type or the mutated X chromosome, thus leaving a mixture of NADPH-competent and -incompetent haematopoietic precursor cells. However, because of the random process of X-chromosome inactivation, X-CGD carriers may show a near-normal or a near-pathological pattern in the expression or activity tests. Thus, a normal pattern does not exclude an individual as an X-CGD carrier. Conversely, females with a near-pathological pattern often present as X-CGD patients.
Oxidase activity or protein expression in single cells
Carrier detection of X-CGD is usually performed by searching for a mosaic pattern of oxidase-positive and -negative neutrophils in the NBT slide test or in the DHR flow-cytometric assay (see sections Superoxide production and Hydrogen peroxide generation). Alternatively, one can perform flow cytometry to detect gp91phox protein expression on the neutrophil surface with the anti-gp91phox monoclonal antibody 7D5 (see section NADPH oxidase component expression). However, it must be kept in mind that up to one-third of all X-linked defects may arise from new mutations in germline cells and will therefore not always be present in the somatic cells of the mother. Thus, failure to define the mother as an X-linked carrier does not disprove the X-linked origin of the disease, or even the possibility of the mother having another child with X-CGD. If a mosaic is found in the mother but no mutation is detectable in CYBB from the patient, the X-linked G6PD gene may carry a mutation.1
Gene sequencing
Once the family-specific mutation is known, it is more reliable to perform carrier detection for any of the CGD subtypes at the DNA level (see section Mutation analysis– Gene sequencing). However, in case the indicator patient has a complete deletion of CYBB (on the X chromosome), the mother cannot be defined as a carrier of this deletion by simple gene sequencing. MLPA or array CGH analysis can then be applied [36,37].
Prenatal diagnosis
Prenatal diagnosis of CGD can be performed by analysis of the NADPH oxidase activity of fetal blood neutrophils [38], but fetal blood sampling cannot be undertaken before 16–18 weeks of gestation. Instead, analysis of DNA from amniotic fluid cells or chorionic villi provides an earlier and more reliable diagnosis for families at risk. In cases where the family-specific mutation is known, this analysis can be performed by PCR amplification and sequencing of the relevant genomic DNA area [39]. Care must be taken to avoid contamination of fetal DNA with maternal DNA; detection of such contamination can be performed by short tandem repeat (STR) analysis. The same strategy as for prenatal diagnosis of X-CGD can be used for prenatal diagnosis of other CGD subtypes [40], although this may be more complicated if the parents each carry different mutations.
In cases where the family-specific mutations are not known, different methods must be applied. Partial or complete gene deletions can be recognized by MLPA or array CGH analysis of genomic DNA, but more subtle abnormalities require the use of allele-specific markers. The MLPA or CGH probes and the allele-specific markers should be chosen in the surroundings of the gene that is supposed to be mutated.
Step-by-step protocols for laboratory diagnostics (short and extensive) are given in Tables 3 and 4 and in Fig. 1.
Table 3.
Short protocol for chronic granulomatous disease (CGD) laboratory diagnostics
| 1. Draw blood from the patient and preferably also from the parents. Always draw blood from a control individual as well. Keep raw data records of each subsequent assay. |
| 2. Determine nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity in blood phagocytes (purified neutrophils, total leucocytes or diluted whole blood). Tests: dihydrorhodamine-1,2,3 (DHR) test [if a fluorescence activated cell sorter (FACS) machine is available] or nitro-blue tetrazolium (NBT) slide test. Stimulus: phorbol myristate acetate (PMA). If DHR is used, be aware of myeloperoxidase (MPO) deficiency, which may cause a false-negative NADPH oxidase activity result. |
| 3. In the DHR test, if the patient's cells show less than 50% of normal NADPH oxidase activity, CGD is likely. In the NBT slide test, if the patient's cells are not or only slightly stained, CGD is likely. Contact a reference centre for further detailed functional and mutational analysis. Females with a clear mixture of DHR- or NBT-positive and -negative cells are usually carriers of X-chromosome-linked CGD (X-CGD). |
Table 4.
Step-by-step protocol for detailed chronic granulomatous disease (CGD) laboratory diagnostics (see also Fig. 1)
| 1. Draw blood from the patient and preferably also from the parents. Always draw blood from a control individual as well. Keep raw data records of each subsequent assay. |
| 2. Determine nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity in blood phagocytes (purified neutrophils, total leucocytes or diluted whole blood). Preferred (semi-) quantitative assays: dihydrorhodamine-1,2,3 (DHR) or Amplex Red. Preferred stimuli: phorbol myristate acetate (PMA) and/or serum-treated zymosan. Optional: zymosan and platelet-activating factor /formyl-methionyl-leucyl-phenylalanine (PAF/fMLP). Preferred qualitative assays: DHR or nitro-blue tetrazolium (NBT) slide test with PMA as the stimulus. If the mother shows a mosaic in one of these last assays, sequence CYBB in the patient; if CYBB is normal, sequence the G6PD gene in the patient. |
| 3. If NADPH oxidase activity in the patient's cells is less than 50% of normal [and myeloperoxidase (MPO) activity is normal]: repeat the assay with a second blood sample, preferably with an alternative assay. Determine NADPH oxidase component expression in lysate from purified neutrophils by Western blotting, on intact neutrophils [with monoclonal antibody (mAb) 7D5 for gp91phox] or in permeabilized neutrophils (p22phox, p40phox, p47phox, p67phox) by fluorescence activated cell sorter (FACS) analysis. |
| 4. If p22phox and gp91phox expression are decreased for more than 50%, sequence CYBB and/or CYBA. If the NADPH oxidase activity in the mother's cells shows a mosaic, start with CYBB. |
| 5. If p67phox or p40phox expression is decreased, sequence NCF2 or NCF4, respectively. |
| 6. If p47phox expression is decreased, perform a gene scan. If only NCF1 pseudogenes are present, the patient is homozygous for delta-GT in NCF1. If one or two NCF1 genes are present, sequence NCF1 cDNA. The mutation must be confirmed in genomic DNA after NCF1-specific amplification. |
| 7. If all NADPH oxidase components are expressed normally and the mother shows a mosaic in the NADPH oxidase activity test: sequence CYBB and – if that reveals no mutation – the G6PD gene in the patient. If the mother shows no mosaic, sequence CYBB, thereafter if necessary CYBA, NCF1, NCF2 or NCF4. Alternatively, one can perform a cell-free assay or transfect Epstein–Barr virus (EBV)-B cells from the patient with wild-type cDNA of the NADPH oxidase components to obtain a clue in which gene the CGD mutation might reside. |
| 8. Mutations must be confirmed by sequencing of parents' DNA. Splice site mutations should be confirmed by cDNA analysis. Carriers of large deletions can be found with multiplex ligase-dependent probe amplification (MLPA) or array comparative genomic hybridization (CGH). |
Acknowledgments
D. R. obtained financial support from the Chronic Granulomatous Disorder Society, London, UK, and from the European Commission E-Rare program (EURO-CGD grant).
Footnotes
Deficiencies in p40phox have been described only once [24] and give rise to a slightly aberrant form of CGD, with a substantial defect in intracellular neutrophil superoxide production during phagocytosis, but unaffected extracellular release of superoxide elicited by phorbol ester or fMLP. Similarly, a defect in Rac2, a small GTPase needed for NADPH oxidase activation, is also extremely rare and induces not only a deficiency in superoxide production with all stimuli tested except phorbol ester (thus leading to CGD), but also in chemotaxis and enzyme release from neutrophils [25]. Finally, CGD-like phenomena are also seen in some patients with a rare, severe form of glucose-6-phosphate dehydrogenase (G6PD) deficiency that manifests not only in the red cells but also in the leucocytes [26]. This deficiency may give rise to lack of sufficient substrate (NADPH) generation for the NADPH oxidase, and thus to a usually mild type of CGD, characterized by a substantial residual neutrophil NADPH oxidase activity with all stimuli tested.
Disclosure
The authors declare no conflicting interests.
Author contributions
D. R. and M. d. B. wrote the paper together.
References
- 1.Roos D, Holland SM, Kuijpers TW. Chronic granulomatous disease. In: Ochs HD, Smith CIE, Puck JM, editors. Primary immunodeficiency diseases. A molecular and genetic approach. 3rd edn. New York: Oxford University Press; 2013. in press. [Google Scholar]
- 2.Reeves EP, Lu H, Jacobs HL, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 3.Chang YC, Segal BH, Holland SM, et al. Virulence of catalase-deficient Aspergillus nidulans in p47-phox–/– mice. Implications for fungal pathogenicity and host defense in chronic granulomatous disease. J Clin Invest. 1998;101:1843–1850. doi: 10.1172/JCI2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messina CG, Reeves EP, Roes J, Segal AW. Catalase-negative Staphylococcus aureus retain virulence in mouse model of chronic granulomatous disease. FEBS Lett. 2002;518:107–110. doi: 10.1016/s0014-5793(02)02658-3. [DOI] [PubMed] [Google Scholar]
- 5.Köker MY, Camcıoğlu Y, Van Leeuwen K, et al. Clinical, functional and genetic characterization of eighty-nine patients in Turkey with chronic granulomatous disease. J Allergy Clin Immunol. 2013;132:1156–1163. doi: 10.1016/j.jaci.2013.05.039. [DOI] [PubMed] [Google Scholar]
- 6.Kuhns DB, Alvord WG, Heller T, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho CML, Vowels MR, Lockwood L, Ziegler JB. Successful bone marrow transplantation in a child with X-linked chronic granulomatous disease. Bone Marrow Transplant. 1996;18:213–215. [PubMed] [Google Scholar]
- 8.Seger RA, Gungor T, Belohradsky BH, et al. Treatment of chronic granulomatous disease with myeloablative conditioning and an unmodified hematopoietic allograft: a survey of the European experience 1985–2000. Blood. 2002;100:4344–4350. doi: 10.1182/blood-2002-02-0583. [DOI] [PubMed] [Google Scholar]
- 9.Åhlin A, Fugeläng J, de Boer M, Ringden O, Fasth A, Winiarski J. Chronic granulomatous disease – haematopoietic stem cell transplantation versus conventional treatment. Acta Paediatr. 2013;102:1087–1094. doi: 10.1111/apa.12384. [DOI] [PubMed] [Google Scholar]
- 10.Ott MG, Schmidt M, Schwarzwälder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 11.Malech HL, Maples PB, Whiting-Theobald N, et al. Prolonged production of NADPH oxidase-corrected granulocytes after gene therapy of chronic granulomatous disease. Proc Natl Acad Sci USA. 1997;94:12133–12138. doi: 10.1073/pnas.94.22.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roos D, de Boer M. Purification and cryopreservation of phagocytes from human blood. Meth Enzymol. 1986;132:225–243. doi: 10.1016/s0076-6879(86)32010-x. [DOI] [PubMed] [Google Scholar]
- 13.Weening RS, Roos D, Loos JA. Oxygen consumption of phagocytizing cells in human leukocyte and granulocyte preparations: a comparative study. J Lab Clin Med. 1974;83:570–577. [PubMed] [Google Scholar]
- 14.Elloumi HZ, Holland SM. Diagnostic assays for chronic granulomatous disease and other neutrophil disorders. Methods Mol Biol. 2007;412:505–523. doi: 10.1007/978-1-59745-467-4_31. [DOI] [PubMed] [Google Scholar]
- 15.Meerhof LJ, Roos D. Heterogeneity in chronic granulomatous disease detected with an improved nitroblue tetrazolium slide test. J Leukoc Biol. 1986;39:699–711. doi: 10.1002/jlb.39.6.699. [DOI] [PubMed] [Google Scholar]
- 16.Roesler J. Remarks on the article Genetics and immunopathology of chronic granulomatous disease by Marie José Stasia and Xing Jun Li. Semin Immunopathol. 2008;30:365. doi: 10.1007/s00281-008-0129-0. [DOI] [PubMed] [Google Scholar]
- 17.Dahlgren C, Karlsson A, Bylund J. Measurement of respiratory burst products generated by professional phagocytes. Methods Mol Biol. 2007;412:349–363. doi: 10.1007/978-1-59745-467-4_23. [DOI] [PubMed] [Google Scholar]
- 18.Emmendörffer A, Nakamura M, Rothe G, Spiekermann K, Lohmann-Matthes M-L, Roesler J. Evaluation of flow cytometric methods for diagnosis of chronic granulomatous disease variants under routine laboratory conditions. Cytometry. 1994;18:147–155. doi: 10.1002/cyto.990180306. [DOI] [PubMed] [Google Scholar]
- 19.Van Pelt LJ, van Zwieten R, Weening RS, Roos D, Verhoeven AJ, Bolscher BG. Limitations on the use of dihydrorhodamine-1,2,3 for flow cytometric analysis of the neutrophil respiratory burst. J Immunol Methods. 1996;191:187–196. doi: 10.1016/0022-1759(96)00024-5. [DOI] [PubMed] [Google Scholar]
- 20.Vowells SJ, Fleisher TA, Sekhsaria S, Alling DW, Maguire TE, Malech HL. Genotype-dependent variability in flow cytometric evaluation of reduced nicotinamide adenine dinucleotide phosphate oxidase function in patients with chronic granulomatous disease. J Pediatr. 1996;128:104–107. doi: 10.1016/s0022-3476(96)70437-7. [DOI] [PubMed] [Google Scholar]
- 21.Mauch L, Lun A, O'Gorman MRG, et al. Chronic granulomatous disease (CGD) and complete myeloperoxidase deficiency both yield strongly reduced dihydrorhodamine 123 test signals but can be easily discerned in routine testing for CGD. Clin Chem. 2007;53:890–896. doi: 10.1373/clinchem.2006.083444. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi A, Yu L, Pötgens AJ, Kuribayashi F, et al. Location of the epitope for 7D5, a monoclonal antibody raised against human flavocytochrome b558, to the extracellular peptide portion of primate gp91phox. Microbiol Immunol. 2001;45:249–257. doi: 10.1111/j.1348-0421.2001.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 23.Köker MY, Sanal O, van Leeuwen K, et al. Four different NCF2 mutations in six families from Turkey and an overview of NCF2 gene mutations. Eur J Clin Invest. 2009;39:942–951. doi: 10.1111/j.1365-2362.2009.02195.x. [DOI] [PubMed] [Google Scholar]
- 24.Matute JD, Arias AA, Wright NA, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambruso DR, Knall C, Abell AN, et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci USA. 2000;97:4654–4659. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Bruggen R, Bautista JM, Petropoulou T, et al. Deletion of leucine-61 in glucose-6-phosphate dehydrogenase leads to chronic nonspherocytic anemia, granulocyte dysfunction, and increased susceptibility to infections. Blood. 2002;100:1026–1030. doi: 10.1182/blood.v100.3.1026. [DOI] [PubMed] [Google Scholar]
- 27.Molshanski-Mor S, Mizrahi A, Ugolev Y, Dahan I, Berdichevsky Y, Pick E. The reductionist approach to the study of NADPH oxidase assembly, or ‘all you wanted to know about cell-free assays but did not dare to ask’. Methods Mol Biol. 2007;412:385–428. doi: 10.1007/978-1-59745-467-4_25. [DOI] [PubMed] [Google Scholar]
- 28.Chanock SJ, Faust LR, Barrett D, et al. O2− production by B lymphocytes lacking the respiratory burst oxidase subunit p47phox after transfection with an expression vector containing a p47phox cDNA. Proc Natl Acad Sci USA. 1992;89:10174–10177. doi: 10.1073/pnas.89.21.10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekker J, de Boer M, Roos D. Gene-scan method for the recognition of carriers and patients with p47-phox-deficient autosomal recessive chronic granulomatous disease. Exp Hematol. 2001;29:1319–1325. doi: 10.1016/s0301-472x(01)00731-7. [DOI] [PubMed] [Google Scholar]
- 30.Roos D, de Boer M, Köker MY, et al. Chronic granulomatous disease caused by mutations other than the common GT deletion in NCF1, the gene encoding the p47phox component of the phagocyte NADPH oxidase. Hum Mutat. 2006;27:1218–1229. doi: 10.1002/humu.20413. [DOI] [PubMed] [Google Scholar]
- 31.Patiño PJ, Perez JE, Lopez JA, et al. Molecular analysis of chronic granulomatous disease caused by defects in gp91-phox. Human Mutat. 1999;13:29–37. doi: 10.1002/(SICI)1098-1004(1999)13:1<29::AID-HUMU3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 32.Di Matteo G, Giordani L, Finocchi A, et al. Molecular characterization of a large cohort of patients with chronic granulomatous disease and identification of novel CYBB mutations: an Italian multicenter study. Mol Immunol. 2009;46:1935–1941. doi: 10.1016/j.molimm.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Hill HR, Augustine NH, Pryor RJ, et al. Rapid genetic analysis of X-linked chronic granulomatous disease by high-resolution melting. J Mol Diagn. 2010;12:368–376. doi: 10.2353/jmoldx.2010.090147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breunis WB, van Mirre E, Bruin M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111:1029–1038. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- 35.Shinawi M, Cheung SW. The array CGH and its clinical applications. Drug Discov Today. 2008;13:760–770. doi: 10.1016/j.drudis.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Stasia MJ, van Leeuwen K, de Boer M, et al. Rare duplication or deletion of exons 6, 7 and 8 in CYBB leading to X-linked chronic granulomatous disease in two patients from different families. J Clin Immunol. 2012;32:653–662. doi: 10.1007/s10875-012-9667-2. [DOI] [PubMed] [Google Scholar]
- 37.Stasia MJ, Mollin M, Martel C, et al. Functional and genetic characterization of two extremely rare cases of Williams–Beuren syndrome associated with chronic granulomatous disease. Eur J Hum Genet. 2013;21:1079–1084. doi: 10.1038/ejhg.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newburger PE, Cohen HJ, Rothchild SB, Hobbins JC, Malawista SE, Mahoney MJ. Prenatal diagnosis of chronic granulomatous disease. N Engl J Med. 1979;300:178–181. doi: 10.1056/NEJM197901253000406. [DOI] [PubMed] [Google Scholar]
- 39.De Boer M, Bolscher BGJM, Sijmons RH, Scheffer H, Weening RS, Roos D. Prenatal diagnosis in a family with X-linked chronic granulomatous disease with the use of the polymerase chain reaction. Prenat Diagn. 1992;12:773–777. doi: 10.1002/pd.1970120910. [DOI] [PubMed] [Google Scholar]
- 40.De Boer M, Singh V, Dekker J, Di Rocco M, Goldblatt D, Roos D. Prenatal diagnosis in two families with autosomal, p47-phox-deficient chronic granulomatous disease due to a novel point mutation in NCF1. Prenat Diagn. 2002;22:235–240. doi: 10.1002/pd.296. [DOI] [PubMed] [Google Scholar]