Abstract
Biotransformation of inorganic arsenic (iAs) is one of the factors that determines the character and magnitude of the diverse detrimental health effects associated with chronic iAs exposure, but it is unknown how iAs biotransformation may impact the epigenome. Here, we integrated analyses of genome-wide, gene-specific promoter DNA methylation levels of peripheral blood leukocytes (PBLs) with urinary arsenical concentrations of subjects from a region of Mexico with high levels of iAs in drinking water. These analyses revealed dramatic differences in DNA methylation profiles associated with concentrations of specific urinary metabolites of arsenic. The majority of individuals in this study had positive indicators of arsenic-related disease, namely pre-diabetes mellitus or diabetes mellitus. Methylation patterns of genes with known associations to diabetes mellitus were associated with urinary concentrations of specific iAs metabolites. Future studies will determine whether these DNA methylation profiles provide mechanistic insight into the development of iAs-associated disease, predict disease risk, and/or serve as biomarkers of iAs exposure in humans.
Keywords: arsenic, epigenome, DNA methylation, arsenic biotransformation
INTRODUCTION
Millions of people worldwide are exposed to concentrations of inorganic arsenic (iAs) in their drinking water that exceed the World Health Organization’s recommended limit of 10 ppb [1, 2]. Chronic iAs exposure has been associated with a variety of adverse health effects in humans, collectively known as arsenicosis. These effects include characteristic skin lesions, cancers of the skin and various internal organs, neurological disorders, cardiovascular disease, and diabetes mellitus (DM) [3]. The mode of action (MOA) of iAs is complex, and the etiology of iAs-associated diseases likely involves multiple mechanisms. Deleterious effects believed to play important roles in iAs-associated disease development include the generation of oxidative stress, the formation of various genetic aberrations, the binding and inhibition of iAs metabolites to enzymes, and perturbation of key signaling pathways [4–7].
Epigenetic alterations are also believed to play an important role in the MOA of iAs. For example, changes in the patterns/levels of DNA methylation, histone post-translational modifications and microRNAs have been observed after iAs exposure in laboratory studies and/or in human populations [8]. These alterations have the potential to greatly impact cellular homeostasis as each of these epigenetic components plays a crucial role in regulating gene expression [9]. The impact on DNA methylation patterns is the most extensively-studied epigenetic alteration associated with arsenic (As) exposure. Importantly, there are several examples in the literature in which alterations in DNA methylation patterns have been implicated as mediators of As toxicity [10–13].
The biotransformation of iAs, which produces trivalent and pentavalent monomethylated and dimethylated arsenicals (MMAs and DMAs, respectively) [14], also plays an important role in the development of iAs-associated disease. The concentrations and proportions of iAs metabolites detected in the urine of chronically-exposed populations can vary considerably between individuals in which total urinary As is comprised of ~10–20% iAs, ~10–20% MMAs, and ~60–80% DMAs [15]. These arsenicals differ in their biological effects. Specifically, methylated trivalent arsenicals, namely monomethylarsonous acid (MMAIII) and dimethylarsinous acid (DMAIII), are the most toxic forms [16, 17]. Therefore, differences in the levels of urinary arsenicals likely influence the risk of iAs-associated disease in exposed individuals. An individual’s capacity to biotransform iAs indeed appears to influence disease risk as high urinary proportions of MMAs and/or high ratios of MMAs/DMAs have been associated with the development of several iAs-associated diseases [18, 19]. The biotransformation of iAs may impact disease development not only by generating highly reactive and toxic metabolites, but through the process of iAs biotransformation itself, which requires S-adenosyl methionine (SAM), the same methyl group source required for DNA methylation [20, 21]. Arsenic-induced alterations in methionine metabolism are one of the major proposed mechanisms by which iAs may alter DNA methylation status. Aside from direct consumption of SAM during iAs biotransformation, iAs exposure can alter methionine metabolism through the consumption of the cellular reductant glutathione (GSH). GSH and SAM metabolic pathways are biochemically linked due to their shared requirement for homocysteine [22]. Exposure to iAs can considerably deplete cellular GSH levels. GSH is consumed during iAs biotransformation [23] and is excreted from the cell as As-glutathione conjugates, which serve as a major defense mechanism against As-induced oxidative stress [24, 25]. Changes in SAM availability due to iAs exposure/biotransformation may therefore perturb DNA methylation patterns and subsequently influence disease risk by altering the expression of key genes. To our knowledge, the relationship between DNA methylation patterns and the urinary concentrations of iAs, MMAs, and DMAs in exposed populations has not been investigated.
Here, we set out to examine the relationship between genome-wide, gene-specific promoter DNA methylation levels of peripheral blood leukocytes (PBLs) and urinary arsenical concentrations in individuals from an endemic arsenicosis population in Zimapán, Mexico [26, 27]. Importantly, half of these individuals have skin lesions characteristic of arsenicosis, and we previously identified genes with differentially methylated promoters in PBLs from these individuals as they relate to skin lesion status [28]. Here, we expand this effort to examine the relationship between PBL DNA methylation status and urinary arsenical levels across these individuals.
MATERIALS AND METHODS
Study Subjects
The subjects in this study are described in detail elsewhere [28]. Briefly, these subjects are part of a larger population from Zimapán, Hildago, Mexico who are exposed to varying concentrations of iAs in their drinking water. Approximately one half of the 46,000 residents living in the area consume drinking water containing higher concentrations of iAs (21–1100 ppb; mean of 110 ppb) than the World Health Organization’s recommended maximum contaminant level of 10 ppb [1, 27]. Spot urine samples were used for arsenical analyses. Total arsenic (tAs) concentrations were measured by hydride generation atomic fluorescence spectrometry (HG-AFS). The concentrations of iAs (trivalent + pentavent), MMAs (trivalent + pentavent), and DMAs (trivalent + pentavalent) were determined by hydride generation atomic absorption spectrometry (HG-AAS) with cryotrapping [27]. The percentage of hemoglobin A1C (HbA1c) in fasting blood of each subject was determined using GDX A1c test cartridges (Cholestech Corp., Hayward, CA) [27].
Association Between Promoter DNA Methylation Levels and Urinary Arsenical Concentrations
Methylated DNA was extracted from peripheral blood leukocytes (PBLs) of 16 females, amplified, and hybridized to Affymetrix Human Promoter 1.0R arrays (Affymetrix, Santa Clara, CA) as previously described [28]. These arrays represent >25,500 human promoter regions, ~14,000 of which contain CpG islands, known targets of DNA methylation. Data were normalized using robust multi-chip average (RMA) and bioinformatically summarized at the CpG island level [29].
Associations between DNA methylation levels and the individual concentrations of iAs, MMAs, DMAs in urine were tested while adjusting for age as a potential confounder. Pearson correlation coefficients were calculated to describe the relationship between urinary arsenical concentrations and promoter DNA methylation levels, and p-values were computed for each correlation coefficient. In order to be considered for further analysis, genes were required to achieve nominal significance (p-value<0.05). Hierarchical clustering was performed using the correlation coefficients of the 812 unique genes that achieved nominal significance for at least one of the arsenical groups using Partek Genomics Suite™ (version 6.5) software (Partek, Inc. St. Louis, MO).
Genes were subsequently analyzed for associated biological functions and canonical pathways using IPA software (Ingenuity Systems, Redwood City, CA). The Ingenuity Knowledge Base within IPA is a literature-based database of molecular interactions and functional annotations based on known relationships between cells, cellular components, drugs and diseases. All p-values were calculated in IPA using a right-tailed Fisher’s exact test, which determines the probability that associated functions and canonical pathways were generated due to chance alone. Individual genes in the DM pathways of interest were considered to have a statistically significant association between promoter DNA methylation levels and concentrations of iAs, MMAs and/or DMAs if they passed the stringent statistical filters of (1) association p-value<0.05 (2) a false discovery rate (FDR) q-value<0.2 [30–32].
RESULTS
Assessment of Urinary Arsenicals and Indicators of Diabetes Mellitus in Study Participants
The concentrations and proportions of iAs, MMAs, and DMAs in urine and percentage of glycosylated hemoglobin (%HbA1c) in blood for each of the 16 study subjects were measured (Table 1). Total As (tAs) measurements ranged between 3.6–31.8 ng As/ml urine across the 16 study subjects with an average of 10.7 ng/ml (Table 1). Measurements of the three major urinary arsenical species were 0.3–4.8 ng iAs/ml, 0.6–6.1 ng MMAs/ml, and 2.3–22.0 ng DMAs/ml. The proportions (% of total urinary As) ranged from 5.3–21.4% for iAs, 10.3–28.9% for MMAs, and 49.3–84.9% for DMAs. The %HbA1c values ranged from 5.4–10.6, with an average of 6.7. These results indicate that eight of the 16 study subjects had positive indicators of diabetes (HbA1c≥6.5%) and seven had positive indicators of pre-diabetes (HbA1c=5.7–6.4%) (Table 1) [33]. Of the eight individuals with iAs-associated skin lesions, five had positive indicators of diabetes and three had positive indicators of pre-diabetes.
TABLE 1.
Urinary Arsenical and Blood HbA1c Measurements in Study Subjects
| Study subject1 |
TAs (ng/ml) |
iAs (ng/ml) |
MMAs (ng/ml) |
DMAs (ng/ml) |
iAs (%TAs) |
MMAs (%TAs) |
DMAs (%TAs) |
%HbA1c |
|---|---|---|---|---|---|---|---|---|
| 1 | 7.8 | 1.5 | 1.7 | 5.3 | 19.3 | 21.2 | 68.0 | 6.2 |
| 2 | 4.3 | 0.6 | 0.6 | 2.3 | 13.0 | 14.9 | 54.9 | 7.2 |
| 3 | 4.6 | 0.7 | 0.6 | 2.3 | 15.4 | 13.1 | 49.3 | 5.4 |
| 4 | 13.0 | 2.3 | 3.1 | 8.0 | 17.9 | 24.2 | 62.0 | 6 |
| 5 | 8.3 | 1.8 | 0.9 | 5.9 | 21.4 | 11.0 | 71.8 | 6.6 |
| 6 | 13.0 | 1.3 | 2.3 | 9.5 | 9.9 | 17.4 | 73.4 | 6 |
| 7 | 5.9 | 0.3 | 1.5 | 4.4 | 5.3 | 25.5 | 74.8 | 6.1 |
| 8 | 7.6 | 1.3 | 2.2 | 4.5 | 16.8 | 28.9 | 59.1 | 6.8 |
| 92 | 3.6 | 0.3 | 0.6 | 3.0 | 9.0 | 16.0 | 84.9 | 6.3 |
| 102 | 5.4 | 0.7 | 1.1 | 4.4 | 13.3 | 19.9 | 81.0 | 6.5 |
| 112 | 31.8 | 4.8 | 6.1 | 22.0 | 15.2 | 19.1 | 69.1 | 8.2 |
| 122 | 14.5 | 1.4 | 1.6 | 11.0 | 9.8 | 10.7 | 75.8 | 6.5 |
| 132 | 20.4 | 1.2 | 2.1 | 16.0 | 6.0 | 10.3 | 78.5 | 10.6 |
| 142 | 8.4 | 0.6 | 0.9 | 7.0 | 7.0 | 10.9 | 82.7 | 6 |
| 152 | 8.4 | 1.6 | 1.2 | 6.7 | 19.7 | 14.2 | 79.5 | 6.2 |
| 162 | 13.9 | 1.7 | 2.0 | 10.2 | 12.4 | 14.2 | 73.4 | 7.8 |
| μ3=10.7 | μ=1.4 | μ=1.4 | μ=7.7 | μ=13.2 | μ=17.7 | μ=71.1 | μ=6.7 |
Genome-wide, Gene-specific DNA Methylation Patterns Associate with Urinary Arsenical Concentrations
Considering the susceptibility to iAs-related diseases has been associated with both the level of iAs exposure and iAs biotransformation capacity [19, 34–39], we examined the relationship between the concentrations of urinary arsenicals and promoter DNA methylation levels across >14,000 genes. Analyses were carried out to identify all genes for which promoter DNA methylation was associated with the urinary concentrations of iAs and the iAs metabolites MMAs and DMAs across the subcohort. For this analysis, a total of 812 unique genes were identified. These corresponded to 455 genes with promoter DNA methylation levels associated with iAs, 556 with MMAs, and 121 with DMAs (Figure 1) (see online supporting material [40]). There was some overlap in terms of common genes between the three arsenical groups, with a minimum overlap of 47% (i.e. 264 of the 556 MMAs-associated genes were also present in the iAs and DMAs groups) and a maximal overlap of 72.7% (88/121) for the DMAs group (Figure 1).
FIGURE 1.
Venn diagram illustrating the number of genes that have a statistically significant association between promoter DNA methylation levels and urinary concentrations of iAs, MMAs and/or DMAs. The five most significant canonical pathways associated with the genes in each group are displayed. Major functions associated with these pathways include: 1cellular growth and development, 2inflammatory response/immune response; 3nuclear receptor signaling, 4cellular stress/injury, 5carbohydrate metabolism, 6intercellular/intracellular signaling, and 7apoptosis.
Functional analyses of the proteins encoded by the differentially methylated genes revealed they represented diverse functions in the cell. The five most significant canonical pathways associated with the total number of genes from each group are indicated in Figure 1. While many of the canonical pathways were distinct for each arsenical class, common themes emerge. For example, genes that play a role in the transforming growth factor-beta (TGF-β) pathway have DNA methylation patterns that are associated with each of the arsenicals. Major functions associated with these canonical pathways include cellular growth and development, inflammatory response/immune response, nuclear receptor signaling, cellular stress/injury, carbohydrate metabolism, intercellular/intracellular signaling, and apoptosis (Figure 1).
Across the cohort, the majority of genes show decreased promoter DNA methylation levels with increasing concentrations of urinary arsenicals for each arsenical group (Figure 2). For example, there were 493 genes out of 556 total genes (or 88.6%) that had decreasing promoter DNA methylation levels with increasing urinary concentrations of MMAs (Figure 2) [40].
FIGURE 2.
Hierarchical clustering of the association coefficients of 812 unique genes that had a statistically significant association between promoter DNA methylation status and concentrations of iAs, MMAs, and/or DMAs in urine. The number of genes that were positively and negatively correlated with concentrations of arsenical is indicated (# positive/# negative) and represented by red and blue tones, respectively.
Association Between DNA Methylation Levels of Diabetes Mellitus-Related Genes and Urinary Arsenicals
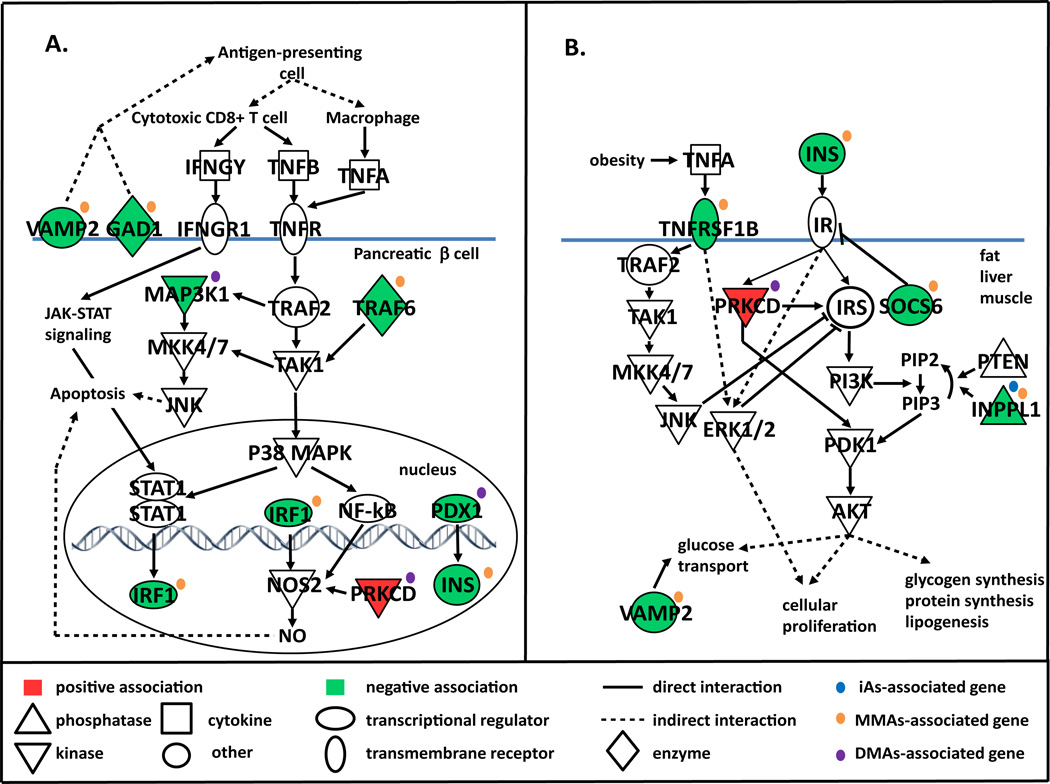
The type 2 DM (T2DM) canonical pathway was among the most significant perturbed pathways associated with urinary concentrations of DMAs (Figure 1). In addition, the type 1 DM (T1DM) pathway was significantly associated with MMAs-associated genes (p=1.0×10−2; results not shown). Together, these genes include those involved in one or more processes associated with DM such as destruction of insulin–producing pancreatic β cells (T1DM and T2DM) and altered insulin signaling in peripheral tissues associated with T2DM (Figure 3A, B, Table 2).
FIGURE 3.
Promoter DNA methylation levels of diabetes mellitus (DM)-associated genes correlated with urinary concentrations (ng/ml) of iAs, MMAs, and/or DMAs. DM-associated genes include those involved in (A) pancreatic β-cell apoptosis particularly related to T1DM and (B) insulin signaling/insulin resistance in peripheral tissues that are primarily associated with T2DM. Genes with promoter DNA methylation levels that had a positive or negative association with urinary arsenical concentrations are shaded in red and green, respectively.
TABLE 2.
Promoter DNA Methylation Levels of Diabetes Mellitus-Related Genes Associated with Urinary Arsenical Concentrations
| Gene symbol |
Gene name | Promoter methylation change with increasing urinary arsenical (ng/ml) |
Arsenical group (ng/ml) |
Association with diabetes mellitus (DM) |
|---|---|---|---|---|
| GAD1 | glutamate decarboxylase 1 | hypomethylation | MMAs | Major β cell autoantigen in T1DM [43] |
| INPPL1 | inositol polyphosphate phosphatase-like 1 | hypomethylation | iAs MMAs | Negative regulator of insulin signaling [46]; expression associated with insulin resistance [70] |
| INS | insulin | hypomethylation | MMAs | Promotes glucose uptake from blood into peripheral tissues [71] |
| IRF1 | interferon regulatory factor 1 | hypomethylation | MMAs | Promotes β cell apoptosis [41] |
| MAP3K1 | mitogen-activated protein kinase kinase kinase 1 | hypomethylation | DMAs | Promotes β cell apoptosis [52] |
| PDX1 | pancreatic and duodenal homeobox 1 | hypomethylation | DMAs | Transcriptional activator of insulin gene; important in β cell differentiation, development, function and survival [50, 51] |
| PRKCD | protein kinase C delta | hypermethylation | DMAs | Promotes β cell apoptosis [53]; regulates insulin signaling [54] |
| TNFRSF1B | tumor necrosis factor receptor superfamily, member 1B | hypomethylation | MMAs | Regulator of TNFa-mediated apoptosis [72]; levels of plasma soluble fraction of receptor positively correlated with insulin resistance [45], severity of diabetic retinopathy [73], and predicting risk of end stage renal disease [74], and chronic kidney disease [75] in diabetics |
| TRAF6 | TNF receptor-associated factor 6 | hypomethylation | MMAs | Promotes β-cell apoptosis [42] |
| SOCS6 | suppressor of cytokine signaling 6 | hypomethylation | MMAs | Regulator of insulin signaling [47, 48] |
| VAMP2 | vesicle-associated membrane protein 2 | hypomethylation | MMAs | Minor antigen in T1DM [44]; involved in translocation of glucose transporter to cell membrane, overexpressed in insulin-resistant tissue [49] |
Two of the MMAs-associated genes are involved in β cell destruction by signaling through two mitogen-activated protein kinase (MAPK) signaling cascades, P38 or c-Jun N-terminal kinases (JNK), or through transcription factor nuclear factor kappa beta (NF-κB). These genes include interferon regulator factor (IRF1) and TNF-receptor-associated factor 6 (TRAF6) [41, 42]. MMAs-associated genes also include the major β cell autoantigen glutamate decarboxylase 1 (GAD1) [43], minor B cell autoantigen vesicle-associated membrane protein 2 (VAMP2) [44] (associated with T1DM) and genes involved in insulin signaling in peripheral tissues, namely tumor necrosis factor receptor superfamily, member 1B (TNFRSF1B), inositol polyphosphate phosphatase-like 1 (INPPL1), suppressor of cytokine signaling 6 (SOC6), and VAMP2 [45–49]. DMAs-associated genes included those involved in insulin signaling and β cell homeostasis such as pancreatic and duodenal homeobox 1 (PDX1) [50, 51]; β cell apoptosis, i.e. mitogen-activated protein kinase kinase kinase 1 (MAP3K1) [52], and both insulin signaling and β cell apoptosis, namely protein kinase C delta (PRKCD) [53, 54]. The DM-related gene associated with urinary iAs concentrations was the insulin signaling regulator INPPL1. With the exception of PRKCD (DMAs-associated gene), the promoter DNA methylation levels of each of these aforementioned DM-related genes decreased with increasing urinary arsenical concentrations (Figure 3, Table 2).
DISCUSSION
In this work, we set out to describe the relationship between promoter DNA methylation levels of PBLs and urinary arsenical concentrations in selected individuals from an arsenicosis endemic population in Zimapán, Mexico. Importantly, urinary arsenical levels and alterations in DNA methylation profiles are implicated in the development of iAs-associated disease. However, to our knowledge, this is the first analysis of the association between genome-wide, gene-specific PBL DNA methylation patterns and the concentrations of urinary arsenicals in an exposed human population.
As expected, the levels and proportions of each urinary arsenical differed among these 16 individuals and were consistent with those of other chronically-exposed populations [15, 55]. Distinct gene sets were identified that showed patterns of DNA methylation associated with iAs, MMAs and/or DMAs concentrations. Most genes showed decreased promoter DNA methylation levels with increasing urinary arsenical concentrations. Interestingly, these patterns of methylation differ from our previous work in which we detailed a general trend of gene specific-hypermethylation associated with skin lesion status [28]. These results underscore that patterns of DNA methylation likely differ when analyzed in the context of exposure or disease.
The results presented here indicate that different urinary arsenical levels, and thus differences in iAs metabolism, are indeed associated with different patterns of gene-specific DNA methylation. Epigenetic alterations have been considered likely crucial events that link toxicant exposure to environmentally-induced disease [56]. Considering the risk of iAs-related disease has been associated with both iAs exposure dose and iAs biotransformation capacity [19, 34–39], these arsenical concentration-specific and arsenical-specific DNA methylation profiles may help inform the risk and/or mechanisms of disease development in iAs-exposed populations.
This aforementioned hypothesis is particularly attractive considering the identification here of altered DNA methylation levels of several DM-related genes associated with concentrations of specific urinary arsenicals. Of note, the majority of study subjects had positive indicators of DM or pre-DM and were from a larger population in the Zimapán and Lagunera regions of Mexico in which the prevalence of DM has been shown to be positively associated with concentrations of iAs in drinking water and DMAIII in urine [27]. Taken together, epidemiological evidence has been historically judged as insufficient or inadequate to establish a causal relationship between iAs exposure and DM development [57–59]. However, other recent studies of chronically-exposed populations have also reported a positive association between T2DM development and tAs concentrations in urine [60] and iAs concentrations in drinking water [61]. Here, we report a dose-response relationship between the promoter DNA methylation levels of several DM-related genes and specific urinary arsenicals, including DMAs. The identified genes are primarily associated with two functions related to DM development, namely pancreatic β cell apoptosis and insulin signaling perturbations in peripheral tissues. These results are consistent with in vitro and in vivo evidence that suggest arsenicals cause diabetogenic effects by targeting both β cell function and insulin-activated signal transduction pathways [62–65].
It is important to note, however, that PBLs are not cells associated with the development of any known iAs-associated disease, and that the DNA methylation profiles of DM-associated genes in PBLs may not reflect patterns observed in cells involved in DM development such as pancreatic β cells. Due to practical limitations associated with examining target cell types across large populations, PBL DNA methylation patterns have been used as biomarkers of toxicant exposure or health outcomes in epidemiological studies, and the value of these PBL methylation profiles has been the subject of considerable discussion [66]. The use of PBLs as intermediate biomarkers of disease risk and toxicant exposure in different target tissues and organs is rapidly developing. For instance, recent publications have reported distinct PBL DNA methylation patterns associated with breast cancer risk [67], insulin resistance [68], and risk of T2DM development [69] in humans. Importantly, the urinary arsenical-specific DNA methylation profiles reported here may be representative of critical factors that link iAs biotransformation to iAs-associated disease as they are enriched for genes with known associations consistent with the disease status (pre-DM/DM) of the majority of study subjects. Future work will expand the number of individuals in the cohort to determine if these PBL methylation profiles may serve as useful biomarkers of iAs exposure or provide insight into the mechanisms and susceptibility of iAs-associated disease.
ACKNOWLEDGMENTS
This work was supported in part by the National Institute of Environmental Health Sciences (P30ES010126, ES019315, ES015326, DK056350, and T32 ES7018). The authors would like to thank Dr. Hemant Kelkar at the Center for Bioinformatics, University of North Carolina at Chapel, for his help with data analysis. This manuscript was reviewed and approved for publication by the National Health and Environmental Effects Research Laboratory of the U.S. Environmental Protection Agency (EPA). Approval does not signify that the contents of this article necessarily reflect the views or policies of the EPA.
Abbreviations
- As
arsenic
- DM
diabetes mellitus
- DMAs
trivalent+pentavalent dimethylated arsenic
- DMAIII
dimethylarsinous acid
- GSH
glutathione
- iAs
inorganic arsenic
- HbA1c
hemoglobin A1c
- MMAs
trivalent+pentavalent monomethylated arsenic
- MMAIII
monomethylarsonous acid
- MOA
mode of action
- PBL
peripheral blood leukocyte
- SAM
S-adenosylmethionine
- T1DM
type 1 diabetes mellitus
- T2DM
type 2 diabetes mellitus
Footnotes
Genome-wide, gene-specific DNA methylation data were submitted to Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/ and are available under accession number GSE26073.
Contributor Information
Kathryn A. Bailey, Email: bailey.kathryn@unc.edu.
Michael C. Wu, Email: mcwu@email.unc.edu.
William O. Ward, Email: ward.william@epamail.epa.gov.
Lisa Smeester, Email: smesta@unc.edu.
Julia E. Rager, Email: jrager@email.unc.edu.
Gonzalo García-Vargas, Email: ggarcia.vargas@gmail.com.
Luz-Maria Del Razo, Email: ldelrazo@cinvestav.mx.
Zuzana Drobná, Email: zuzana_drobna@med.unc.edu.
Miroslav Stýblo, Email: styblo@med.unc.edu.
Rebecca C. Fry, Email: rfry@unc.edu.
REFERENCES
- 1.World Health Organization. Geneva, Switzerland: WHO Press; 2006. Guidelines for drinking water quality. [Google Scholar]
- 2.Centeno JA, Tseng CH, Van der Voet GB, Finkelman RB. Global impacts of geogenic arsenic: a medical geology research case. Ambio. 2007;36(1):78–81. doi: 10.1579/0044-7447(2007)36[78:giogaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Rahman MM, Ng JC, Naidu R. Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ. Geochem. Health. 2009;31(Suppl 1):189–200. doi: 10.1007/s10653-008-9235-0. [DOI] [PubMed] [Google Scholar]
- 4.Jomova K, Jenisova Z, Feszterova M, Baros S, Liska J, Hudecova D, Rhodes CJ, Valko M. Arsenic: toxicity, oxidative stress and human disease. J. Appl. Toxicol. 2011;31(2):95–107. doi: 10.1002/jat.1649. [DOI] [PubMed] [Google Scholar]
- 5.Kitchin KT, Wallace K. The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J. Inorgan. Biochem. 2008;102(3):532–539. doi: 10.1016/j.jinorgbio.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Druwe IL, Vaillancourt RR. Influence of arsenate and arsenite on signal transduction pathways: an update. Arch. Toxicol. 2010;84(8):585–596. doi: 10.1007/s00204-010-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossman TG, Klein CB. Genetic and epigenetic effects of environmental arsenicals. Metallomics. 2011;3(11):1135–1141. doi: 10.1039/c1mt00074h. [DOI] [PubMed] [Google Scholar]
- 8.Bailey KA, Fry RC. Arsenic-Induced Changes to the Epigenome. In: Sahu SC, editor. Toxicology and Epigenetics. United Kingdom: Wiley; 2012. pp. 149–190. [Google Scholar]
- 9.Haluskova J. Epigenetic studies in human diseases. Folia Biol. 2010;56(3):83–96. [PubMed] [Google Scholar]
- 10.Chen H, Liu J, Zhao CQ, Diwan BA, Merrick BA, Waalkes MP. Association of c-myc overexpression and hyperproliferation with arsenite-induced malignant transformation. Toxicol. Appl. Pharmacol. 2001;175(3):260–268. doi: 10.1006/taap.2001.9253. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, Barrett JC, Tsutsui T. Transformation by inorganic arsenic compounds of normal Syrian hamster embryo cells into a neoplastic state in which they become anchorage-independent and cause tumors in newborn hamsters. Int. J. Cancer. 2002;99(5):629–634. doi: 10.1002/ijc.10407. [DOI] [PubMed] [Google Scholar]
- 12.Chai CY, Huang YC, Hung WC, Kang WY, Chen WT. Arsenic salts induced autophagic cell death and hypermethylation of DAPK promoter in SV-40 immortalized human uroepithelial cells. Toxicol. Lett. 2007;173(1):48–56. doi: 10.1016/j.toxlet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Cui X, Wakai T, Shirai Y, Yokoyama N, Hatakeyama K, Hirano S. Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells. Hum. Pathol. 2006;37(3):298–311. doi: 10.1016/j.humpath.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol. Appl. Pharmacol. 2001;176(2):127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- 15.Vahter M. Mechanisms of arsenic biotransformation. Toxicol. 2002;181–182:211–217. doi: 10.1016/s0300-483x(02)00285-8. [DOI] [PubMed] [Google Scholar]
- 16.Kitchin KT, Ahmad S. Oxidative stress as a possible mode of action for arsenic carcinogenesis. Toxicol. Lett. 2003;137(1–2):3–13. doi: 10.1016/s0378-4274(02)00376-4. [DOI] [PubMed] [Google Scholar]
- 17.Styblo M, Del Razo LM, Vega L, Germolec DR, LeCluyse EL, Hamilton GA, Reed W, Wang C, Cullen WR, Thomas DJ. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch. Toxicol. 2000;74(6):289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 18.Tseng CH. Metabolism of inorganic arsenic and non-cancerous health hazards associated with chronic exposure in humans. J. Environ. Biol. 2007;28(2 Suppl):349–357. [PubMed] [Google Scholar]
- 19.Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 2007;25(1):1–22. doi: 10.1080/10590500701201695. [DOI] [PubMed] [Google Scholar]
- 20.Mass MJ, Wang L. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat. Res. 1997;386(3):263–277. doi: 10.1016/s1383-5742(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl. Acad. Sci. USA. 1997;94(20):10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochem. 2000;39(42):13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 23.Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp. Biol. Med. 2007;232(1):3–13. [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie EM. Arsenic-glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs) J. Inorg. Biochem. 2012;108:141–149. doi: 10.1016/j.jinorgbio.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu M, Hochadel JF, Fulmer BA, Waalkes MP. Effect of glutathione depletion and metallothionein gene expression on arsenic-induced cytotoxicity and c-myc expression in vitro. Toxicol. Sci. 1998;45(2):204–211. doi: 10.1006/toxs.1998.2539. [DOI] [PubMed] [Google Scholar]
- 26.Valenzuela OL, Drobna Z, Hernandez-Castellanos E, Sanchez-Pena LC, Garcia-Vargas GG, Borja-Aburto VH, Styblo M, Del Razo LM. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol. Appl. Pharmacol. 2009;239(2):200–207. doi: 10.1016/j.taap.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Razo LM, Garcia-Vargas GG, Valenzuela OL, Castellanos EH, Sanchez-Pena LC, Currier JM, Drobna Z, Loomis D, Styblo M. Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapan and Lagunera regions in Mexico. Environ. Health. 2011;10:73. doi: 10.1186/1476-069X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeester L, Rager JE, Bailey KA, Guan X, Smith N, Garcia-Vargas G, Del Razo LM, Drobna Z, Kelkar H, Styblo M, Fry RC. Epigenetic changes in individuals with arsenicosis. Chem Res Toxicol. 2011;24(2):165–167. doi: 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. (Methodological) 1995;57:289–300. [Google Scholar]
- 31.Efron B. Size, power and false discovery rates. Ann. Stat. 2007;35:1351. [Google Scholar]
- 32.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B. (Methodological) 2002;64:479–498. [Google Scholar]
- 33.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2012;35(Suppl 1):S64–S71. doi: 10.2337/dc12-s064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Li B, Xu Y, Wang Y, Jin Y, Itoh T, Yoshida T, Sun G. Arsenic methylation capacity and its correlation with skin lesions induced by contaminated drinking water consumption in residents of chronic arsenicosis area. Environ. Toxicol. 2011;26(2):118–123. doi: 10.1002/tox.20535. [DOI] [PubMed] [Google Scholar]
- 35.Huang YK, Huang YL, Hsueh YM, Yang MH, Wu MM, Chen SY, Hsu LI, Chen CJ. Arsenic exposure, urinary arsenic speciation, and the incidence of urothelial carcinoma: a twelve-year follow-up study. Cancer Causes Control. 2008;19(8):829–839. doi: 10.1007/s10552-008-9146-5. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida T, Yamauchi H, Fan Sun G. Chronic health effects in people exposed to arsenic via the drinking water: dose-response relationships in review. Toxicol. Appl. Pharmacol. 2004;198(3):243–252. doi: 10.1016/j.taap.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Hsieh YC, Lien LM, Chung WT, Hsieh FI, Hsieh PF, Wu MM, Tseng HP, Chiou HY, Chen CJ. Significantly increased risk of carotid atherosclerosis with arsenic exposure and polymorphisms in arsenic metabolism genes. Environmental research. 2011;111(6):804–810. doi: 10.1016/j.envres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Chiou HY, Huang WI, Su CL, Chang SF, Hsu YH, Chen CJ. Dose-response relationship between prevalence of cerebrovascular disease and ingested inorganic arsenic. Stroke. 1997;28(9):1717–1723. doi: 10.1161/01.str.28.9.1717. [DOI] [PubMed] [Google Scholar]
- 39.Tseng WP. Effects and dose--response relationships of skin cancer and blackfoot disease with arsenic. Environ. Health Perspect. 1977;19:109–119. doi: 10.1289/ehp.7719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Online supporting information available at: http://www.bios.unc.edu/research/epigenetics
- 41.Suk K, Kim S, Kim YH, Kim KA, Chang I, Yagita H, Shong M, Lee MS. IFN-gamma/TNF-alpha synergism as the final effector in autoimmune diabetes: a key role for STAT1/IFN regulatory factor-1 pathway in pancreatic beta cell death. J. Immunol. 2001;166(7):4481–4489. doi: 10.4049/jimmunol.166.7.4481. [DOI] [PubMed] [Google Scholar]
- 42.Mandrup-Poulsen T. Apoptotic signal transduction pathways in diabetes. Biochem. Pharmacol. 2003;66(8):1433–1440. doi: 10.1016/s0006-2952(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 43.Yoon JW, Jun HS. Cellular and molecular pathogenic mechanisms of insulin-dependent diabetes mellitus. Ann. N. Y. Acad. Sci. 2001;928:200–211. doi: 10.1111/j.1749-6632.2001.tb05650.x. [DOI] [PubMed] [Google Scholar]
- 44.Hirai H, Miura J, Hu Y, Larsson H, Larsson K, Lernmark A, Ivarsson SA, Wu T, Kingman A, Tzioufas AG, Notkins AL. Selective screening of secretory vesicle-associated proteins for autoantigens in type 1 diabetes: VAMP2 and NPY are new minor autoantigens. Clin Immunol. 2008;127(3):366–374. doi: 10.1016/j.clim.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Real JM, Broch M, Ricart W, Casamitjana R, Gutierrez C, Vendrell J, Richart C. Plasma levels of the soluble fraction of tumor necrosis factor receptor 2 and insulin resistance. Diabetes. 1998;47(11):1757–1762. doi: 10.2337/diabetes.47.11.1757. [DOI] [PubMed] [Google Scholar]
- 46.Clement S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, Sasaki T, Penninger J, Doherty M, Malaisse W, Dumont JE, Le Marchand-Brustel Y, Erneux C, Hue L, Schurmans S. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409(6816):92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 47.Howard JK, Flier JS. Attenuation of leptin and insulin signaling by SOCS proteins. Trends Endocrinol. Metab. 2006;17(9):365–371. doi: 10.1016/j.tem.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Lebrun P, Van Obberghen E. SOCS proteins causing trouble in insulin action. Acta Physiol (Oxf) 2008;192(1):29–36. doi: 10.1111/j.1748-1716.2007.01782.x. [DOI] [PubMed] [Google Scholar]
- 49.Maier VH, Melvin DR, Lister CA, Chapman H, Gould GW, Murphy GJ. v- and t-SNARE protein expression in models of insulin resistance: normalization of glycemia by rosiglitazone treatment corrects overexpression of cellubrevin, vesicle-associated membrane protein-2, and syntaxin 4 in skeletal muscle of Zucker diabetic fatty rats. Diabetes. 2000;49(4):618–625. doi: 10.2337/diabetes.49.4.618. [DOI] [PubMed] [Google Scholar]
- 50.Babu DA, Deering TG, Mirmira RG. A feat of metabolic proportions: Pdx1 orchestrates islet development and function in the maintenance of glucose homeostasis. Mol. Gene.t Metab. 2007;92(1–2):43–55. doi: 10.1016/j.ymgme.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimoto K, Polonsky KS. Pdx1 and other factors that regulate pancreatic beta-cell survival. Diabetes Obes. Metab. 2009;11(Suppl 4):30–37. doi: 10.1111/j.1463-1326.2009.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mokhtari D, Myers JW, Welsh N. The MAPK kinase kinase-1 is essential for stress-induced pancreatic islet cell death. Endocrinology. 2008;149(6):3046–3053. doi: 10.1210/en.2007-0438. [DOI] [PubMed] [Google Scholar]
- 53.Cantley J, Boslem E, Laybutt DR, Cordery DV, Pearson G, Carpenter L, Leitges M, Biden TJ. Deletion of protein kinase Cdelta in mice modulates stability of inflammatory genes and protects against cytokine-stimulated beta cell death in vitro and in vivo. Diabetologia. 2011;54(2):380–389. doi: 10.1007/s00125-010-1962-y. [DOI] [PubMed] [Google Scholar]
- 54.Brand C, Cipok M, Attali V, Bak A, Sampson SR. Protein kinase Cdelta participates in insulin-induced activation of PKB via PDK1. Biochem. Biophys. Res. Commun. 2006;349(3):954–962. doi: 10.1016/j.bbrc.2006.08.100. [DOI] [PubMed] [Google Scholar]
- 55.Valenzuela OL, Borja-Aburto VH, Garcia-Vargas GG, Cruz-Gonzalez MB, Garcia-Montalvo EA, Calderon-Aranda ES, Del Razo LM. Urinary trivalent methylated arsenic species in a population chronically exposed to inorganic arsenic. Environ. Health. Perspect. 2005;113(3):250–254. doi: 10.1289/ehp.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, Saffery R. Prospects for epigenetic epidemiology. American journal of epidemiology. 2009;169(4):389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang CF, Chen YW, Yang CY, Tsai KS, Yang RS, Liu SH. Arsenic and diabetes: current perspectives. Kaohsiung J Med Sci. 2011;27(9):402–410. doi: 10.1016/j.kjms.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Tseng CH, Tseng CP, Chiou HY, Hsueh YM, Chong CK, Chen CJ. Epidemiologic evidence of diabetogenic effect of arsenic. Toxicol. Lett. 2002;133(1):69–76. doi: 10.1016/s0378-4274(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 59.Navas-Acien A, Silbergeld EK, Streeter RA, Clark JM, Burke TA, Guallar E. Arsenic exposure and type 2 diabetes: a systematic review of the experimental and epidemiological evidence. Environ. Health Perspect. 2006;114(5):641–648. doi: 10.1289/ehp.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coronado-Gonzalez JA, Del Razo LM, Garcia-Vargas G, Sanmiguel-Salazar F, Escobedo-de la Pena J. Inorganic arsenic exposure and type 2 diabetes mellitus in Mexico. Environ. Res. 2007;104(3):383–389. doi: 10.1016/j.envres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 61.Islam MR, Khan I, Hassan SM, McEvoy M, D'Este C, Attia J, Peel R, Sultana M, Akter S, Milton AH. Association between type 2 diabetes and chronic arsenic exposure in drinking water: A cross sectional study in Bangladesh. Environ. Health. 2012;11:38. doi: 10.1186/1476-069X-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paul DS, Harmon AW, Devesa V, Thomas DJ, Styblo M. Molecular mechanisms of the diabetogenic effects of arsenic: inhibition of insulin signaling by arsenite and methylarsonous acid. Environ. Health Perspect. 2007;115(5):734–742. doi: 10.1289/ehp.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul DS, Hernandez-Zavala A, Walton FS, Adair BM, Dedina J, Matousek T, Styblo M. Examination of the effects of arsenic on glucose homeostasis in cell culture and animal studies: development of a mouse model for arsenic-induced diabetes. Toxicol. Appl. Pharmacol. 2007;222(3):305–314. doi: 10.1016/j.taap.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu J, Woods CG, Yehuda-Shnaidman E, Zhang Q, Wong V, Collins S, Sun G, Andersen ME, Pi J. Low-level arsenic impairs glucose-stimulated insulin secretion in pancreatic beta cells: involvement of cellular adaptive response to oxidative stress. Environ. Health Perspect. 2010;118(6):864–870. doi: 10.1289/ehp.0901608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davila-Esqueda ME, Morales JM, Jimenez-Capdeville ME, De la Cruz E, Falcon-Escobedo R, Chi-Ahumada E, Martin-Perez S. Low-level subchronic arsenic exposure from prenatal developmental stages to adult life results in an impaired glucose homeostasis. Exp. Clin. Endocrinol. Diabetes. 2011;119(10):613–617. doi: 10.1055/s-0031-1287782. [DOI] [PubMed] [Google Scholar]
- 66.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6(7):828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X, Gammon MD, Hernandez-Vargas H, Herceg Z, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. DNA methylation in peripheral blood measured by LUMA is associated with breast cancer in a population-based study. FASEB J. 2012;26(6):2657–2666. doi: 10.1096/fj.11-197251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes. 2012;61(2):542–546. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, Wainstein J, Friedlander Y, Levy-Lahad E, Glaser B, Hellman A. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum. Mol. Genet. 2012;21(2):371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baumgartener JW. SHIP2: an emerging target for the treatment of type 2 diabetes mellitus. Curr. Drug Targets. Immune Endocrin. Metabol. Disord. 2003;3(4):291–298. doi: 10.2174/1568008033340144. [DOI] [PubMed] [Google Scholar]
- 71.Aronoff S, Berkowitz K, Shreiner B, Want L. Glucose metabolism and regulation: beyond insulin and glucagon. Diabetes Spectr. 2004;17(3):183–190. [Google Scholar]
- 72.Benjafield AV, Glenn CL, Wang XL, Colagiuri S, Morris BJ. TNFRSF1B in genetic predisposition to clinical neuropathy and effect on HDL cholesterol and glycosylated hemoglobin in type 2 diabetes. Diabetes Care. 2001;24(4):753–757. doi: 10.2337/diacare.24.4.753. [DOI] [PubMed] [Google Scholar]
- 73.Kuo JZ, Guo X, Klein R, Klein BE, Cui J, Rotter JI, Ipp E, Chen YD. Systemic soluble tumor necrosis factor receptors 1 and 2 are associated with severity of diabetic retinopathy in hispanics. Ophthalmology. 2012;119(5):1041–1046. doi: 10.1016/j.ophtha.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J. Am. Soc. Nephrol. 2012;23(3):507–515. doi: 10.1681/ASN.2011060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gohda T, Niewczas MA, Ficociello LH, Walker WH, Skupien J, Rosetti F, Cullere X, Johnson AC, Crabtree G, Smiles AM, Mayadas TN, Warram JH, Krolewski AS. Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol. 2012;23(3):516–524. doi: 10.1681/ASN.2011060628. [DOI] [PMC free article] [PubMed] [Google Scholar]