Abstract
In this study, novel peptides (NIPP-1, NIPP-2) derived from Navicula incerta (microalgae) protein hydrolysate were explored for their inhibitory effects on collagen release in hepatic fibrosis with the investigation of its underlying mechanism of action. TGF-β1 activated fibrosis in LX-2 cells was examined in the presence or absence of purified peptides NIPP-1 and NIPP-2. Besides the mechanisms of liver cell injury, protective effects of NIPP-1 and NIPP-2 were studied to show the protective mechanism against TGF-β1 stimulated fibrogenesis. Our results showed that the core protein of NIPP-1 peptide prevented fibril formation of type I collagen, elevated the MMP level and inhibited TIMP production in a dose-dependent manner. The treatment of NIPP-1 and NIPP-2 on TGF-β1 induced LX-2 cells alleviated hepatic fibrosis. Moreover, α-SMA, TIMPs, collagen and PDGF in the NIPP-1 treated groups were significantly decreased. Therefore, it could be suggested that NIPP-1 has potential to be used in anti-fibrosis treatment.
Keywords: microalgae, hepatic stellate cells, hepatic fibrosis, LX-2, liver injury
INTRODUCTION
Hepatic fibrosis is a common response to various chronic hepatic injuries, characterized by the deposition of extracellular matrix (ECM). The key event in hepatic fibro-genesis is the activation of hepatic stellate cells (HSC) due to altered circumstances (1). Furthermore, hepatic fibrosis is a reversible wound healing response to chronic liver injury due to a variety of insults, including viral hepatitis (especially hepatitis B and C), alcohol abuse, drugs, metabolic diseases, autoimmune attack of hepatocytes and congenital abnormalities (2,3). The accumulation of ECM in HSC is caused by an increased synthesis of ECM by activated fibroblasts. Although the mechanisms for the pathological activation of fibroblasts in HSC are not completely understood, much evidence suggests that transforming growth factor (TGF) β and platelet-derived growth factor (PDGF) play important roles for activation of fibroblasts (1). However, accumulation of ECM results from both increased synthesis and decreased degradation. The factors responsible for ECM remodeling have been identified as a family of zinc-dependent enzyme matrix metalloproteinase (MMPs), their inhibitors, tissue inhibitor of metalloproteinases (TIMPs) and several converting enzymes. In human liver diseases, down-regulation of MMP-1 and up-regulation of MMP-2 and MMP-9 were observed (4), allowing degradation of normal matrix in the subendothelial space and its sub-stitude by fibrillar collagens. The activated MMPs are regulated in part by TIMPs (TIMP-1 and TIMP-2) (5). Accumulating evidence suggests that hepatic fibrosis is reversible and therefore recovery from cirrhosis may be possible. Increased collagenolytic activity is a major mechanism of fibrosis resolution.
LX-2 cells are a low-passaged human hepatic stellate cell line derived from normal human stellate cells that are spontaneously immortalized (4). LX-2 cells were selected by their ability to grow under low serum conditions. The cells exhibit the typical features of HSCs in primary culture, expressing desmin and glial acidic fibrillary protein, and responding to platelet-derived growth factor BB and TGF-β1.
Nowadays, microalgae provide numerous commercial applications. Moreover, microalgae are cultivated as a source of highly valuable molecules. Many studies have shown the interest of marine microalgae protein hydrolysates as functional foods and flavour enhancers. Recently, among marine organisms, microalgae was studied as a potent source of food additive, nutraceutical and pharmaceuticals. According to criteria of nutritional quality and cost, many marine organism sources have been investigated for the production of protein hydrolysates in functional foods (6). However, despite a variety of biological functions of microalgae, effective application of microalgae is still limited because of their inefficient cultivation and high production cost.
In the present study, TGF-β1 activated fibrosis in LX-2 cells was examined in the presence or absence of purified peptides from microalgae, Navicula incerta. Besides the mechanisms of liver cell injury, purified peptides were studied to show their protective mechanism against TGF-β1 stimulated fibrogenesis.
MATERIALS AND METHODS
Materials
The nuclear inhibitor of protein phosphatase (NIPP)-1 and NIPP-2 were prepared as described below, dissolved in distilled water, and stored in aliquots at −20ºC. All chemicals required for purification, including hydrolysate enzymes, were obtained from Sigma Chemical Co. (St. Louis, MO, USA) and Novo Nordisk (Bagsvaerd, Denmark). The MTT [3-(4,5-dimethyl-2-yl)-2,5-diphenylte-trazolium bromide] reagent and monobromobimane were purchased from Molecular Probes (Eugene, OR, USA). The Griess reagent and TPA (phorbol 12-myristate 13-acetate) were purchased from Sigma Chemical Co. Human recombinant TGF-β1 was from Biovision (Milpitas, CA, USA). Goat polyclonal anti-decorin, goat polyclonal anti-collagen type III antibody and peroxidase-conjugated anti-goat secondary antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Mouse monoclonal anti-α-SMA antibody was purchased from Sigma Chemical Co. Rabbit monoclonal anti-phospho-Smad2 antibody was purchased from Cell Signaling Technology (Danvers, MA, USA). BCA protein assay kit, electrophoresis reagents and goat anti-rabbit peroxidase IgG were purchased from Pierce Biotechnology Inc. (Rockford, IL, USA).
Primary and secondary antibodies used for Western blot analysis were purchased from Santa Cruz Biotechnology Inc. and Amersham Pharmacia Biosciences (Piscataway, NJ, USA), respectively. TRIzol reagent, oligo (dT) 15 primer, reverse transcriptase and Taq DNA polymerase were purchased from Promega (Madison, WI, USA). SB203580 and SN50 were obtained from Calbiochem (San Diego, CA, USA). Dulbecco’s modified eagle medium (DMEM), trypsin/EDTA, and fetal bovine serum (FBS) were purchased from Gibco BRL (Carlsbad, CA, USA) and Life Technologies (Carlsbad, CA, USA). All other chemicals and solvents were of analytical grade.
Preparation and identification of NIPP-1, NIPP-2
The benthic diatom, Navicula incerta (strain KMMCC B-001) used in this study was generously provided by Korea Marine Microalgae Culture Center. Culture medium was maintained in standard F/2 (Guillard’s) medium. Cell wall was broken down by enzyme Tunicase FN and hydrolyzed in 50ºC for 2 h. In brief, freeze dried Navicula incerta (100 g) was homogenized and enzymatically hydrolyzed with commercial enzymes (alcalase, neutrase, papain, pepsin, pronase, E trypsin and α-chymotrypsin). Substrate and enzyme were mixed at enzyme : substrate ratio of 1:100 (w/w) (7).
Lyophilized hydrolysates were loaded onto HiPrep 16/10 CM FF ion-exchange column (GE Healthcare, Uppsala, Sweden) equilibrated with 20 mM sodium acetate buffer (pH 4.0), and eluted with a linear gradient of NaCl (0~2 M) in the same buffer at a flow rate of 62 mL/h using fast protein liquid chromatography (FPLC). The UV absorbance at 280 nm was monitored in each 4 mL fractions, then pooled and lyophilized immediately. The lyophilized fractions were further purified on a Primesphere 10 C18 (20×250 mm) column permeation reversed-phase high-performance liquid chromatography (RPHPLC) with a linear gradient of acetonitrile (0~35% in 30 min) containing 0.1% trifluoroacetic acid (TFA) at a flow rate of 1.0 mL/min. Then the accurate molecular masses and amino acid sequences of purified peptides were ascertained by quadrupole time-of-flight (Q-Tof) mass spectroscopy (Micromass, Altrincham, UK) coupled to an electrospray ionization (ESI) source (8).
Culture of hepatic stellate cells LX-2 and transforming growth factor-beta1 (TGF- β1) treatment
LX-2, an immortalized human HSC line, was provided by Dr. S. L. Friedman, Mount Sinai School of Medicine, NY. Details for the generation of LX-2 have been described previously. Cells were cultured and maintained in DMEM supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% FBS and maintained at 37ºC under a humidified atmosphere with 5% CO2. The medium was changed 48 h after plating. Sequentially, the medium containing adenoviruses was removed and replaced with 0.8 mL/well (12-well dish) or 2.5 mL/dish (60-mm dish) of serum-free DMEM with human recombinant TGF-β1 (final concentration 2 ng/mL). The cultures of 12-well and 60-mm dishes were incubated for 24 or 48 h, respectively. Recombinant human TGF-β1 concentration to be used in experiments was determined by PCR assay by treating cells with different known concentrations.
Cell proliferation and cytotoxicity
MTT assay was used to determine the effect of TGF-β1 on the proliferation of human LX-2 cells. LX-2 cells were seeded (5×104 cell/well) into 96-well tissue culture plates containing 100 μL DMEM with 10% FBS. After 24 h of incubation (37ºC, 5% CO2), the medium was carefully removed and 100 μL fresh medium containing TGF-β1 (2 ng/mL) with various concentrations of NIPP-1 or NIPP-2 peptides added into the wells. After 48 h of incubation, 20 μL of MTT dye solution was added to each well. After 4 h incubation, 200 μL of solubilization/stop solution was added for dissolving the formazan crystals and the absorbance was read using GENios− Multifunction Microplate Reader (Tecan, Switzerland, UK) at 540 nm. Absorbance values were the mean±SE of triplicates for each treatment. The cells in only controls and compound controls were included.
For the cytotoxicity determination studies, the colorimetric [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] MTT assay was performed. The human LX-2 cells cultured in DMEM and treated with the different concentrations of purified peptides for 48 h in a humidified 5% CO2 environment at 37ºC. Sequentially, 20 μL of MTT dye solution was added to each well. After 4 h incubation, 200 μL of solubilization/stop solution was added for dissolving the formazan crystals and the absorbance was read using GENios− Multifunction Microplate Reader (Tecan) at 540 nm.
Measurement of procollagen type I produced by fibroblasts
Measuring procollagen type I C-peptide is the standard to detect type I collagen synthesis activity. Therefore, type I collagen production by fibroblasts was assessed by measuring the procollagen type I C-peptide concentration by enzyme-linked immunosorbent assay (ELISA) using a procollagen type I C-peptide kit (Takara Shuzo Co., ltd, Kyoto, Japan) as described. In brief, fibroblasts were seeded into 96-well tissue culture plates at a density of 4×104 cells/well and cultured at 37ºC in a 5% CO2 humidified incubator in DMEM containing 10% heat-inactivated fetal calf serum (FCS) until confluent. The medium was then replaced with FCS-free DMEM, and after 24 h of serum deprivation, tryptase was added as described in manufacturer’s instructions. As controls, heparin at concentrations from 0.01 to 10 mg/mL or serum-free medium was used in the collagen synthesis assay. After culturing for 24 h at 37ºC in the incubator, procollagen type I C-peptide concentrations in the culture supernatants of the fibroblasts were measured.
Sircol collagen assay
Cells were seeded into 6-well plates at a density 5×104 cells/well and cultured for 2 days. Total collagen content was determined by assaying total soluble collagen with a Sircol Collagen Assay Kit (Biocolor, County Antrim, UK) according to the manufacturer’s instructions. Each sample was incubated for 24 h at 4ºC with stirring. After centrifugation, 100 mL of each supernatant was assayed. One mL of Sircol dye reagent that binds to collagen was added to each sample and the solutions were mixed for 30 min. After centrifugation, the pellet was suspended in 1 mL of the alkali reagent included in the kit and read at 540 nm with a spectrophotometer. Collagen standard solution was utilized to construct a standard curve.
Indirect immunofluorescence staining of cultured cells
Following 2 days of culture, the media was removed from cultured HSC. For immunofluorescence staining, cells were fixed in 3% formaldehyde (Sigma Chemical Co.) in PBS for 10 minutes at room temperature followed by three washes with PBS, and then permeabilized with 0.3% Triton X-100 in PBS for 10 minutes. Subsequently, cells were stained with anti-actin, and anti-α-smooth muscle actin (anti-α-SMA) (IgG1, Sigma Chemical Co.) primary antibodies diluted in PBS-Tween 0.1% for 1 hour at room temperature. Samples were then incubated with FITC-conjugated anti-goat antibodies (IgG2a specific; Invitrogen, Carlsbad, CA, USA) for 1 h at room temperature. After washing in PBS, images were observed with a fluorescence microscope (CTR 6000, Leica, Wetzlar, Germany) at 200× magnification.
Western blot analysis
Whole cell protein extraction and Western blotting were performed as previously described (8). Briefly, cells were lysed with RIPA lysis buffer. Equal amounts of protein were separated on 10% sodium dodecyl sulfate (SDS)- polyacrylamide gels, and transferred onto nitrocellulose membranes. After incubation with the appropriate primary antibody, the membranes were incubated for 1 h at room temperature with a secondary antibody conjugated to horseradish peroxidase. Following three washes in tris-buffered saline-Tween (TBST), immunoreactive bands were visualized using ECL detection system (Amersham Pharmacia Biosciences), according to the manufacturer’s instructions. Western blots were visualized using an LAS30001 Luminescent image analyzer and protein expression was quantified by MULTI GAUGE V3.0 software (Fujifilm Life Science, Tokyo, Japan).
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
RNA isolation and RT-PCR analysis were performed according to standard procedures. Total RNA was extracted from LX-2 cells after treatment with NIPP-1 and NIPP-2. For the extraction, the cells were lysed with Trizol1 and centrifuged at 12,000 rpm for 15 min at 25ºC following the addition of chloroform. Isopropanol was added to the supernatant at a 1:1 ratio and the RNA pellet was obtained by centrifugation. After washing with ethanol, extracted RNA was solubilized in diethyl pyrocarbonate-treated RNAse-free water and quantified by measuring the absorbance at 260 nm using the GENios− multifunction microplate reader (Tecan). Equal amounts of RNA (1 μg) were reverse transcribed in a mastermix containing 1× reverse transcriptase (RT) buffer, 1 mM dNTPs, 500 ng of oligo (dT) 15 primers, 140 U of murine Moloney leukaemia virus (MMLV) reverse transcriptase and 40 U of RNase inhibitor, for 45 min at 42ºC. PCR was carried out in an automatic Whatman thermocycler (Biometra, Göttingen, Germany) to amplify TIMPs, MMPs, α-SMA, type 1 collagen, TGF-β1 and PDGF. Primer sequences used to amplify the desired cDNA were as follows:
TIMP-1 forward and reverse primers: 5′-TGACATCCGGTTCGTCTACA-3′ and 5′-TGATGTGCAAGAGTCCATCC-3′; TIMP-2 forward and reverse primers: 5′-AAGCGGTCAGTGAGAAGGAA-3′ and 5′-GGGGGCCGTGTAGATAAACT-3′; MMP-2 forward and reverse primers: 5′-CCCCAAGCTCATCGCAGAT-3′ and 5′-GGTCCACGACGGCATCC-3′; MMP-9 forward and reverse primers: 5′-CACTGTCCACCCCTCAGAGC-3′ and 5′-CACTTGTCGGCGATAAGG-3′; Type 1 collagen forward and reverse primers: 5′-CAGTCGCTTCACCTACAGCA-3′ and 5′-GGTGGAGGGAGTTTACACGA-3′; α-SMA forward and reverse primers: 5′-AGGCACCCCTGAACCCCAA-3′ and 5′-CAGCACCGCCTGGATAGCC-3′; TGF-β1 forward and reverse primers: 5′-CGACTGCCAAATGAAGAGGACC-3′ and 5′-AAACCTGAGCCAGAACCTGACG-3′; PDGF forward and reverse primers: 5′-CCAGAAGCCATCAGCAGCAAG-3′ and 5′-AGGCCCTGAGAGATCTGTGG-3′; and GAPDH forward and reverse primers: 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′ and 5′-CATGTAGGCCATGAGGTCCACCAC-3′. The reactions were performed with primers under the same conditions described above. The amplification cycles were 95ºC for 30 s, 55ºC for 1 min, and 72ºC for 45 s. After 35 cycles, the PCR products were separated by electrophoresis on a 1.5% agarose gel for 45 min at 100 V. Gels were then stained with 1 mg/mL ethidium bromide visualized by UV light using AlphaEase− gel image analysis software (Alpha Innotech, San Leandro, CA, USA).
Statistical analysis
Each value was expressed as means±SEM (n=3). The statistical significance of differences was analyzed by Student’s t-test using SPSS (Chicago, IL, USA).
RESULTS
Chemical analysis of NIPP- 1 and NIPP-2
The NIPP-1 and NIPP-2 were purified from the hydrolysate of microalgae, Navicula incerta using chromatographic methods, combining fast protein liquid chromatography (FPLC) on a HiPrep 16/10 CM FF ion-exchange column and repeated reversed-phase high-performance liquid chromatography (RPHPLC) on a Primesphere 10 C18 (20×250 mm, Phenomenex Co., Ltd., Torrance, CA, USA) column (8). The purity of Src homology region 2 domain-containing phosphatase-1 (SHP-1) was over 99% according to the assessment of RPHPLC and N-terminal sequence analysis. The molecular mass of the N. incerta hydrolysis, which isolated novel peptides NIPP-1 and NIPP-2, was determined to be 1,909 Da by Q-TOF mass spectroscopy coupled to an electrospray ionization (ESI) source. The amino acid sequence of the purified peptides were analysed as; NIPP-1 (Pro-Gly-Trp-Asn-Gln-Trp-Phe-Leu) with molecular mass 1,171 Da, and NIPP-2 (Val-Glu-Val-Leu-Pro-Pro-Ala-Glu-Leu) with molecular mass 1,108 Da (8).
Cell proliferation and cytotoxicity
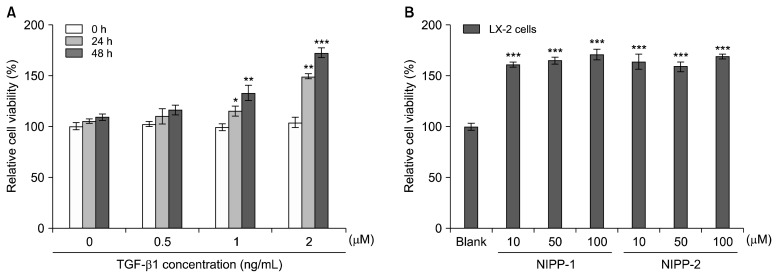
Effects of TGF-β1 on LX-2 cell proliferation and cell viability were examined by the MTT assay (Fig. 1). TGF-β1 was shown to increase LX-2 cell proliferation. In the absence of TGF-β1, no significant effect on LX-2 cell proliferation was observed. As shown in Fig. 1A, the effect of TGF-β1 on LX-2 cell proliferation seemed to be correlated with the dose and the effects seemed to increase with the time of incubation. The cell proliferation at 1 and 2 ng/mL concentrations of TGF-β1 was also observed at 24 and 48 h. However, TGF-β1 at 0 and 0.5 ng/mL concentrations did not affect proliferation evidently.
Fig. 1.
Effect of various concentrations of TGF-β on LX-2 cell proliferation at 48 h after incubation (A). Effects of NIPP-1 and NIPP-2 on the viability of LX-2 cells (B). Relative cell viability was assessed by the MTT assay, as described. Results of three independent experiments were averaged and shown as percentage cell viability compared with the viability of untreated control cells. Each value was expressed as the mean±SD of triplicate experiments. *P<0.05, **P<0.01 and ***P<0.001 as compared with blank.
Cytotoxicity of NIPP-1 and NIPP-2 were tested at three concentrations (10, 50 and 100 μM) using the MTT assay in cultured LX-2 cells. As depicted in Fig. 1B, neither NIPP-1 nor NIPP-2 showed significant (P< 0.05) cytotoxicity on LX-2 cells at all tested concentrations after 48 h of treatment. Non-toxic concentrations ranging from 10–100 μM/mL were selected for both peptides for further analysis.
Measurement of procollagen type I produced by fibroblasts
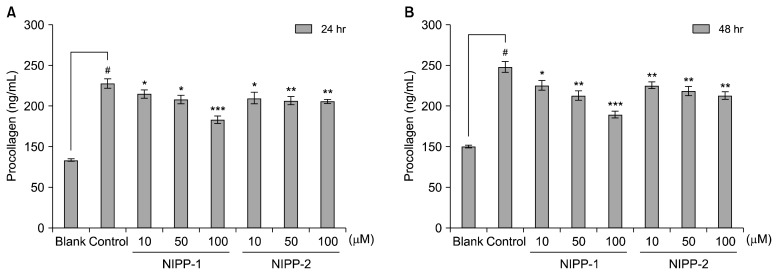
Soluble collagen assay was carried out to strengthen the data of the procollagen assay, which showed the inhibiting effect of NIPP-1 and NIPP-2 on the amount of cellular procollagen. Soluble collagen assay was performed for both supernatant and intracellular amounts of soluble collagen. Data gained from the soluble collagen assay correlated with the previous results in that both NIPP-1 and NIPP-2 decreased the soluble collagen in both supernatant and cells (Fig. 2). Ethanol addition increased the procollagen amount to 214 ng/mL and 237 ng/mL after 24 and 48 h incubation, respectively. NIPP-1 treatment decreased the elevated procollagen amount as much as 160 ng/mL at the highest concentration (100 μM) after 24 h incubation and 167 ng/mL after 48 h (Fig. 2A). On the other hand, NIPP-2 treatment lowered the procollagen amount to 187 ng/mL with 24 h treatment and 195 ng/mL after 48 h incubation at the concentration of 100 μM (Fig. 2B). Parallel to previous data, NIPP-1 showed higher activity than NIPP-2 for both supernatant and intracellular collagen amounts in a dose-dependent manner.
Fig. 2.
Effects of NIPP-1 and NIPP-2 on procollagen synthesis in LX-2 cells. 10% FBS was used for the tests to eliminate the interference of serum with the produced protein. Procollagen type I C-peptide assay was done after 24 h (A) and 48 h (B) of treatment. Each value was expressed as the mean±SD of triplicate experiments. *P<0.05, **P<0.01 and ***P<0.001 as compared TGF-β1 treated cells. #P<0.001 as compared with TGF-β1 non-treated cells.
Sircol collagen assay
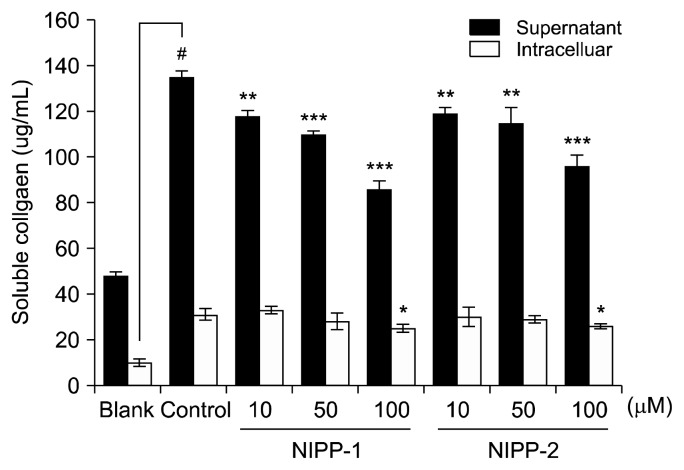
Procollagen production of LX-2 cells as a response to NIPP-1 and NIPP-2 was confirmed by procollagen type-1 C-peptide assay after 24 and 48 h treatment. Both peptides decreased the procollagen amount in a dose-dependent manner (Fig. 3). Soluble collagen amount was elevated to 135 μg/mL and 31 μg/mL found in supernatant and intracellular, respectively, after ethanol addition. NIPP-1 treatment lowered the elevated soluble collagen amount in supernatant to 86 μg/mL while NIPP-2 treatment decreased the supernatant soluble collagen amount to 96 μg/mL at the concentration of 100 μM. However, both NIPP-1 and NIPP-2 treatment failed to change in-tracellular soluble collagen amount. In detail, 100 μM NIPP-1 and NIPP-2 treatments kept the intracellular collagen amount at 25 μg/mL and 26 μg/mL respectively. Also, 48 h results showed that both NIPP-1 and NIPP-2 exert their protective effects against procollagen production in a time-dependent manner. Comparison of both 24 and 48 h results proved that NIPP-1 decreased the procollagen amount significantly than NIPP-2 did in both dose and time-dependent experiments.
Fig. 3.
Effects of NIPP-1 and NIPP-2 peptides on soluble collagen production in primary cultured LX-2 cells. The soluble collagen produced by the cells was not sufficient to confirm the difference, therefore only the tendency was considered. Each value was expressed as the mean±SD of triplicate experiments. *P<0.05, **P<0.01 and ***P<0.001 as compared with control. #P<0.001 as compared with blank.
Indirect immunofluorescence and cell imaging
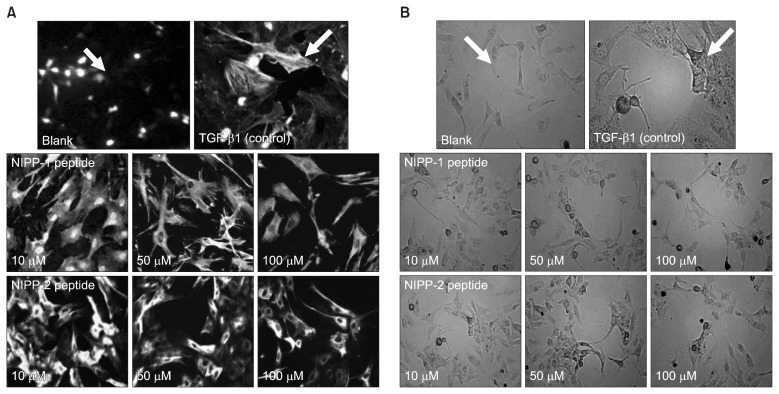
Indirect immunofluorescence was performed to observe the fibrosis inhibitory effect of NIPP-1 and NIPP-2. Images taken from the immunofluorescence assay clearly demonstrated the inhibitory activity of NIPP-1 and NIPP-2 (Fig. 4A). According to density of fluorescent light emitted from intracellular actin, NIPP-1 was observed to be more active than NIPP-2. Both peptides exerted a clear dose-dependent inhibition. However, at same concentration levels, NIPP-1 inhibited fibrosis significantly more than NIPP-2 and at the concentration of 100 μM, NIPP-1 inhibited the fibrogenesis almost to the level of 70% of control.
Fig. 4.
Effect on α-SMA expression in HSCs (LX-2) fibroblasts demonstrated by immunofluorescent cell images (A) and cell invasion (B). Cells were stained with the following primary antibodies: anti-actin, α–smooth muscle antibody (IgG1, Sigma). After 48 hr incubations, the cells were photographed under a phase contrast microscope (×20). Blank: non-stimulated, Control and Samples: TGF-β1-stimulated.
To show the effect of NIPP-1 and NIPP-2 on TGF-β1- induced cell fibroblasts visually, images of LX-2 cells (Fig. 4B) exposed to TGF-β1 and NIPP-1 and -2 were taken by a phase contrast microscope. Images of cell shapes tend to show fibroblast-like appearances (more spindle-like and spread) and high density after TGF-β1 initiation. However, images of peptide treated wells show decrease in spread, shape and density of cells. In addition, comparison of image data demonstrate similar anti-fibrosis effects of NIPP-1 and NIPP-2 where NIPP-1 inhibited the fibrogenesis slightly more than NIPP-2.
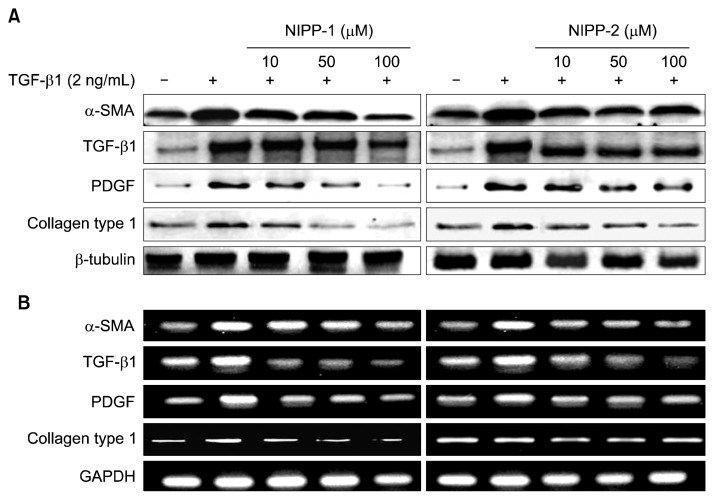
Effects of NIPP-1 and NIPP-2 on α-SMA, TGF-β1, collagen and PDGF expressions in LX-2 cells stimulated by TGF-β1
In LX-2 cells, TGF-β1 effects of NIPP-1 and NIPP-2 on the levels of α-SMA, TGF-β1, collagen and PDGF protein and mRNA expressions were determined by western blot analysis and RT-PCR, respectively (Fig. 5). TGF-β1 is the key mediator in human fibrogenesis. In HSCs, TGF-β1 favors the transition to myofibroblast-like cells, stimulates the synthesis of ECM proteins, and inhibits their degradation through the TGF-β1 signaling pathway. Thus, TGF-β1 stimulates cell proliferation through the PDGF-mediated pathway. Also, growth factor-stimulated ECM expansion directly increases collagen. In the western blot analysis, TGF-β1 treatment sufficiently increased α-SMA, TGF-β1, collagen type 1 and PDGF protein expressions compared to the non-treated groups. The stimulated α-SMA, TGF-β1 and PDGF protein expression levels were evidently decreased with NIPP-1 treatment (10, 50 and 100 μM/mL) for 48 h (Fig. 5A) while, NIPP-2 treatment did not show any significant decrease in protein expressions even at higher concentrations. However, in case of collagen type 1 both NIPP-1 and NIPP-2 treatment at different concentrations from 10 to 100 μM decreased the expression of type 1 collagen levels in LX-2 cells.
Fig. 5.
Effect of NIPP-1 and NIPP-2 treatment on α-smooth muscle actin (α-SMA), TGF-β, PDGF, collagen type 1 protein expressions (A) and mRNA levels (B) in HSCs stimulated with TGF-β1. LX-2 cells were treated with NIPP-1 and NIPP-2 in the presence or absence of 2 ng/mL TGF-β1 for 48 h.
RT-PCR analysis was conducted to find the effects of NIPP-1 and NIPP-2 peptides on mRNA expression levels of α-SMA, TGF-β1, collagen and PDGF genes in LX-2 cells stimulated by TGF-β1. The NIPP-1 and NIPP-2 peptide treatments for 48 h inhibited the TGF-β1 stimulated expression of α-SMA, TGF-β1, collagen type 1 and PDGF mRNA. Collective analysis of this data showed that both peptides have inhibitory activity against gene expressions of these proteins. Similar to western blot analysis results, NIPP-2 did not show significant activity even though the concentration was increased up to 100 μM. However, NIPP-1 treatment decreased the gene expression significantly (Fig. 5B).
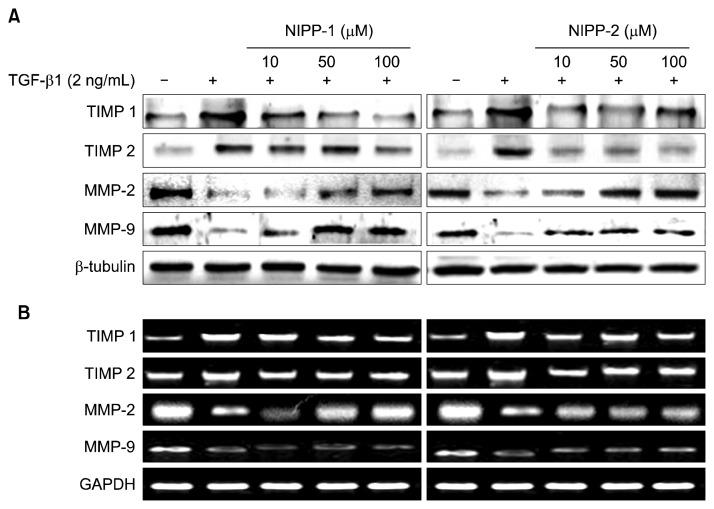
Effects on NIPP-1 and NIPP-2 of TIMPs and MMPs expressions in LX-2 cells stimulated with TGF-β1
TIMP-1 is a specific inhibitor of matrix metalloproteinases which acts as the key regulator of MMP activity and ECM degradation. Therefore, TIMPs are key mediators for decreasing collagen synthesis through inhibiting collagenase enzyme activity. Hence, expressions of TIMPs are directly related to fibrosis inhibition. The expression of TIMP-1 in LX-2 cells treated with TGF-β1 in the absence or presence of NIPP-1 and NIPP-2 was similar to that of TIMP-2. Expression of TIMP-1 mRNA in activated HSCs was attenuated by NIPP-1 and NIPP-2. Both NIPP-1 and -2 peptides decreased TIMP-1 and -2 protein transcriptions and gene expressions in a dose dependent-manner in activated HSCs, which were observed by western blot and RT-PCR analysis (Fig. 6). For both peptides, RT-PCR and Western blot analysis demonstrated similar patterns. However, parallel to previous results, NIPP-2 activity was not as significant as NIPP-1, which clearly decreased both protein and gene expressions.
Fig. 6.
Effect of NIPP-1 and NIPP-2 treatment on TIMPs and MMPs protein expressions (A) and mRNA levels (B) in HSCs stimulated with TGF-β1. LX-2 cells were treated with NIPP-1 and NIPP-2 in the presence or absence of 2 ng/mL TGF-β1 for 48 h.
MMPs have important roles in degrading collagen fibers. Thus, MMP activity is directly related to fibrosis in HSCs. Alcoholic hepatic fibrosis causes elevated levels of TIMPs which directly lowers the MMP levels. Therefore, elevation of MMPs in a fibrosis condition is crucial for improved alcoholic liver toxicity.
To examine the effect of NIPP-1 and NIPP-2 peptides on TGF-β1 induced specific MMP-2 and -9 expressions, LX-2 cells were incubated with various concentrations of NIPP-1 and NIPP-2 peptides which were previously stimulated with TGF- β1. Collectively, protein levels of MMP-2 and -9, demonstrated by western blot analysis, followed the same pattern with mRNA expression levels. NIPP-1 efficiently elevated MMP levels at all concentrations. Different concentrations of NIPP-1 ranging from 10 to 100 μM/mL showed a marked elevation of MMP-2 mRNA and protein levels in a dose dependent manner while elevating the MMP-9 levels as well, however in a slightly decreased manner. NIPP-2 treatment only increased the protein and mRNA expressions of MMP-2 and MPP-9, whereby MMP-2 levels were increased more than MMP-9.
DISCUSSION
Hepatic stellate cells (HSCs) have been regarded as a key cellular source of ECM in hepatic fibrosis. In the normal liver, HSCs are in the quiescent state, but upon injury, HSCs become activated and begin to proliferate (9). LX-2, a human HSC cell line used in the present study, retains key features of HSCs and provides a valuable in vitro model in the study of liver disease. Increased fibrogenesis is the most direct way in which HSCs contribute to hepatic fibrosis (10).
TGF-β1 is a potent fibrogenic factor affecting a wide spectrum of cellular processes including differentiation, proliferation, apoptosis and migration. Activation of stellate cells contributes to hepatic fibrosis and TGF-β1 is the most potent fibrogenic factor. Several studies have reported that modulation of oxygen tension was recognized as an important regulator of gene activation of TGF-β1 in rat brain and liver, and human fibroblast and hepatoma cells. TGF-β1 takes part in initiation and maintenance of fibrogenesis in the liver (10,11). In our experiments, TGF-β1 was used as the stimulator of HSCs for the formation of an in vitro fibrogenic model. Under treatment with TGF-β1, HSCs had significant changes in proliferation, gene and protein expressions. Therefore, specific inhibition of TGF-β1 seems to be a promising approach for anti-fibrosis (11,12).
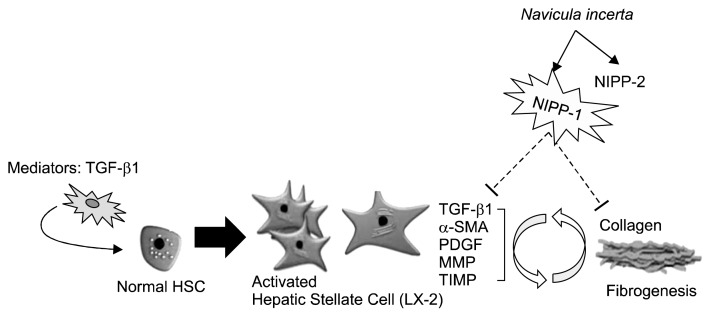
In this study, we have shown that LX-2 cells proliferate under stimulation by TGF-β1, suggesting the possibility of using NIPP-1 and NIPP-2 isolated from micro-algae, Navicula incerta to prevent and reduce hepatic fibrosis through directly binding TGF-β1 and partially blocking its signal transduction. A major group of enzymes responsible for ECM degradation is the MMP family (2). TIMPs, specific inhibitors of MMPs, have been identified as the important regulators of MMP activity and ECM degradation (13). Expression of TIMP-1 and MMP-2 is increased in hepatic fibrosis, which reflects the changes of hepatic fibrosis (14,15). In our study, Western blot and RT-PCR results showed that NIPP-1 significantly inhibited mRNA expression of TIMP-1 and MMP-2, which were up-regulated under TGF-β1 stimulation, suggesting that NIPP-1 has the ability of modulating or attenuating TGF-β1 activity (Fig. 7). Furthermore, Western blotting results also demonstrated that the increased expression level of type I collagen stimulated by TGF-β1 was reduced by NIPP-1. In addition, the expression of α-SMA, the marker of HSC activation, was remarkably increased by exogenous TGF-β1. NIPP-1 inhibited the elevation of α-SMA, indicating further that NIPP-1 may arrest HSC activation. Our results showed that the core protein of NIPP-1 peptide prevented fibril formation of α-SMA, type I collagen, MMPs and TIMPs in a dose-dependent manner.
Fig. 7.
Potential acting mechanism of NIPP-1 peptide on TGF-β1 stimulated HSCs (LX-2). Evidently, NIPP-1 peptide normalize the elevated levels of fibrosis mediators (TGF-β, PDGF, TIMPs and collagen) in TGF-β1 stimulated HSCs (LX-2).
Microalgae are able to enhance the nutritional content of conventional food preparations and hence, positively affect the health of humans and animals (16) due to their original chemical composition. The high protein content of various microalgal species is one of the main reasons to consider them as an unconventional source of protein (17). In addition, the amino acid pattern of almost all algae compares favorably with that of other food proteins (18). As the cells are capable of synthesizing all amino acids, they can provide the essential ones to humans and animals (19). However, to completely characterize the protein and determine the amino acid content of microalgae, information on the nutritive value of the protein and the degree of availability of amino acids should be given. Many metabolic studies have confirmed the capacities of microalgae as a novel source of protein: the average quality of most of the algae examined is equal or even superior to that of other conventional high-quality plant proteins (20).
Therefore, our study suggests NIPP-1 as a highly potent bioactive peptide to be utilized as a food additive, nutraceutical and pharmaceutical agent.
ACKNOWLEDGMENTS
This research was supported by a grant from Marine Bioprocess Research Center of the Marine Biotechnology Program funded by the Ministry of Oceans and Fisheries, Republic of Korea.
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 2.Czochra P, Klopcic B, Meyer E, Herkel J, Garcia-Lazaro JF, Thieringer F, Schirmacher P, Biesterfeld S, Galle PR, Lohse AW, Kanzler S. Liver fibrosis induced by hepatic overexpression of PDGF-β in transgenic mice. J Hepatol. 2006;45:419–428. doi: 10.1016/j.jhep.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Felver ME, Mezey E, McGire M, Mitchell MC, Herlong HF, Veech GA, Veech RL. Plasma tumor necrosis factor alpha predicts decreased long-term survival in severe alcoholic hepatitis. Alcohol Clin Exp Res. 1990;14:255–259. doi: 10.1111/j.1530-0277.1990.tb00482.x. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL. The cellular basis of hepatic fibrosis– Mechanisms and treatment strategies. N Engl J Med. 1993;328:1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- 5.Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21:437–451. doi: 10.1055/s-2001-17558. [DOI] [PubMed] [Google Scholar]
- 6.Spolaore P, Joannis-Cassan C, Duran E, Isambert A. Commercial applications of microalgae. J Biosci Bioeng. 2006;101:87–96. doi: 10.1263/jbb.101.87. [DOI] [PubMed] [Google Scholar]
- 7.Kang KH, Qian ZJ, Ryu BM, Kim SK. Characterization of growth and protein contents from microalgae Navicula incerta with the investigation of antioxidant activity of enzymatic hydrolysates. Food Sci Biotechnol. 2011;20:183–191. [Google Scholar]
- 8.Kang KH, Qian ZJ, Ryu BM, Karadeniz F, Kim DK, Kim SK. Antioxidant peptides from protein hydrolysate of microalgae Navicula incerta and their protective effects in HepG2/CYP2E1 cells induced by ethanol. Phytother Res. 2012;26:1555–1563. doi: 10.1002/ptr.4603. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Friedman SL. Liver fibrogenesis and the role of hepatic stellate cells: new insights and prospects for therapy. J Gastroenterol Hepatol. 1999;14:618–633. doi: 10.1046/j.1440-1746.1999.01928.x. [DOI] [PubMed] [Google Scholar]
- 10.Gressner AM, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J Cell Mol Med. 2006;10:76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanzler S, Lohse AW, Keil A, Henninger J, Dienes HP, Schirmacher P, Rose-John S, zum Buschenfelde KH, Blessing M. TGF beta1 in liver fibrosis: an inducible transgenic mouse model to study liver fibrogenesis. Am J Physiol. 1999;276:1059–1068. doi: 10.1152/ajpgi.1999.276.4.G1059. [DOI] [PubMed] [Google Scholar]
- 12.Iredale JP. Hepatic stellate cell behavior during resolution of liver injury. Semin Liver Dis. 2001;21:427–436. doi: 10.1055/s-2001-17557. [DOI] [PubMed] [Google Scholar]
- 13.Reeves HL, Friedman SL. Activation of hepatic stellate cells–a key issue in liver fibrosis. Front Biosci. 2002;7:808–826. doi: 10.2741/reeves. [DOI] [PubMed] [Google Scholar]
- 14.Shi YF, Zhang Q, Cheung PY, Shi L, Fong CC, Zhang Y, Tzang CH, Chan BP, Fong WF, Chun J, Kung HF, Yang M. Effects of rhDecorin on TGF-beta1 induced human hepatic stellate cells LX-2 activation. Biochimica Biophysica Acta. 2006;1760:1587–1595. doi: 10.1016/j.bbagen.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Svegliati-Baroni G, D’Ambrosio L, Ferreti G, Casini A, Di Sario A, Salzano R, Ridolfi F, Saccomanno S, Jezequel AM, Benedetti A. Fibrogenic effect of oxidative stress on rat hepatic stellate cells. Hepatology. 1998;27:720–726. doi: 10.1002/hep.510270313. [DOI] [PubMed] [Google Scholar]
- 16.Borowitzka MA. Microalgae as sources of pharmacetuicals and other biologically active compounds. J Appl Phycol. 1995;7:3–15. [Google Scholar]
- 17.Brown MR, Jeffrey SW, Volkman JK, Dunstan GA. Nutritional properties of microalgae for mariculture. Aquaculture. 1997;151:315–331. [Google Scholar]
- 18.Brown MR, Jeffreyi SW. The amino acid and gross composition of marine diatoms potentially useful for mariculture. J Appl Phycol. 1995;7:521–527. [Google Scholar]
- 19.Lupo MP. Antioxidants and vitamins in cosmetics. Clin Dermatol. 2001;19:467–473. doi: 10.1016/s0738-081x(01)00188-2. [DOI] [PubMed] [Google Scholar]
- 20.Sakata K. Antioxidants from marine organisms. In: Hiramatsu M, Yoshikawa T, Inoue M, editors. Food and Free Radicals. Springer Publishing; Tokyo, Japan: 2006. pp. 85–100. [Google Scholar]