SUMMARY
Megakaryocyte morphogenesis employs a “hypertrophy-like” developmental program, dependent on P-TEFb kinase activation and cytoskeletal remodeling. P-TEFb activation classically occurs by a feedback regulated process of signal-induced, reversible release of active Cdk9-cyclin T modules from large inactive 7SK snRNP complexes. Here we have identified an alternative pathway of irreversible P-TEFb activation in megakaryopoiesis, mediated by dissolution of the 7SK snRNP complex. In this pathway calpain 2 cleavage of the core 7SK snRNP component MePCE promoted P-TEFb release and consequent upregulation of a cohort of cytoskeleton remodeling factors, including α-actinin-1. In a subset of human megakaryocytic leukemias, the transcription factor GATA1 undergoes truncating mutation (GATA1s). Here we linked the GATA1s mutation to defects in megakaryocytic upregulation of calpain 2 and of P-TEFb-dependent cytoskeletal remodeling factors. Restoring calpain 2 expression in GATA1s-mutant megakaryocytes rescued normal development, implicating this morphogenetic pathway as a target in human leukemogenesis.
Keywords: megakaryopoiesis, P-TEFb, 7SK snRNP, calpain 2, GATA1s mutant
INTRODUCTION
Mammalian hematopoietic differentiation proceeds by a series of binary decisions yielding progenitors of increasingly limited developmental potential, with the megakaryocyte lineage emerging from a bipotent megakaryocyte-erythroid progenitor (MEP). Megakaryocytic and erythroid cells, despite common origins, shared transcription factors and shared signaling pathways, differ profoundly in their developmental programs. Erythroid morphogenesis occurs through progressive reduction in cell size accompanied by nuclear condensation and ultimately extrusion. Megakaryocytic morphogenesis contrastingly involves marked expansion in cell mass combined with acquisition of a lobulated, polyploid nucleus containing up to 32-64N. In this regard, megakaryopoiesis bears resemblance to the program of cardiomyocyte hypertrophy, in which pressure overload elicits cellular enlargement and polyploidization (Liu et al., 2010).
Mechanistically, the developmental morphogenesis of megakaryocytes and the hypertrophic response of cardiomyocytes share key regulatory elements. At the transcriptional level, both programs rely on a complex of serum response factor (SRF) with myocardin-related transcription factors (Cheng et al., 2009; Halene et al., 2010; Kuwahara et al., 2010; Nelson et al., 2005; Smith et al., 2012), as well as on MEF2C (Gekas et al., 2009; Munoz et al., 2009). At the signaling level, both programs require high amplitude activation of the P-TEFb kinase pathway (Elagib et al., 2008; Sano et al., 2002). A critical distinction is that cardiomyocyte hypertrophy consists of a reversible response to a pathologic stimulus while normal megakaryocyte morphogenesis represents an irreversible, terminal program largely driven by cell-intrinsic mechanisms.
Cellular P-TEFb kinase activity is tightly regulated, with the majority of the Cdk9-cyclin T kinase modules sequestered in large inactive complexes containing the 7SK snRNA, HEXIM1, MePCE, LARP7, and additional factors (Barboric et al., 2009; Jeronimo et al., 2007; Peterlin and Price, 2006; Price, 2008; Xue et al., 2010). Several stimuli, including hypertrophic agonists (e.g. endothelin-1), UV irradiation, HIV-1 Tat, and hexamethylene bisacetamide activate P-TEFb by inducing the release of Cdk9-cyclin T from the 7SK snRNP complex (Chen et al., 2008; Contreras et al., 2007; Krueger et al., 2010; Sano et al., 2002; Sano and Schneider, 2004). These stimuli variously trigger signaling via Gq, calcineurin, PP1α, and PI3K, ultimately leading to remodeling of the 7SK snRNP that promotes dissociation of Cdk9-cyclin T and HEXIM1 away from the stable core complex of 7SK, MePCE, and LARP7. Once activated, P-TEFb promotes transcriptional elongation by phosphorylating RNA polymerase II and associated pausing factors. Feedback autoregulation results from the rapid and potent induction of HEXIM1 transcription (Bartholomeeusen et al., 2012; Garriga et al., 2010; He et al., 2006), effectively driving resequestration of Cdk9-cyclin T back into an inactive 7SK snRNP complex (Bartholomeeusen et al., 2012; Zhou et al., 2012).
GATA1, a master transcriptional regulator of megakaryocyte and erythroid differentiation, physically and functionally interacts with P-TEFb in hematopoietic cells (Bottardi et al., 2011; Elagib et al., 2008). Somatic mutations yielding an N-terminal truncated, “short” GATA1 protein (GATA1s) occur in virtually all megakaryocytic neoplasms associated with Down syndrome (Wickrema and Crispino, 2007). In knock-in mice, the mutant GATA1s induces transient megakaryocytic hyperproliferation and maturational defects during fetal liver hematopoiesis (Li et al., 2005). Megakaryocytic hyperproliferation and aberrant differentiation have also been elicited by P-TEFb inhibiton in adult mice with megakaryocytic GATA1 deficiency, supporting the notion of a GATA1-P-TEFb megakaryopoietic pathway that might be affected in Down syndrome neoplasms (Elagib et al., 2008).
In the current study, we have identified a megakaryopoietic P-TEFb activation pathway characterized by downregulation of the 7SK snRNP core components MePCE, LARP7, and 7SK snRNA. The protease calpain 2 critically participated in this pathway, undergoing recruitment to P-TEFb, targeting MePCE for proteolysis, and promoting P-TEFb-dependent megakaryocyte morphogenesis. Downstream of P-TEFb in this pathway were identified a cohort of coregulated cytoskeletal remodeling factors involved in execution of the morphogenetic program. In a large panel of human megakaryocytic leukemias, decreased calpain 2 levels significantly correlated with the presence of the GATA1s mutation. In addition, murine fetal liver megakaryocytes from GATA1s knockin mice displayed defects in upregulation of calpain 2 and of downstream cytoskeletal remodeling factors. Lentiviral restoration of calpain 2 expression specifically ameliorated developmental defects in GATA1s knockin fetal megakaryocytes. These findings thus support a megakaryocyte morphogenetic pathway involving GATA1, calpain 2, P-TEFb, and the actin cytoskeleton. Perturbations of this pathway may play a role in the pathogenesis of Down syndrome megakaryocytic neoplasms.
RESULTS
Global P-TEFb Activation in Megakaryopoiesis
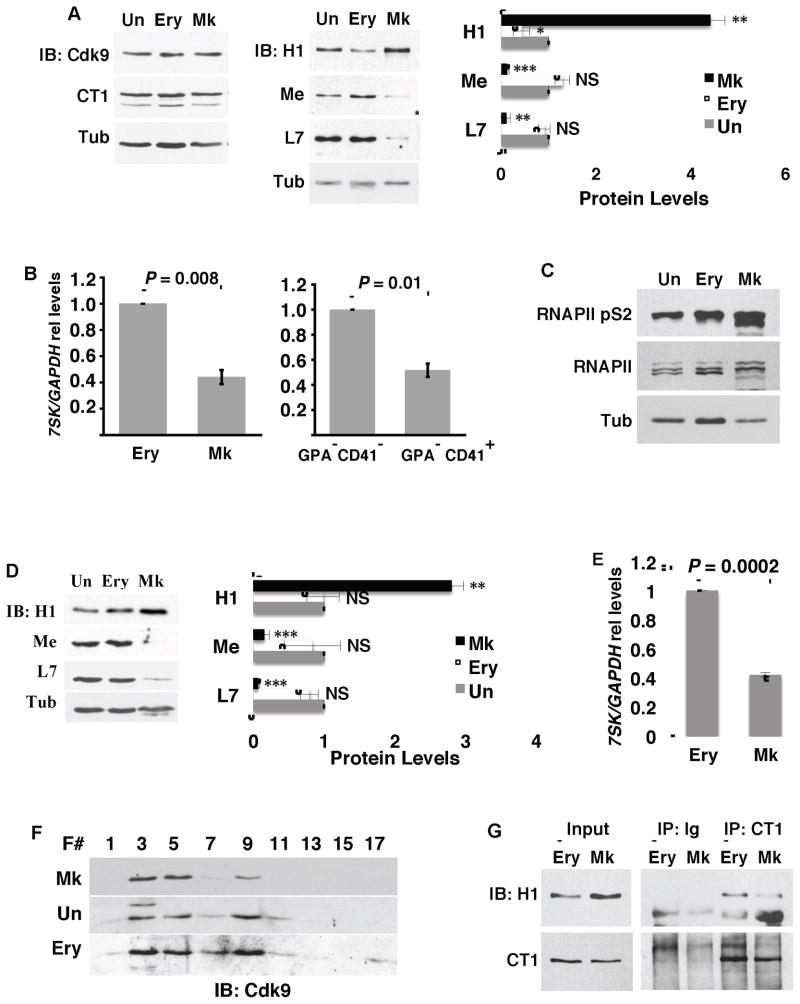
Previous work has suggested a critical role for high-amplitude P-TEFb activation in megakaryocyte differentiation and divergence from the erythroid lineage (Elagib et al., 2008). To examine the mechanistic basis for this activation, 7SK snRNP complex components were quantified in megakaryocytic, erythroid, and undifferentiated cells derived from primary human hematopoietic progenitors. The principal P-TEFb factors in hematopoietic cells, Cdk9 and cyclin T1, showed similar protein levels in megakaryocytic (Mk), undifferentiated (Un) and erythroid (Ery) cells (Figure 1A). By contrast, megakaryocytic cells specifically downregulated all of the components of the recently-defined (Barboric et al., 2009; Xue et al., 2010) 7SK snRNP core complex: MePCE (Me), LARP7 (L7), and the 7SK snRNA (Figures 1A and 1B). Additionally, megakaryocytic cells displayed enhanced phosphorylation of RNA polymerase II carboxy terminal domain serine 2 (RNAPII S2), a specific target of P-TEFb phosphorylation (Peterlin and Price, 2006) (Figure 1C). Concomitant with downregulation of the 7SK inhibitory scaffold, megakaryocytes specifically upregulated HEXIM1, reflecting increased cellular P-TEFb activity (Bartholomeeusen et al., 2012; Garriga et al., 2010; He et al., 2006) (Figure 1A). The megakaryocytic induction of HEXIM1 occurred at the mRNA level (Figure S1A) and was prevented by the Cdk9 inhibitor flavopiridol (FP) and by shRNA knockdown of Cdk9 (Figure S1B). MEPCE mRNA levels showed no significant decline during megakaryocytic differentiation, suggesting regulation of this factor at the protein level (Figure S1C).
Figure 1. Megakaryocytic Downregulation of 7SK snRNP Core Components and Associated P-TEFb Release.
(A) Analysis of 7SK snRNP protein components. (Left and middle panels) Primary human progenitors either undifferentiated (Un) or cultured for 5 days in erythroid (Ery) or megakaryocytic (Mk) medium underwent immunoblot for Cdk9, cyclin T1 (CT1), HEXIM1 (H1), MePCE (Me), LARP7 (L7), and tubulin (Tub). (Right panel) Scanning densitometry from three independent experiments conducted as in the middle panel. Results represent mean ± SEM for signals relative to those in undifferentiated cells. In addition, all signals are normalized to tubulin. * P < 0.05; ** P < 0.01; *** P < 0.005; NS, not significant.
(B) (Left panel) Relative 7SK levels in human progenitors cultured as in (A). (Right panel) Relative 7SK levels in sorted CD41− GPA− double negative and CD41+ GPA− (Mk) cells from megakaryocytic culture. Graphs represent mean ± SEM of 7SK normalized to GAPDH in three independent experiments.
(C) Phosphorylation of RNA polymerase II carboxy terminal domain serine 2 (RNAPII pS2). Human CD34+ progenitors cultured 3 days in expansion medium (Un) or 6 days in erythroid or megakaryocyte medium underwent immunoblotting of whole cell lysates for RNAPII pS2, total RNAPII, or tubulin.
(D and E) HPC7 cells either undifferentiated (Un) or grown in Ery or Mk medium underwent analysis as in (A) and (B). Graphs both represent mean ± SEM for three independent experiments. *** P < 0.005.
(F and G) Megakaryocytic P-TEFb release. Extracts from HPC7 cells cultured as in (D) were subjected to glycerol gradient fractionation and immunoblotting for Cdk9 (F) or cyclin T1 immunoprecipitation followed by immunoblot for HEXIM1 (G). F # = fraction number. See also Figure S1.
Similar studies were carried out on a non-transformed murine hematopoietic cell line, HPC7, which retains cytokine-responsive multilineage differentiation potential (Pinto do O et al., 1998). These cells underwent rapid and efficient erythroid or megakaryocytic differentiation in response to 48 hours treatment with erythropoietin (Ery) or thrombopoietin (Mk), respectively (Figure S1D). As with primary human progenitors, HPC7 megakaryocytic differentiation specifically correlated with downregulation of the 7SK snRNP core components and upregulation of HEXIM1 (Figures 1D and 1E, Figure S1E). In addition, the HPC7 cells did not downregulate cyclin T1 or Cdk9 during megakaryocytic differentiation (Figure S1F). Glycerol gradient analysis of Cdk9 distribution between large inactive and small active complexes (Sedore et al., 2007) revealed 80% of megakaryocytic Cdk9 to be within the small complex (Fraction 5). In erythroid and undifferentiated cells, by contrast, the majority of Cdk9 (~70%) resided in the large complex (Fraction 9) (Figure 1F). (Fraction 3 contained insoluble debris and represents background.) Another validated approach toward assessment of intracellular P-TEFb status consists of immunoprecipitation to determine association with the repressor HEXIM1, an interaction dependent on the integrity of the 7SK snRNP core complex (Chen et al., 2008). This approach confirmed a decrease in cyclin T1-HEXIM1 complexes in HPC7 cells undergoing megakaryocytic differentiation, as compared with cells either undergoing erythroid differentiation (Figure 1G) or maintained undifferentiated (see Figure 2C). The results in Figure 1 thus suggest a lineage-specific mechanism of P-TEFb activation in megakaryopoiesis, during which the dissolution of the 7SK snRNP is accompanied by large-scale release of active P-TEFb.
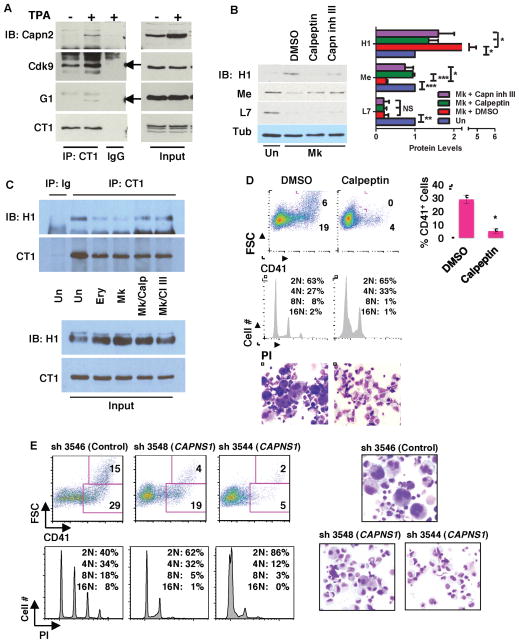
Figure 2. Calpain Contribution to Megakaryocytic P-TEFb Activation and Differentiation.
(A) Analysis of Calpain 2 association with P-TEFb. Extracts from K562 cells treated with either DMSO (−) or 25nM TPA (+), underwent immunoprecipitation (IP) with antibody to cyclin T1 (CT1) or control IgG followed by immunoblotting for calpain 2 (Capn2), Cdk9 (arrow), GATA-1 (G1) (arrow), and cyclin T1 (CT1).
(B) Effects of calpain inhibition on megakaryocytic MePCE downregulation and HEXIM1 upregulation. Primary human progenitors either undifferentiated (Un) or cultured in megakaryocytic medium (Mk) with DMSO, 40μM calpeptin or 20μM calpain inhibitor III (Capn inh III), underwent immunoblot and densitometry as in Figure 1A. Results from three independent experiments are shown as mean ± SEM for signals relative to those in undifferentiated cells. In addition, all signals are normalized to tubulin. * P < 0.05; ** P < 0.01; *** P < 0.005; NS, not significant.
(C) Effects of calpain inhibition on P-TEFb dissociation from HEXIM1. Extracts from HPC7 cells grown 48 hours in expansion (Un), erythroid (Ery), or megakaryocytic (Mk) medium underwent immunoprecipitation for cyclin T1 (CT1) followed by immunoblotting for HEXIM1 (H1) and CT1. The cells undergoing megakaryocytic culture were treated in the final 16 hours with either DMSO, 50μM calpeptin (Calp), or 25μM Capn inh III (CI III).
(D) Effects of calpain inhibition on megakaryocytic differentiation. Primary human progenitors grown for 6 days in megakaryocytic medium with DMSO or 40μM calpeptin were analyzed by flow cytometry for CD41 expression, and DNA content by propidium iodide staining (PI). Cell morphology was assessed by light microscopy of Wright-stained cytospins (200X). Graph represents mean ± SEM for CD41 expression in three independent experiments; * P < 0.05.
(E) Role of calpain S1 in megakaryocytic differentiation. Primary human progenitors transduced with lentiviral shRNA constructs targeting Calpain S1 (CAPNS1) underwent megakaryocytic culture for 5 days followed by analysis as in (C). For documentation of knockdown see Figure S2E. See also Figure S2.
Calpain Contribution to Megakaryocytic P-TEFb Activation and Differentiation
Mapping of the mammalian transcriptional protein interactome has identified calpain subunits (S1 and 2) as components of an extended 7SK snRNP network (Jeronimo et al., 2007), suggesting a role for proteolysis in remodeling of this complex. To examine the association of calpain with P-TEFb in a cell line model of megakaryocyte differentiation, K562 cells before and after phorbol ester (TPA) treatment underwent immunoprecipitation of endogenous cyclin T1 (CT1). As described for GATA1 (Elagib et al., 2008), calpain 2 displayed inducible recruitment to P-TEFb in association with differentiation induction (Figure 2A). By contrast, Cdk9 coprecipitated with cyclin T1 both before and after induction. Immunoprecipitation of endogenous HEXIM1 also revealed calpain 2 binding that was enhanced by TPA treatment, suggesting inducible recruitment of calpain 2 to the 7SK snRNP complex during megakaryocytic differentiation (Figure S2A).
Evidence for lineage-selective, endogenous calpain activation was provided by immunoblot detection of a characteristic 190 kd filamin A cleavage fragment (Xu et al., 2010), emerging during megakaryocytic but not erythroid differentiation of primary progenitors (Figure S2B). shRNA knockdowns confirmed that this cleavage depended on calpain 2 expression (see Figure 3E). Calpastatin, the endogenous calpain inhibitor with numerous isoforms (Takano et al., 1993; Wendt et al., 2004), showed strong erythroid upregulation of large (~110 kD) and small (~70 kD) variants with minimal megakaryocytic upregulation of a large variant; by contrast calpain 2 showed megakaryocytic but not erythroid upregulation (Figure S2C). To assess the contribution of calpain activity to the megakaryocytic remodeling of the 7SK snRNP (see Figure 1), calpain protease inhibitors were added to primary megakaryocytic cultures. Notably, these inhibitors prevented both the downregulation of MePCE and the upregulation of HEXIM1, but had no effect on LARP7 (Figures 2B and S2D). In addition, short term administration of the inhibitors prevented the dissociation of HEXIM1 from P-TEFb, as determined by coimmunoprecipitation analysis of HPC7 extracts (Figure 2C).
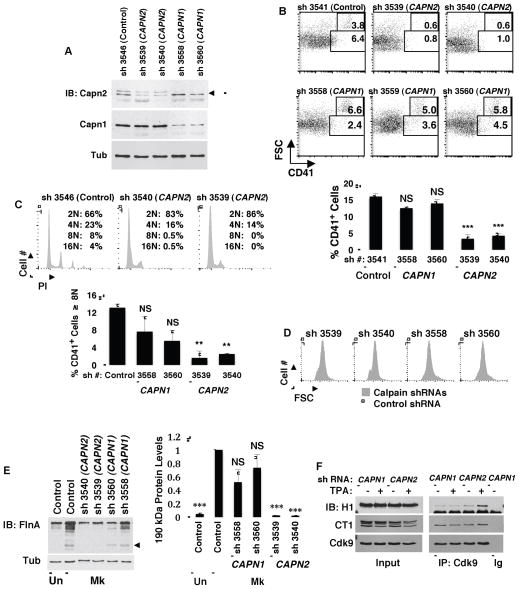
Figure 3. Implication of Calpain 2 as the Isoenzyme Involved in Megakaryocytic Differentiation.
(A) Specificity of calpain 1 and 2 knockdowns. Primary human progenitors transduced with lentiviral shRNA constructs targeting calpain 1 (CAPN1) or 2 (CAPN2) were analyzed by immunoblot. Arrow: calpain 2; band above arrow: lot-dependent cross-reactivity of antibody to calpain 1; smear below arrow: most likely autolyzed calpain 2.
(B and C) Relative contributions of calpain 1 versus 2 to megakaryocytic differentiation. Progenitors transduced with lentiviral shRNA constructs as in (A) were analyzed for megakaryocytic differentiation as in Figure 2E. Graphs represent mean ± SEM for CD41 expression or for percentage of CD41+ cells with DNA content of ≥ 8N, with both graphs derived from three independent experiments; ** P < 0.01; *** P < 0.005; NS, not significant.
(D) Relative contributions of calpain 1 versus 2 to cellular enlargment during megakaryopoiesis. Forward scatter (FSC) profiles of cells subjected to shRNA knockdowns and culture as in (B).
(E) Relative contributions of calpain 1 versus 2 to filamin A cleavage associated with megakaryocytic differentiation. (Left panel) Human progenitors transduced with shRNA constructs as (A) underwent megakaryocytic culture followed by immunoblotting for filamin A (FlnA). Arrow indicates 190 kd cleavage fragment. (Right panel) Densitometric analysis of 190 kd cleavage fragment from three independent experiments performed as in Left panel. Graph represents mean ± SEM; *** P < 0.005.
(F) Relative contributions of calpain 1 versus 2 to HEXIM1 dissociation from P-TEFb. K562 cells expressing shRNAs knocking down either calpain 1 or calpain 2 underwent induction with TPA (25nM 48 hours) followed by immunoprecipitation of Cdk9 and immunoblotting for HEXIM1 (H1), cyclin T1 (CT1) and Cdk9.
Participation of calpain in megakaryocytic differentiation was supported by the repressive effect of the inhibitors on cellular enlargement, polyploidization, and CD41 upregulation (Figures 2D, and S2E). This finding raised the possibility that the block in MePCE downregulation associated with calpain inhibition might occur secondary to impaired megakaryocytic differentiation. To rule out this possibility, progenitors were allowed first to undergo megakaryocytic differentiation and were subsequently treated for only 18 hours with calpain inhibitors. This short-course treatment reversed MePCE downregulation in the absence of any effects on megakaryocytic differentiation, consistent with a direct influence of calpain activity on MePCE levels (Figure S2D).
Multiple approaches were taken to confirm the contribution of calpain to megakaryocytic differentiation. Because calpain inhibitors also target cathepsin proteases, progenitors underwent control treatment with cathepsin inhibitors, which did not affect megakaryocytic differentiation (data not shown). In addition, we subjected progenitors to lentiviral shRNA knockdown of calpain S1, the regulatory subunit required for calpain activity. Two different shRNAs diminished calpain S1 levels by ~3-fold (Figure S2F), and both blocked megakaryocytic differentiation in a manner similar to the calpain inhibitors (Figure 2E). The block in megakaryopoiesis caused by calpain inhibition was not associated with major impairments in viability or with redirection of cells down the erythroid lineage (data not shown).
To further validate calpain involvement in megakaryopoiesis, in vivo gene deletion was conducted by crossing CAPNS1f/f (S1f/f) mice (Tan et al., 2006) with a deleter strain expressing Cre recombinase driven by the megakaryoctyic PF4 promoter (Tiedt et al., 2007). Despite evidence for incomplete excision in marrow megakaryocytic cultures (data not shown), CAPNS1f/f;PF4-Cre (S1f/f;PF4-Cre) mice had significant thrombocytopenia accompanied by normal red and white cell counts (Figure S2G). In addition, CAPNS1f/f;PF4-Cre marrows showed defective megakaryocytic maturation in ex vivo cultures, reflected by impairments in upregulation of CD41/CD42 and in morphogenesis (Figure S2H and data not shown).
Implication of Calpain 2 as the Isozyme Mediating Megakaryopoietic P-TEFb Activation
The principal hematopoietic calpains that depend on calpain S1 for their activity consist of calpain 1 and calpain 2. To determine which of these isozymes might participate in regulating megakaryopoiesis, each underwent targeting by shRNAs in primary human progenitors, with ~2-fold, isozyme-specific knockdowns obtained for both (Figure 3A). The calpain 2 knockdowns significantly impaired megakaryopoiesis including CD41 upregulation, polyploidization and cellular enlargement, while calpain 1 knockdowns had no effects distinct from those of transducing control vector (Figures 3B–D). In addition, calpain 2, and not calpain 1, contributed to the filamin A cleavage observed during megakaryopoiesis (Figure 3E). shRNA knockdowns in K562 cells ± TPA were conducted to compare contributions of calpain 1 versus calpain 2 to the dissociation of HEXIM1 from P-TEFb; knockdown of calpain 2 notably enhanced HEXIM1 association with P-TEFb, particularly in TPA treated cells (Figure 3F). As a leukemic cell line, K562 cells undergo incomplete megakaryocytic differentiation with TPA induction and, unlike non-transformed cells, lack dynamic regulation of HEXIM1 expression.
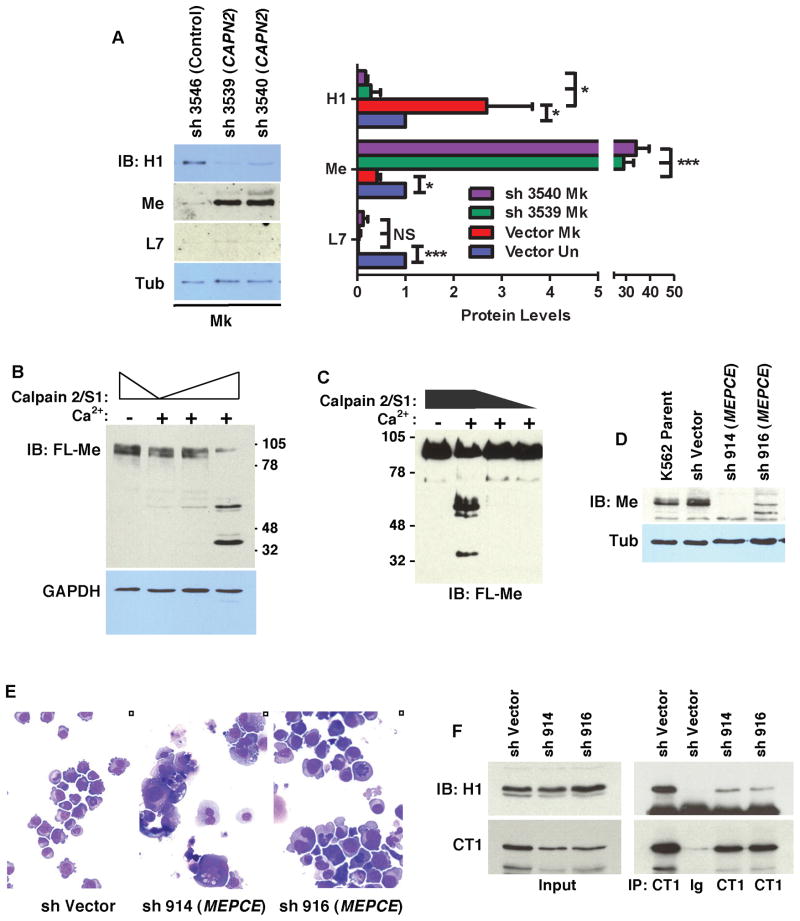
Calpain 2 involvement in megakaryopoietic P-TEFb activation was supported by a consistent decrease in HEXIM1 expression associated with its knockdown (Figure 4A and S3A), but not with calpain 1 knockdown (not shown). In addition, MePCE showed dramatic upregulation in cells subjected to calpain 2 knockdown (Figures 4A and S3A), suggestive of direct proteolytic targeting. This notion was further supported by cell-free assays in which recombinant calpain 2/S1 cleaved MePCE in a calcium-dependent manner (Figures 4B and 4C). In these assays, calpain cleavage of MePCE in cellular extracts occurred with similar efficiency with or without RNase A addition, and MePCE isolated in complex with cyclin T1 underwent efficient digestion by calpain 2/S1 (Figures S3B–C). These findings suggest calpain cleavage of MePCE may occur in the context of the 7SK snRNP complex. By contrast, calpain 2 knockdown failed to reverse megakaryocytic repression of LARP7 (Figure 4A and S3A), as observed with calpain inhibitors (see Figure 2B), consistent with a distinct, calpain-independent mechanism regulating this component of the 7SK snRNP. These results thus provide evidence for a developmental pathway in which calpain 2-dependent destruction of MePCE contributes, along with LARP7 downregulation, to ongoing P-TEFb activation, which in turn promotes megakaryocyte differentiation. Additional support for this pathway came from shRNA-mediated MePCE knockdown, which induced megakaryocyte morphogenesis in K562 cells and promoted dissociation of HEXIM1 from P-TEFb (Figures 4D–F).
Figure 4. Involvement of Calpain 2 in Megakaryocytic Downregulation of MePCE and Consequences of MePCE Downregulation.
(A) Effects of calpain 2 knockdown on MePCE (Me), HEXIM1 (H1) and LARP7 (L7) levels. Cells transduced and cultured as in Figure 3B underwent immunoblotting followed by scanning densitometry. Graph depicts mean ± SEM for three independent experiments. * P < 0.05; ** P < 0.01; *** P < 0.005; NS, not significant.
(B) In vitro analysis of MePCE cleavage by calpain 2. Extracts from HEK293T cells transfected with FLAG-MePCE (FL-Me) expression vector were incubated with 50 to 200ng purified calpain 2/S1 ± 2 mM CaCl2 (Ca2+) followed by immunoblot for FL-Me and GAPDH.
(C) In vitro analysis of MePCE cleavage by calpain 2. Purified FL-Me protein from HEK293T transfectants was subjected to in vitro cleavage and immunoblot as in (B).
(D–E) K562 cells transduced with shRNA constructs targeting MePCE underwent immunoblotting in (D). Cell morphology was assessed by light microscopy on Wright-stained cytospins (200X) in (E).
(F) Effect of MePCE downregulation on P-TEFb interaction with HEXIM1. Extracts from K562 cells subjected to shRNA knockdown of MePCE underwent immunoprecipitation of cyclin T1 followed by immunoblotting for HEXIM1 (H1) and cyclin T1 (CT1) See also Figure S3.
Calpain 2 and P-TEFb Regulation of Megakaryocytic Cytoskeleton Remodeling Factors
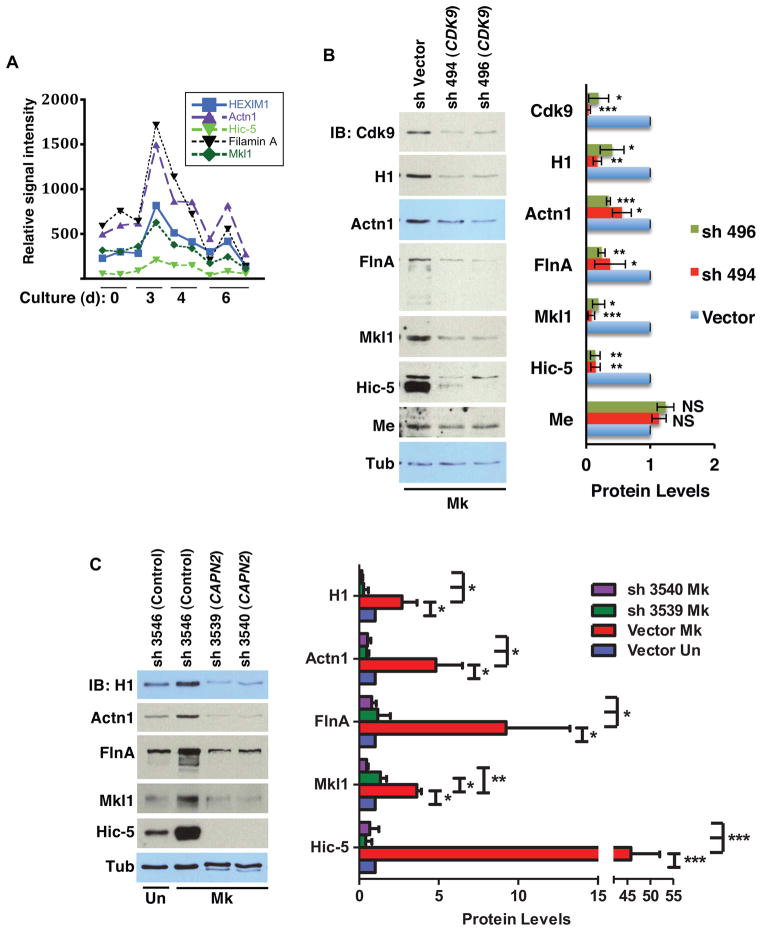
To characterize downstream factors regulated by calpain-P-TEFb signaling during megakaryopoiesis, in silico analysis was applied to identify genes whose expression covaried with HEXIM1 during megakaryocytic differentiation (see Supplemental Experimental Procedures). HEXIM1 was selected as the prototype based on its documented control by P-TEFb (Garriga et al., 2010; He et al., 2006) (see Figure S1B) and its calpain-dependent upregulation during megakaryopoiesis (see Figure 2B). Analysis of a dataset derived from purified megakaryocytic progenitors at four distinct stages of development (Chen et al., 2007) revealed a group of four actin-associated, cytoskeleton remodeling factors whose mRNA levels paralleled that of HEXIM1 during megakaryopoiesis (Figure 5A). These factors, like HEXIM1, all displayed upregulation during megakaryocytic but not erythroid differentiation of primary progenitors (Figures S4A–B). Their regulation by P-TEFb was confirmed by Cdk9 knockdown in primary megakaryocytic cultures. Notably, the levels of all four factors, as well as of HEXIM1, decreased with knockdown of Cdk9, while the levels of MePCE and tubulin remained largely unaffected (Figure 5B). Cdk9 knockdown in undifferentiated cells did affect expression of these factors but not to the same extent as in the megakaryocytic knockdown (Figure S4C). Thus, Cdk9 likely plays a more prominent role in megakaryocytic upregulation of these factors than in their basal expression. Note that the relatively high Hic-5 expression seen in undifferentiated cells subjected to lentiviral transduction (Figure S4C), versus that in untransduced cells (Figure S4A), results from the extended culture period required for transduction and selection. Calpain 2 regulation of these cytoskeletal remodeling factors in megakaryocytes was also demonstrated by shRNA knockdowns in primary human progenitors (Figure 5C).
Figure 5. P-TEFb and Calpain 2 Regulate a Cohort of Megakaryocytic Cytoskeletal Remodeling Factors.
(A) Expression patterns of gene cohort and HEXIM1 during megakaryocytic differentiation. Plotted are normalized signals obtained from GEO DataSet Record GDS2521 comparing relative mRNA levels from murine fetal liver megakaryocytic progenitors at various developmental stages.
(B) Assessment of P-TEFb influence on expression of factors identified in (A). (Left panel) Primary human progenitors transduced with shRNA constructs targeting Cdk9 were cultured in megakaryocytic medium followed by immunoblot with the indicated antibodies. (Right panel) Densitometry derived from three independent experiments conducted as in Left panel showing mean ± SEM for relative protein levels normalized to Tubulin. * P < 0.05; ** P < 0.01; *** P < 0.005; NS, not significant. HEXIM1 (H1), α-actinin 1 (Actn1), filamin A (FlnA), MePCE (Me), tubulin (Tub).
(C) Assessment of calpain 2 influence on expression of factors identified in (A). Progenitors subjected to calpain 2 knockdown were analyzed as in (B). (Right panel) Densitometry derived from three independent experiments conducted as in Left panel showing mean ± SEM for relative protein levels normalized to Tubulin. * P < 0.05; ** P < 0.01; *** P < 0.005. See also Figure S4.
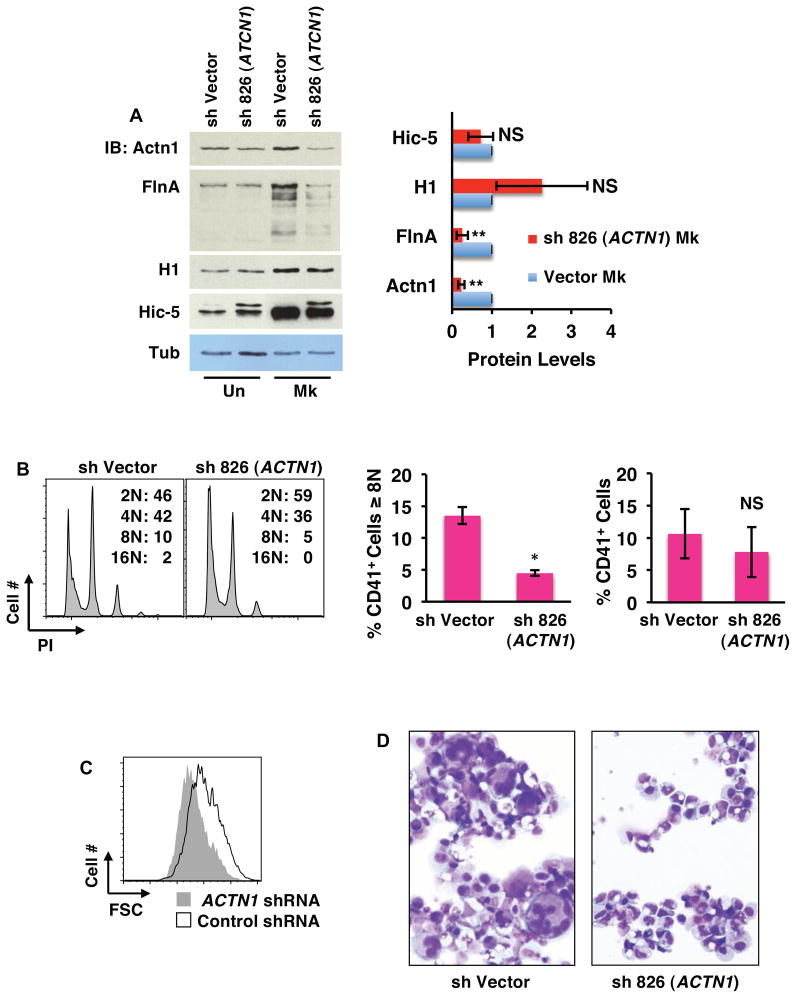
Two of the four factors, Mkl1 and filamin A, have been previously implicated in megakaryocyte development including polyploidization (Begonja et al., 2011; Cheng et al., 2009). One of the factors, α-actinin, has been shown to regulate cytokinesis both in yeast and in mammalian cells (Mukhina et al., 2007; Wu et al., 2001). Notably, overexpression of α-actinin consistently causes a failure in cytokinesis leading to polyploidization. As megakaryocytic endomitosis arises from a failure in cytokinesis (Lordier et al., 2008), we examined the contribution of α-actinin upregulation to megakaryocyte polyploidization and enlargement. Knockdown of α-actinin 1 in primary progenitors did not affect levels of HEXIM1 or Hic-5 but did diminish levels of filamin A, without affecting its proteolytic cleavage (Figure 6A). α-actinin thus does not exert feedback regulation of P-TEFb or calpain but may interact with filamin A. Phenotypically, α-actinin 1 knockdown prevented megakaryocytic polyploidization and enlargement but minimally affected CD41 upregulation (Figures 6B–D and S5A; see Figures S5B–E for results with additional shRNA construct). These results therefore implicate α-actinin 1 as an important target downstream of calpain-P-TEFb signaling in megakaryocytic morphogenesis.
Figure 6. Contribution of α-actinin-1 to Megakaryocytic Enlargement and Polyploidization.
(A) Effects of megakaryocytic knockdown of α-actinin-1 on filamin A and Hic-5 expression. Primary human progenitors transduced with shRNA targeting α-actinin-1 (ACTN1) were analyzed as in Figure 5B. (Right panel) Densitometry derived from three independent experiments conducted as in Left panel showing mean ± SEM for relative protein levels normalized to Tubulin. * P < 0.05; ** P < 0.01; NS, not significant. HEXIM1 (H1), α-actinin 1 (Actn1), filamin A (FlnA), tubulin (Tub).
(B–D) Effects of α-actinin-1 knockdown on megakaryocyte morphogenesis. Progenitors transduced with shRNA targeting α-actinin-1 underwent megakaryocyte morphogenesis culture followed by flow cytometry analysis as in Figure 3B–D. Cell morphology was assessed by light microscopy of Wright-stained cytospins (200X). Graphs in (B) show mean ± SEM for three independent experiments. * P < 0.05; NS, not significant. See also Figure S5.
Calpain 2 Dysregulation in Association with the GATA1s Mutation
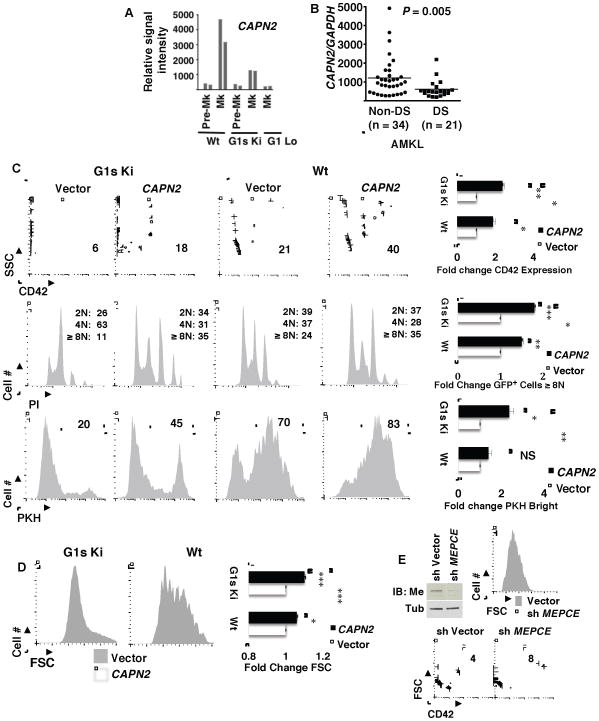
In genetic complementation studies comparing effects of introducing wild type versus mutant GATA1 alleles into GATA1-deficient megakaryocytes, calpain 2 was previously identified as a GATA1 target gene with a distinct pattern of regulation. Specifically, calpain 2 showed strong induction by wild type GATA1 but poor induction by GATA1s, while most other GATA1 target genes studied showed equivalent regulation by both alleles (Muntean and Crispino, 2005). These findings suggested that the GATA1s mutation may interfere with calpain 2 upregulation during megakaryopoiesis. Independent support for this notion came from our analysis of the gene expression dataset of Li et al. (GEO GDS1316), comparing fetal liver megakaryocytes from wild type versus GATA1s knock in mice (G1s Ki) (Li et al., 2005). This dataset clearly confirmed an association between the GATA1s mutation and defective megakaryocytic upregulation of calpain 2, with no abnormalities seen in calpain 1 expression (Figure 7A and S6A). Megakaryocytes deficient in overall GATA1 expression, from the G1 Lo strain, also showed markedly diminished calpain 2 expression. To assess potential consequences of the calpain 2 deficiency in G1s Ki megakaryocytes, this dataset was further analyzed for expression patterns of the calpain 2 regulated cytoskeletal remodeling factors (see Figure 5C). Notably, all four factors showed profiles nearly identical to that of calpain 2, including defective upregulation in G1s Ki and G1 Lo megakaryocytes (Figure S6B). Thus, the calpain 2 deficiency associated with GATA1s may impair the developmental induction of key morphogenetic mediators.
Figure 7. Calpain 2 Deficiency Occurs in Megakaryocytic Cells with Mutant GATA1s and Contributes to Aberrant Megakaryopoiesis.
(A) Expression patterns of calpain 2 (CAPN2) in developing fetal liver megakaryocytes from wild type (Wt), GATA1s knockin (G1s Ki), and megakaryocytic GATA1 knockout (G1 Lo) mice. Plotted are normalized signals obtained from GEO DataSet Record GDS1316 comparing relative mRNA levels from murine fetal liver pre-megakaryocytes (Pre-Mk) and megakaryocytes (Mk) of the indicated strains. Paired bars represent two independent experiments.
(B) Calpain 2 expression in human megakaryocytic proliferative disorders bearing the GATA1s mutation. Plotted are normalized signals obtained from GEO DataSet Record GSE4119 comparing relative mRNA levels from cases of acute megakaryoblastic leukemia occurring in patients with Down syndrome (DS-AMKL) vs cases unassociated with Down syndrome (Non-DS-AMKL).
(C–D) Effects of lentiviral-mediated calpain 2 restoration on megakaryocytic differentiation in fetal liver progenitors with GATA1s mutation. Day 13.5 fetal liver progenitors from G1s Ki and wild type (Wt) mice underwent transduction with calpain 2 or control lentiviral expression constructs, followed by megakaryocytic culture and flow cytometric analysis. GFP+ transduced cells were analyzed for expression of CD42, ploidy (PI), megakaryocytic growth arrest (PKH26 retention in CD41+ cells), and megakaryocytic cell size (FSC in CD41+ cells). Each of the four graphs in C–D represents mean ± SEM for three independent experiments. Graphic results are presented as fold change relative to vector transduced cells within each strain; * P < 0.05; ** P < 0.01; *** P < 0.005; NS, not significant.
(E) Impact of MePCE knockdown on megakaryocyte differentiation in G1S Ki fetal liver cells. Embryonic day 13.5 fetal liver cells were transduced with lentiviral shRNA constructs, selected in puromycin, subjected to Mk culture, and analyzed for size (FSC) and CD42 upregulation. Immunoblot on left shows knockdown of MePCE in MEL cells transduced with the shRNA to MEPCE (A5). See also Figure S6.
To determine whether calpain 2 deficiency also correlated with the GATA1s mutation in patient samples, we analyzed the gene expression dataset of Bourquin et al. (GEO GSE4119) comparing Down syndrome-associated (DS) versus non-Down syndrome-associated (non-DS) megakaryoblastic neoplasms (Bourquin et al., 2006). In this dataset, the GATA1s mutation occurred in all DS cases and in none of the non-DS cases. Notably, the DS acute megakaryoblastic leukemia (DS-AMKL) cases expressed significantly less calpain 2 than did non-DS AMKL cases (Figure 7B). Because of clinical and molecular heterogeneity within AMKL, we also compared two well-defined infantile megakaryoblastic neoplasms: DS transient myeloproliferative disorder (DS-TMD) versus AMKL with the t(1;22) chromosomal abnormality. Again, significantly less calpain 2 expression occurred in the DS samples (Figure S6C), confirming a clinical correlation between the GATA1s mutation and calpain 2 deficiency. TP53BP2, a control gene neighboring calpain 2 on chromosome 1, showed no differences in expression between the DS-TMD versus t(1;22) cases (data not shown).
Contribution of Calpain 2 Deficiency to GATA1s-associated Dysmegakaryopoiesis
The GATA1s mutation causes abnormalities in fetal liver megakaryopoiesis including excessive cellular proliferation and impaired upregulation of several differentiation markers (Li et al., 2005). Consistent with these prior findings, we observed defects in CD42 upregulation, polyploidization, and cellular enlargement in G1s Ki fetal liver megakaryocytes (Figures 7C–D, compare Wt vector vs G1s Ki vector; Figure S6E). Immunoblot analysis confirmed diminished calpain 2 protein expression in G1s Ki megakaryocytes, with no alteration in calpastatin levels (Figure S6D). Consistent with its regulation of P-TEFb activity, calpain 2 deficiency in G1s Ki megakaryocytes correlated with global deficiency in phosphorylation on serine 2 within the carboxy terminal domain of RNA polymerase II (Figure S6D). To determine whether calpain 2 deficiency contributes to GATA1s-associated dysmegakaryopoiesis, fetal liver cells transduced with control or calpain 2-expressing lentivirus underwent megakaryocytic culture and analysis. Transduction of G1s Ki cells with calpain 2-expressing lentivirus significantly enhanced several differentiation parameters within the CD41+ megakaryocytic population: CD42 expression, polyploidization, proliferation arrest reflected by PKH26 dye retention, and cellular enlargement (Figures 7C–D). By comparison, non-transduced (GFP−) megakaryocytic cells derived from control vector versus calpain 2 lentiviral exposures showed no differences in these parameters (data not shown). Wild type fetal liver megakaryocytes transduced with calpain 2 lentivirus also showed increases in CD42 expression, cellular enlargement, and polyploidization (see Figures 7C–D). However, the impact of enforcing calpain 2 expression in wild type cells was significantly less than in G1s Ki cells (see graphs in Figures 7C–D). With regard to proliferation, enforced calpain 2 had no significant effect in wild type cells, in contrast to its arresting effect in G1s Ki cells (Figure 7C). A protease-deficient calpain 2 mutant (C105S) completely failed to rescue cellular enlargement in G1s Ki fetal liver progenitors and relatively weakly augmented CD42 levels, the latter effect possibly due to calpastatin sequestration (Figures S6F–G). To determine whether MePCE downregulation might contribute to the rescue conferred by enforced calpain 2 expression, G1s Ki fetal liver cells underwent shRNA-mediated MePCE knockdown, which enhanced cellular enlargement and CD42 expression (Figure 7F). In aggregate, these results support calpain 2 deficiency, and consequent failure of MePCE downregulation, as factors contributing to the defective fetal megakaryopoiesis associated with the GATA1s mutation (see pathway diagram in Figure 8).
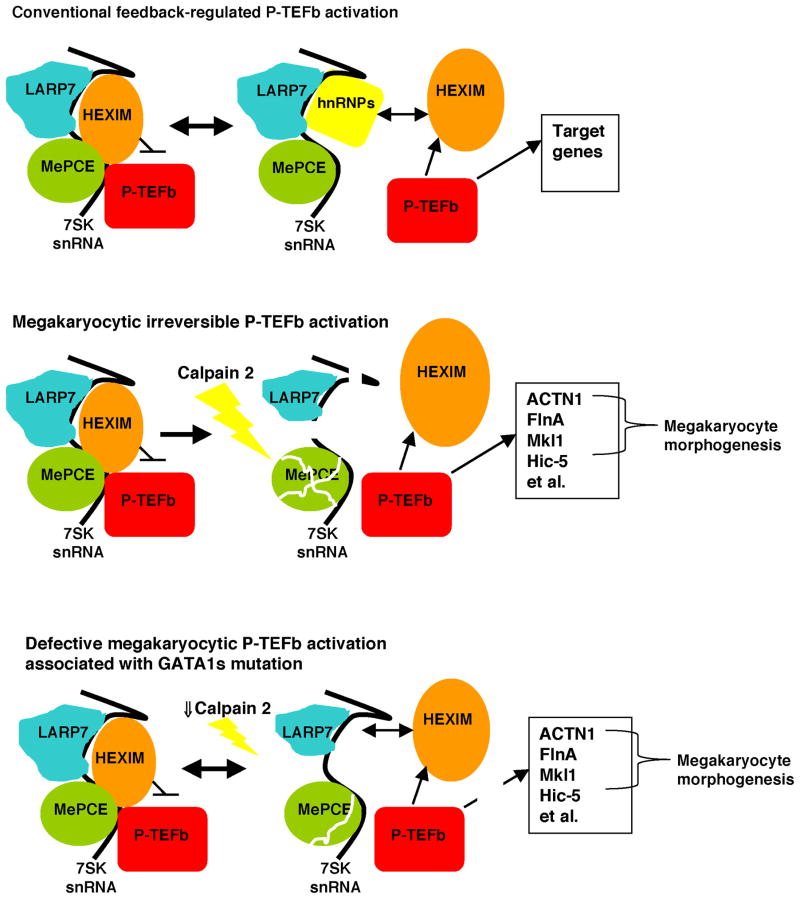
Figure 8. A Model Depicting Pathway for P-TEFb Activation in Megakaryopoiesis and Perturbation of this Pathway by a Leukemogenic GATA1 Mutation.
In the top panel, non megakaryocytic cells such as erythroblasts show reversible, feedback regulated P-TEFb activation in which P-TEFb and HEXIM1 reversibly associate with the 7SK snRNP. In this scenario, the majority of P-TEFb resides within the large inactive complex.
In the middle panel, megakaryocytes show global and irreversible P-TEFb activation. In this scenario, calpain 2 undergoes upregulation and activation, promoting 7SK snRNP destruction through direct proteolysis of MePCE. An additional contribution derives from calpain-independent LARP7 downregulation. In the absence of the 7SK snRNP, chronic unopposed P-TEFb activation drives transcription of a cohort of genes that includes cytoskeletal remodeling factors that promote megakaryocyte morphogenesis and HEXIM1.
In the bottom panel, megakaryocytic upregulation of calpain 2 is impaired in cells bearing the leukemogenic GATA1s mutation. These cells show impairment in the destruction of the 7SK snRNP, retain capacity for feedback inhibition of P-TEFb, and are compromised in the activation of key P-TEFb target genes.
DISCUSSION
The hypertrophic response in cardiomyocytes is driven by P-TEFb activation through a mechanism involving dissociation of Cdk9-Cyclin T complexes from the 7SK snRNP (Sano et al., 2002). How hypertrophic signaling induces this dissociation remains unclear but may involve a conformational shift in the 7SK snRNP complex elicited by the phosphatase activity of calcineurin (Chen et al., 2008). Recent studies have revealed a contribution of calpain activity to cardiomyocyte hypertrophy in diabetic mice, and calpain activation also maintains cardiomyocyte integrity during hemodynamic stress (Li et al., 2011; Taneike et al., 2011). This latter function appears to occur through a membrane repair mechanism but may also involve coordinated programming of cytoskeletal remodeling (Yamaguchi et al., 2012).
In megakaryopoiesis, the morphogenetic program appears to be initiated by calpain 2-mediated proteolysis of MePCE, a critical 7SK snRNP core component known to stabilize the 7SK snRNA (Barboric et al., 2009; Xue et al., 2010). Megakaryocytic calpain activation correlates with upregulation of calpain 2 but must also involve specialized signaling via calcium influx, upon which calpain function is absolutely dependent (Campbell and Davies, 2012). An additional contribution to differential calpain activation in megakaryocytic versus erythroid cells likely resides in the erythroid-specific upregulation of calpastatin, the calpain inhibitor (Figure S2C). During megakaryopoiesis, calpain-induced downregulation of the 7SK snRNA, a required cofactor for HEXIM repression of Cdk9 (Michels et al., 2004; Yik et al., 2003), uncouples P-TEFb from repressive feedback mediated by continually rising HEXIM1 levels. In addition to calpain-dependent downregulation of MePCE, a distinct mechanism involving calpain-independent downregulation of LARP7, another 7SK snRNP core component, probably contributes as well to megakaryocytic downregulation of 7SK snRNA. Mechanisms for LARP7 downregulation could potentially involve alternative proteolytic pathways as well as transcriptional and post-transcriptional repression. Thus, multiple pathways during megakaryopoiesis likely cooperate in promoting global and irreversible shifting of P-TEFb toward its active form, thereby driving progression along a developmental pathway of continual cellular enlargement (see Figure 8). Whether megakaryocytic P-TEFb activation reinforces the very pathways that promote its activation is an intriguing topic for future investigation.
Normal megakaryopoiesis comprises, among many changes, the coordinated upregulation of cytoskeletal regulatory proteins that promote the dramatic cellular enlargement. Comparisons of transcriptional profiles from discrete developmental stages of megakaryopoiesis show activation of cytoskeletal-associated genes to occur early, during the MK-3 stage of Chen et al., concomitant with activation of genes in the m-calpain (calpain 2) pathway (Chen et al., 2007). Similarly, analysis of genes upregulated during polyploidization has identified a large cohort of cytoskeletal factors, including ACTN1 (encoding α-actinin-1) (Raslova et al., 2007). Factors previously known to contribute to megakaryocytic cytoskeletal remodeling include SRF and Mkl1 (also known as MAL or MRTF-A), which together transactivate genes contributing to actin stress-fiber formation (Gilles et al., 2009; Halene et al., 2010).
In the current study, four cytoskeletal regulatory factors were found to be controlled by the calpain 2-P-TEFb signaling pathway. One of these factors, Mkl1, itself regulates expression of cytoskeletal genes during megakaryopoiesis. Three of the factors, α-actinin, Hic-5, and Mkl1, have been implicated in cardiomyocyte hypertrophy (Kuwahara et al., 2010; Ridinger et al., 2009; Yund et al., 2009). Two of the factors, α-actinin and filamin A, function in actin cross-linkage, leading to F-actin stabilization (Mukhina et al., 2007; Nakamura et al., 2011), and our data suggest that α-actinin may regulate filamin A levels in megakaryocytes (Figure 6A). An interesting property shared by Mkl1, Hic-5, and filamin A consists of their dual functions as actin-binding factors in the cytosol and transcriptional regulators in the nucleus (Gilles et al., 2009; Heitzer and DeFranco, 2006; Nakamura et al., 2011), potentially allowing for cytoskeletal-nuclear cross-talk during megakaryopoiesis. Studies of megakaryopoiesis in knockout mice have demonstrated participation of both Mkl1 and filamin A in polyploidization (Begonja et al., 2011; Cheng et al., 2009). Our knockdown experiments in primary human progenitors identify α-actinin-1 as an additional factor whose upregulation contributes to megakaryocytic polyploidization and enlargement (Figures 6B–D and S5C–E). Future experiments will address whether Hic-5 contributes to megakaryocyte morphogenesis and the extent to which these cytoskeletal factors engage in cross-talk.
The leukemia-associated GATA1 mutant, GATA1s, causes developmental abnormalities including hyperproliferation in fetal liver megakaryocytic progenitors (Li et al., 2005). The transcriptional defects in GATA1s responsible for these abnormalities have been attributed to a loss of repressive control over oncogenic factors such as c-Myc, c-Myb, PU.1, Ikaros, GATA-2, and an array of E2F target genes (Klusmann et al., 2010; Li et al., 2005). The defects in GATA1s function may arise from loss of a binding domain for pRb and E2F (Kadri et al., 2009; Klusmann et al., 2010). Our data suggest that loss of an activation function, namely upregulation of calpain 2, also represents a critical defect in GATA1s. Previous data show GATA1s to retain normal activation capability for most megakaryocytic target genes examined, with the exception of calpain 2 and GP1bα (Muntean and Crispino, 2005). The ability of enforced calpain 2 expression to induce CD42 upregulation in G1s Ki fetal liver megakaryocytes (Figure 7C) suggests that GP1bα may lie downstream of calpain 2 in GATA1 regulation of megakaryopoiesis. The likelihood that defective signaling via calpain 2 to P-TEFb contributes to DS megakaryocytic neoplasia is supported by the deficiency of calpain 2 expression in DS-AMKL/TMD specimens and by studies in which P-TEFb inhibition in mice elicits a megakaryoblastic disorder resembling DS-TMD (Elagib et al., 2008). Future studies will examine the interplay of pRb-E2F, Calpain 2-P-TEFb, and other hypertrophic pathways in normal and neoplastic megakaryopoiesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Purified primary human CD34+ hematopoietic progenitors were cultured in serum free unilineage media as described (Elagib et al., 2008). Undifferentiated cells (Un) were cultured in prestimulation medium 72 hours. Erythroid (Ery) and megakaryocytic (Mk) cells were cultured in their respective media for the indicated durations following an initial prestimulation phase. Protease inhibitors (EMD Chemicals Inc., Gibbstown, NJ, USA) dissolved in DMSO or DMSO only were added to cultures as indicated. K562 and HEK293T cells were cultured under standard conditions (Elagib et al., 2008). HPC7 cells were maintained in growth medium (IMDM, 5% FBS, 100 ng/ml SCF) as described (Pinto do O et al., 1998); for differentiation induction the cells were cultured 48 hours in either megakaryocyte medium (IMDM, 5% FBS, 2.5 ng/ml SCF, 50 ng/ml TPO, 10 ng/ml IL-6) or erythroid medium (IMDM, 5% FBS, 20 ng/ml SCF, 4 U/ml EPO). Fetal liver cells from day 13.5 mouse embryos were dissaggregated, depleted of red cells, and then expanded in a 1:1 mixtue of RPMI 1640 and DMEM supplemented with 100 ng/ml SCF, 20 ng/ml IL-3, 2 U/ml erythropoietin, and 1% Nutridoma™ (Roche, Indianapolis, IN, USA). To promote megakaryopoiesis, the cells were transferred to RPMI 1640 with 10% fetal bovine serum, 25 ng/ml SCF, and 10 ng/ml TPO. To prevent shedding of the CD42 antigen (Nishikii et al., 2008) the medium was supplemented after the initial 24 hours with 20 μM GM6001 (EMD Chemicals Inc.). Megakaryocytic cultures derived from marrow of 5-FU treated mice were performed as described by Hamlett et al. (Hamlett et al., 2008).
Cell Transduction and Transfection
For lentiviral shRNA-mediated knockdown, pLKO.1-derived constructs targeting human mRNAs of interest were purchased from Open Biosystems (Huntsville, AL). For lentiviral expression of calpain 2, a full-length rat cDNA was inserted upstream of an IRES element previously introduced into pWPXLD, a parent vector provided by Didier Trono (École Polytechnique Fédéral de Lausanne, Lausanne, Switzerland). Construct packaging by HEK293T cotransfection with pCMV-dR8.74 plus pMD2.G was followed by spinoculation of target cells (Elagib et al., 2008). Cells transduced with shRNA constructs underwent selection with puromycin, 2 μg/ml during an initial 72 hour expansion phase and 1 μg/ml during the subsequent differentiation phase. FLAG-MePCE plasmid was a gift from Dr. Matjaz Barboric (University of Helsinki, Helsinki, Finland) (Barboric et al., 2009).
Immunoprecipitation, Immunoblot, and RNA quantitation
Immunoprecipitation of endogenous P-TEFb and HEXIM1 from HPC7 and K562 cellular extracts, followed by immunoblot detection was performed as described (Elagib et al., 2008). For direct immunoblot analysis of cells, whole cell lysates in 1X SDS-PAGE loading buffer supplemented with protease and phosphatase inhibitors underwent shearing of DNA prior to standard analysis (Elagib et al., 2008). Densitometry data were acquired on a GS800 calibrated densitometer (Bio-Rad, Hercules, CA) and analyzed with Quantity One software (Bio-Rad). For quantiation of HEXIM1, MEPCE, and 7SK, total cellular RNA isolated using the RNeasy Plus Mini Kit (QIAGEN Inc., Valencia, CA, USA) underwent conversion to cDNA using the iScript kit (Bio-Rad Life Science, Hercules, CA, USA) followed by quantitative PCR on the iCycler platform using iQ SYBR Green Supermix (Bio-Rad). Relative transcript levels were calculated using GAPDH normalization with the ΔΔCT formula.
Glycerol Gradient Analysis
HPC7 extracts (in 150 mM NaCl, 2 mM MgCl2, 10 mM HEPES, 1 mM DTT, 1 mM PMSF, 20 μM calpain inhibitor III, EDTA-free protease inhibitor cocktail [Roche], 40 U/ml RNAse inhibitor [Promega], 0.5% NP-40) were loaded on 10 – 45% glycerol gradients prepared in extraction buffer. The gradients were spun in a SW-41 rotor at 40,000g for 16 hours at 4°C, and resultant fractions underwent SDS-PAGE-immunoblot.
In Vitro MePCE Cleavage Assays
FLAG-MePCE purified from HEK293T transfectants, or unpurified extracts, underwent incubation in a 30 μl reaction volume with 50–200 ng His6-Calpain 2/S1 complex purified from E. coli (EMD Chemicals Inc.). Where indicated, CaCl2 was added to a final concentration of 2 mM. After incubation at 30° C for 15 minutes, the reaction was stopped by addition of an equal volume of 2X SDS-PAGE loading buffer with 100 μM calpain inhibitor III, followed by immunoblot analysis with anti-FLAG and anti-MePCE antibodies.
Flow Cytometry and Cell Sorting
CD41 and GPA in human progenitors, CD41 in murine progenitors, and ploidy in both species were assessed as described (Elagib et al., 2008). For antibody detection of murine CD42b, cells were stained with phycoerythrin-conjugated anti-GPIbα (Emfret Analytics, Wurzburg, Germany). Dye dilution analysis of proliferation was performed on fetal liver cells after spinoculation and involved washing of cells followed by incubation with the red membrane dye PKH26 (Sigma-Aldrich, St. Louis, MO, USA). After stopping the staining reaction with 1% BSA, cells underwent washing followed by culture for 72 hours in megakaryocytic medium. Proliferation of transduced megakaryocytes was analyzed by gating on GFP+ CD41+ cells. For cell sorting, progenitors costained for CD41 and GPA were fractionated on a FACSVantage SE Turbo Sorter with DIVA Option (Becton Dickinson).
Mice
Mice with megakaryocytic deletion of Calpain S1 were derived from the capn4PZ strain (Tan et al., 2006). The PF4-Cre transgenic strain was purchased as C57BL/6-Tg(Pf4-cre)Q3Rsko/J (The Jackson Laboratory, Bar Harbor, ME, USA). The G1S Ki mice consisted of the the GATA1ΔE2 strain (Li et al., 2005). Blood obtained by retro-orbital phlebotomy was analyzed for complete blood count values (CBC) on a Hemavet 950FS analyzer (Drew Scientific).
Statistical Analysis
Statstical comparisons between 2 groups employed Student’s t-test (two-tailed, unpaired), and comparisons among >2 groups employed ANOVA.
Supplementary Material
HIGHLIGHTS.
Megakaryocytic P-TEFb activation occurs due to dissolution of the 7SK snRNP complex
Calpain 2 cleavage of 7SK snRNP factor MePCE drives megakaryocytic P-TEFb activation
Calpain 2/P-TEFb promote morphogenesis by upregulating cytoskeletal remodelers
Calpain 2 deficiency underlies dysmegakaryopoiesis seen with leukemic GATA1 mutation
Acknowledgments
This paper is dedicated to the memory of Marcia Finberg. We thank Dr. Matjaz Barboric for the FLAG-MePCE expression construct, Dr. Leif Carlsson for HPC7 cells, Dr. Didier Trono for lentiviral plasmids, and Daniel Matson for generous assistance with glycerol gradient analysis. For helpful discussions and advice, thanks to Drs. B. Matija Peterlin, David Price, Prabu Reddi, and Vijay Sankaran. This work was supported by the following NIH grants: CA100057, DK079924, and DK090926.
Footnotes
Supplemental Information includes seven figures and Supplmental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. BET bromodomain inhibition activates transcription via a transient release of P-TEFb from 7SK snRNP. J Biol Chem. 2012 doi: 10.1074/jbc.M112.410746. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begonja AJ, Hoffmeister KM, Hartwig JH, Falet H. FlnA-null megakaryocytes prematurely release large and fragile platelets that circulate poorly. Blood. 2011;118:2285–2295. doi: 10.1182/blood-2011-04-348482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottardi S, Zmiri FA, Bourgoin V, Ross J, Mavoungou L, Milot E. Ikaros interacts with P-TEFb and cooperates with GATA-1 to enhance transcription elongation. Nucl Acids Res. 2011;39:3505–3519. doi: 10.1093/nar/gkq1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, Baruchel A, Cave H, Dastugue N, Hasle H, Kaspers GL, Lessard M, Michaux L, Vyas P, van Wering E, Zwaan CM, Golub TR, Orkin SH. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci USA. 2006;103:3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RL, Davies PL. Structure-function relationships in calpains. Biochem J. 2012;447:335–351. doi: 10.1042/BJ20120921. [DOI] [PubMed] [Google Scholar]
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hu M, Shivdasani RA. Expression analysis of primary mouse megakaryocyte differentiation and its application in identifying stage-specific molecular markers and a novel transcriptional target of NF-E2. Blood. 2007;109:1451–1459. doi: 10.1182/blood-2006-08-038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E-c, Luo Q, Bruscia EM, Renda MJ, Troy JA, Massaro SA, Tuck D, Schulz V, Mane SM, Berliner N, Sun Y, Morris SW, Qiu C, Krause DS. Role for MKL1 in megakaryocytic maturation. Blood. 2009;113:2826–2834. doi: 10.1182/blood-2008-09-180596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PloS Pathogens. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib KE, Mihaylov IS, Delehanty LL, Bullock GC, Ouma KD, Caronia JF, Gonias SL, Goldfarb AN. Cross-talk of GATA-1 and P-TEFb in megakaryocyte differentiation. Blood. 2008;112:4884–4894. doi: 10.1182/blood-2008-03-145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Xie H, Obradovic Z, Grana X. Selective control of gene expression by CDK9 in human cells. J Cell Physiol. 2010;222:200–208. doi: 10.1002/jcp.21938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C, Rhodes KE, Gereige LM, Helgadottir H, Ferrari R, Kurdistani SK, Montecino-Rodriguez E, Bassel-Duby R, Olson EN, Krivstov AV, Armstrong S, Orkin SH, Pellegrini M, Mikkola HKA. MEF2C is a lineage-restricted target of Scl/Tal1 and regulates megakaryopoiesis and B-cell homeostasis. Blood. 2009;113:3461–3471. doi: 10.1182/blood-2008-07-167577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles L, Bluteau D, Boukour S, Chang Y, Zhang Y, Robert T, Dessen P, Debili N, Bernard OA, Vainchenker W, Raslova H. MAL/SRF complex is involved in platelet formation and megakaryocyte migration by regulating MYL9 (MLC2) and MMP9. Blood. 2009;114:4221–4232. doi: 10.1182/blood-2009-03-209932. [DOI] [PubMed] [Google Scholar]
- Halene S, Gao Y, Hahn K, Massaro S, Italiano JE, Schulz V, Lin S, Kupfer GM, Krause DS. Serum response factor is an essential transcription factor in megakaryocytic maturation. Blood. 2010;116:1942–1950. doi: 10.1182/blood-2010-01-261743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlett I, Draper J, Strouboulis J, Iborra F, Porcher C, Vyas P. Characterization of megakaryocyte GATA1-interacting proteins: the corepressor ETO2 and GATA1 interact to regulate terminal megakaryocytic maturation. Blood. 2008;112:2738–2749. doi: 10.1182/blood-2008-03-146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol Cell Biol. 2006;26:7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer MD, DeFranco DB. Hic-5/ARA55, a LIM domain-containing nuclear receptor coactivator expressed in prostate stromal cells. Cancer Res. 2006;66:7326–7333. doi: 10.1158/0008-5472.CAN-05-2379. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri Z, Shimizu R, Ohneda O, Maouche-Chretien L, Gisselbrecht S, Yamamoto M, Romeo PH, Leboulch P, Chretien S. Direct binding of pRb/E2F-2 to GATA-1 regulates maturation and terminal cell division during erythropoiesis. PloS Biology. 2009;7:1–15. doi: 10.1371/journal.pbio.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Godinho FJ, Heitmann K, Maroz A, Koch ML, Reinhardt D, Orkin SH, Li Z. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24:1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS ONE. 2010;5:1–12. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K, Kinoshita H, Kuwabara Y, Nakagawa Y, Usami S, Minami T, Yamada Y, Fujiwara M, Nakao K. Myocardin-related transcription factor A is a common mediator of mechanical stress- and neurohumoral stimulation-induced cardiac hypertrophic signaling leading to activation of brain natriuretic peptide gene expression. Mol Cell Biol. 2010;30:4134–4148. doi: 10.1128/MCB.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ma J, Zhu H, Singh M, Hill D, Greer PA, Arnold JM, Abel ED, Peng T. Targeted inhibition of calpain reduces myocardial hypertrophy and fibrosis in mouse models of type 1 diabetes. Diabetes. 2011;60:2985–2994. doi: 10.2337/db10-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yue S, Chen X, Kubin T, Braun T. Regulation of cardiomyocyte polyploidy and multinucleation by cyclinG1. Circ Res. 2010;106:1498–1506. doi: 10.1161/CIRCRESAHA.109.211888. [DOI] [PubMed] [Google Scholar]
- Lordier L, Jalil A, Aurade F, Larbret F, Larghero J, Debili N, Vainchenker W, Chang Y. Megakaryocyte endomitosis is s failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood. 2008;112:3164–3174. doi: 10.1182/blood-2008-03-144956. [DOI] [PubMed] [Google Scholar]
- Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding o the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhina S, Wang Y-l, Murata-Hori M. Alpha-actinin is required for tightly regulated remodeling of the actin cortical network during cytokinesis. Dev Cell. 2007;13:554–565. doi: 10.1016/j.devcel.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JP, Collao A, Chiong M, Maldonado C, Adasme T, Carrasco L, Ocaranza P, Bravo R, Gonzalez L, Diaz-Araya G, Hidalgo C, Lavandero S. The transcription factor MEF2C mediates cardiomyocyte hypertrophy induced by IGF-1 signaling. Biochem Biophys Res Commun. 2009;388:155–160. doi: 10.1016/j.bbrc.2009.07.147. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Crispino JD. Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood. 2005;106:1223–1231. doi: 10.1182/blood-2005-02-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins organizers of cell structure and function. Cell Adh Mig. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TJ, Balza R, Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J Mol Cell Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Nishikii H, Eto K, Tamura N, Hattori K, Heissig B, Kanaji T, Sawaguchi A, Goto S, Ware J, Nakauchi H. Metalloproteinase regulation improves in vitro generation of efficacious platelets from mouse embryonic stem cells. J Exp Med. 2008;205:1917–1927. doi: 10.1084/jem.20071482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pinto do OP, Kolterud A, Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized Steel factor-dependent multipotent hematopoietic precursors. EMBO J. 1998;17:5744–5756. doi: 10.1093/emboj/17.19.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Raslova H, Kauffmann A, Sekkai D, Ripoche H, Larbret F, Robert T, Tronik Le Roux D, Kroemer G, Debili N, Dessen P, Lazar V, Vainchenker W. Interrelation between polyploidization and megakaryocytic differentiation: a gene profiling approach. Blood. 2007;109:3225–3234. doi: 10.1182/blood-2006-07-037838. [DOI] [PubMed] [Google Scholar]
- Ridinger H, Rutenberg C, Lutz D, Buness A, Petersen I, Amann K, Maercker C. Expression and tissue localization of beta-catenin, alpha-actinin and chondroitin sulfate proteoglycan 6 is modulated during rat and human left ventricular hypertrophy. Exp Mol Pathol. 2009;86:23–31. doi: 10.1016/j.yexmp.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- Sano M, Schneider MD. Cyclin-dependent kinase-9 an RNAPII kinase at the nexus of cardiac growth and death cascades. Circ Res. 2004;95:867–876. doi: 10.1161/01.RES.0000146675.88354.04. [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucl Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EC, Thon JN, Devine MT, Lin S, Schulz VP, Guo Y, Massaro SA, Halene S, Gallagher P, Italiano JE, Krause DS. MKL1 and MKL2 play redundant and crucial roles in megakaryocyte maturation and platelet formation. Blood. 2012;120:2317–2329. doi: 10.1182/blood-2012-04-420828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E, Nosaka T, Lee WJ, Nakamura K, Takahashi T, Funaki M, Okada H, Hatanaka M, Maki M. Molecular diversity of calpastatin in human erythroid cells. Arch Biochem Biophys. 1993;303:349–354. doi: 10.1006/abbi.1993.1294. [DOI] [PubMed] [Google Scholar]
- Tan Y, Dourdin N, Wu C, De Veyra T, Elce JS, Greer PA. Conditional disruption of ubiquitous calpains in the mouse. Genesis. 2006;44:297–303. doi: 10.1002/dvg.20216. [DOI] [PubMed] [Google Scholar]
- Taneike M, Mizote I, Morita T, Watanabe T, Hikoso S, Yamaguchi O, Takeda T, Oka T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Takeda J, Mochizuki N, Komuro I, Otsu K. Calpain protects the heart from hemodynamic stress. J Biol Chem. 2011;286:32170–32177. doi: 10.1074/jbc.M111.248088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109:1503–1506. doi: 10.1182/blood-2006-04-020362. [DOI] [PubMed] [Google Scholar]
- Wendt A, Thompson VF, Goll DE. Interaction of calpastatin with calpain: a review. Biol Chem. 2004;385:465–472. doi: 10.1515/BC.2004.054. [DOI] [PubMed] [Google Scholar]
- Wickrema A, Crispino JD. Erythroid and megakaryocytic transformation. Oncogene. 2007;26:6803–6815. doi: 10.1038/sj.onc.1210763. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Bahler J, Pringle JR. Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell. 2001;12:1061–1077. doi: 10.1091/mbc.12.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, Teng L, Ingber DE, Mammoto A, Kumar R, Alaoui-Jamali MA. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207:2421–2437. doi: 10.1084/jem.20100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucl Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi O, Taneike M, Otsu K. Cooperation between proteolytic systems in cardiomyocyte recycling. Cardiovasc Res. 2012;96:46–52. doi: 10.1093/cvr/cvs236. [DOI] [PubMed] [Google Scholar]
- Yik JHN, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Yund EE, Hill JA, Keller RS. Hic-5 is required for fetal gene expression and cytoskeletal organization of neonatal cardiac myocytes. J Mol Cell Cardiol. 2009;47:520–527. doi: 10.1016/j.yjmcc.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.