Abstract
Accurate segmentation is an important preprocessing step for measuring the internal deformation of the tongue during speech and swallowing using 3D dynamic MRI. In an MRI stack, manual segmentation of every 2D slice and time frame is time-consuming due to the large number of volumes captured over the entire task cycle. In this paper, we propose a semi-automatic segmentation workflow for processing 3D dynamic MRI of the tongue. The steps comprise seeding a few slices, seed propagation by deformable registration, random walker segmentation of the temporal stack of images and 3D super-resolution volumes. This method was validated on the tongue of two subjects carrying out the same speech task with multi-slice 2D dynamic cine-MR images obtained at three orthogonal orientations and 26 time frames. The resulting semi-automatic segmentations of 52 volumes showed an average dice similarity coefficient (DSC) score of 0.9 with reduced segmented volume variability compared to manual segmentations.
Keywords: Tongue, segmentation, random walker, deformable registration, super-resolution reconstruction
1. INTRODUCTION
The mortality due to tongue cancer is lower than other cancers, but the incidence of oral cancer has increased in the last four decades. Generally, treatment with surgical ablation of tongue tumor (glossectomy) and chemo-radiotherapy may lead to speech and swallowing complications, thus affecting the quality of the patient’s life. Therefore, understanding the relationship between tumor, tongue structure and function becomes crucial for diagnosis, surgical planning and outcomes, and scientific studies. However, the ability to measure speech or swallowing dysfunction in these patients has been limited and is largely semi-quantitative. Characterization of tongue motion is challenging because the tongue deforms rapidly over a wide range with complex interactions between multiple muscles to produce fast and accurate movements [1]. Currently, there is no tool to directly characterize tongue motion and function with respect to surgical approach and reconstruction procedures, or chemo-radiation treatment in these patients [2].
Magnetic resonance imaging (MRI) plays an important role in the analysis of the structure and function of the tongue due to its excellent soft tissue contrast. In particular, fast MR imaging with tagging and target tracking capability allows quantitative analysis of tongue motion while carrying out a specific speech or swallowing task. 3D dynamic MRI (alternatively, 4D-MRI) is desirable because it yields volumetric images that change over time. However, there still exist undesirable trade-offs between temporal resolution and signal-to-noise ratio (SNR), i.e., image quality [3]. Consequently, the majority of dynamic MR imaging techniques involve sequential multi-slice 2D image acquisition as it is readily suited to minimize intra-scan motion while maintaining high spatio-temporal resolution [3-6]. There have been numerous attempts to compute 3D motion using tagged-MRI, mostly for cardiac motion analysis [4-7]. Several well-established algorithms such as the harmonic phase (HARP) tracking algorithm [4, 5] and incompressible deformation estimation algorithm (IDEA) [6] enabled a computation of 2D and 3D motion fields from tagged-MR data. We have recently proposed a workflow (see Fig. 1) using HARP and IDEA to analyze 3D motion of the tongue from multi-slice dynamic cine- and tagged-MRI [8, 9].
Fig. 1.
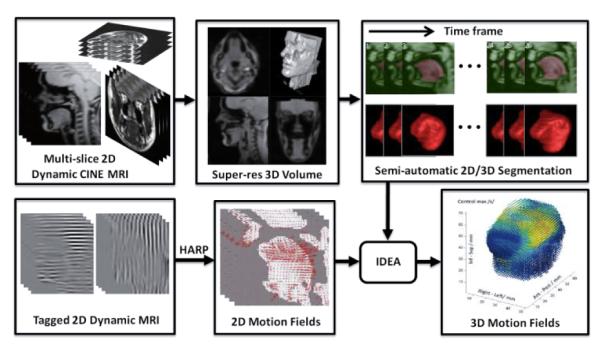
3D tongue motion analysis workflow based on multi-slice dynamic cine- and tagged-MRI.
Image segmentation of the target anatomical structures is a fundamental and challenging problem in the MR-based analysis. 3D motion analysis requires properly segmented tongue volume masks on which the 3D motion field is computed. Although there are numerous methods available for a single MR image/volume segmentation [10], there is no systematic and efficient approach to segment time-varying volumes for motion analysis. Therefore, the user has to segment individual slices or volumes at every time frame using a manual or semi-automatic method. This is time-consuming due to the large number of images or volumes throughout the entire task cycle; in our experiments, there are 26 volumes per second.
This paper proposes a semi-automatic segmentation method, which bridges the gap between fast 4D-MR image acquisition and established 2D/3D motion analyses to complete the dynamic MR-based tongue motion analysis workflow. The proposed method computes a tongue mask at every time frame with minimal user input, thus significantly alleviating the segmentation burden for the user.
2. METHODS
2.1. Image acquisition
Multi-slice 2D dynamic cine-MRI and tagged-MRI datasets are acquired from the subject using exactly the same orientation, spatial and temporal parameters in the axial, coronal, and sagittal orientations while the subject repeats a speech task. The slice image repetitions are sorted based on the speech phase, and averaged to produce an averaged multi-slice 2D dynamic MR image sequence at three orthogonal orientations and multiple time frames. A typical number of slices in each orientation of the cine- and tagged-MR datasets in our experiments is 10-12 axial, 9-14 coronal and 7 sagittal. The tagged images contain horizontal and vertical tags over 26 time frames. Each image is 128×128 pixels with a pixel size of 1.875×1.875 mm2, and both slice-thickness and tag spacing are 6 mm.
2.2. Super-resolution volume reconstruction
These multi-slice 2D dynamic MR scans provide high in-plane resolution (1.875 mm), but relatively poor through-plane (slice-selection direction) resolution (6.0 mm). Consequently, each dataset by itself is not sufficient for volumetric image processing and analysis such as segmentation, registration, and 3D motion analysis. To overcome this limitation, we derive a high-resolution, isotropic 3D volume from the three orthogonal 2D multi-slice image stacks using a super-resolution reconstruction technique developed by our group [11]. We first generate isotropic volumes by upsampling each stack in the through-plane direction using a fifth-order B-spline interpolation. We choose a target volume (in this study, sagittal) and register the other two volumes (axial and coronal) to the target. We register by translating in the 3 degrees-of-freedom using mutual information as a similarity measure because the currently implemented HARP tracking and IDEA algorithms can only accommodate translational motion of the tagged image plane. Slight intensity differences between the three registered volumes are corrected by using a spline-based intensity regression method that uses local intensity matching [11]. A single super-resolution volume of 128×128×128 voxels with an isotropic voxel size of 1.875 mm is reconstructed by averaging these intensity-corrected registered volumes. The super-resolution reconstruction is computed at every time frame to form a high-quality 3D dynamic MRI. The super-resolution volume not only provides a 3D volume with higher spatial resolution, but also reduces the blurring artifact caused by the slight misalignment between multi-slice images at three orientations through registration process. Therefore, direct segmentation of the 3D super-resolution volume yields an improved segmentation outcome compared to each 2D slice image segmentation followed by merging them into a 3D mask.
2.3. Random walker segmentation
We use the random walker (RW) segmentation algorithm [12] for segmenting both 2D cine images and 3D super-resolution volumes due to its attractive features such as fast computation, flexibility, ease of user interaction, and ability to produce an arbitrary segmentation with enough interaction. RW is a robust, graph-based, semi-automatic algorithm to find a globally optimal probabilistic solution for multi-label, interactive image segmentation. A user specifies a small number of pixels with user-defined labels as seeds (in our case, on the tongue and the background). Each unlabeled pixel is assigned to the label with the greatest probability that a random walker starting at this pixel will reach one of the seeds with this label. An interactive segmentation method is desirable for our application because the user often has to segment the region where there is no obvious image contrast and sometimes needs to edit the segmentation results.
We define a graph that consists of a pair G = (V,E) with vertices (or nodes) v ∈ V and edges e ∈ E. We use a typical Gaussian weighting to each edge eij given by wij = exp{–β(gi – gj)2} where gi indicates the image intensity at pixel i and β is a free parameter for which we used the same value as in [12]. The RW probabilities are found by minimizing the combinatorial Dirichlet problem , where x is a real-valued vector defined over the set of nodes and L represents the combinatorial Laplacian matrix [12].
2.4. Temporal stack segmentation
In a single cine-MRI dataset, there are many temporal stacks that must be segmented, one for each slice, each orientation, and each time frame. All together, there are about 800 images (~10 slices × 3 orientations × 26 time frames), which can be parsed into ~30 temporal stacks (~30 slices/time frame, each with 26 time frames) or 26 super-resolution volumes (1 volume/time frame × 26 time frames). It is challenging to segment all of the obtained 2D cine images (or the super-resolution volumes) due to the amount of data. Therefore, we propose here a systematic approach to segment the original 2D temporal stacks at once based on a minimal set of user-placed seeds to get 2D tongue masks.
A single temporal stack is smoother between adjacent time frames than between adjacent slices due to the relatively large slice spacing. For each slice, we use time as the third dimension instead of through-plane direction to form a 3D stack volume (2D target view + time). We segment this 3D stack volume using RW segmentation. For each slice, seeds need to be input at only one time frame and then propagated to 3-4 other distributed time frames by B-spline deformable registration (see Fig. 2(a)). The user-given and propagated seeds are then used to segment the 3D temporal stack volume using RW (Fig. 2(b)). The process is repeated for different slices at any orientations. Note that we only need to process several user-chosen slices (in this study, we only use 7 sagittal slices) that are well-spread over the target volume because these 3D temporal stack segmentations are used to segment 3D super-resolution volumes of all 26 time frames.
Fig. 2.
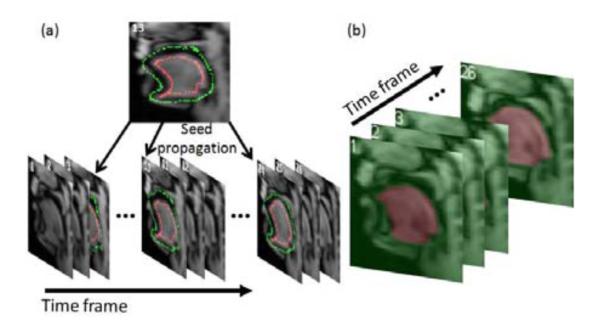
Temporal stack segmentation of a set of sagittal images in a single slice. (a) Seeds provided by the user at time frame 13 (red: tongue, green: background), are propagated to different time frames (3, 10, 24). (b) 3D temporal stack segmentation by RW using the seeds in (a).
2.5. Super-resolution volume segmentation
3D temporal stack segmentations (7 sagittal slices in our case) are used to generate seeds for the segmentation of 26 super-resolution volumes. Since the 2D cine images are sub-planes of the super-resolution volume, seeds on the 2D cine images can be directly imported to the corresponding slice images of the 3D super-resolution volume. For the time frames where no seeds are provided, seeds are extracted from the segmented 2D masks. To remove possible segmentation errors near the boundary and reliably extract seeds from the segmented 2D mask, the segmented mask M is first eroded using a disk structuring element D. Eroded mask Me for each label is computed by
where E is a Euclidean space, Ds is a translation of D by the vector s, i.e., Ds = {x + s|x ∈ D}, ∀s ∈ E. Boundary and the skeleton of the eroded mask for each label l are then extracted. Image skeleton is computed by the medial axis transform. Seeds for each label are created by the union of the points on the boundary and the skeleton of the eroded mask:
where i is the slice index and Nl is the number of labels.
Once all the seeds are imported and extracted from the 2D cine images, the super-resolution volume at each time frame is segmented by RW using these seeds. Figure 3 shows an example of seeds extracted to a sagittal slice of a super-resolution volume from a 2D segmented mask. Figure 4 shows two example super-resolution volume segmentations performed on time frame 13 (seeds are provided) and 20 (seeds are extracted from 2D cine segmented masks).
Fig. 3.
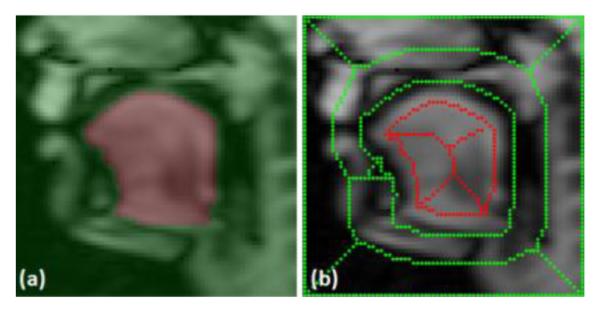
Seed extraction from 2D cine to 3D super-resolution volume. (a) Segmented 2D cine image. (b) Corresponding sagittal slice of the super-resolution volume overlaid with extracted seeds.
Fig. 4.
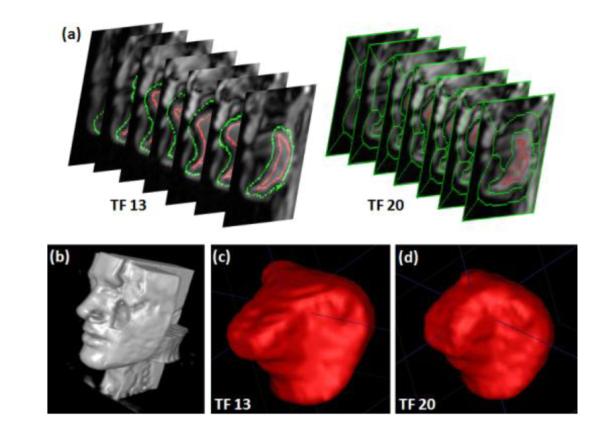
Example segmentations of super-resolution volumes of tongue. (a) User-given seeds imported to the super-resolution volume at time frame 13 (left) and the seeds extracted from 2D sagittal temporal stack segmentations at time frame 20 (right). (b) Surface rendering of a super-resolution reconstruction. The tongue is located in middle of the volume where axial, coronal, sagittal images are intersecting (c) Surface rendering of the segmented tongue at time frame 13 and (d) at time frame 20 among 26 super-resolution volume segmentations.
3. RESULTS
We evaluated the proposed methods on two normal volunteers who performed the same speech task. Each subject repeated the sound “asouk” and multi-slice cine- and tagged-MR images (128×128 pixels, a pixel size of 1.875×1.875 mm2) were acquired. A user-chosen ROI of 70×70 pixels on each slice was used for segmentation. Subject 1 data had 12 axial, 14 coronal and 7 sagittal slice images, and the subject 2 data had 10 axial, 9 coronal and 7 sagittal slice images. There were 26 time frames for both data sets. An isotropic super-resolution volume (128×128×128 voxels, voxel size of 1.875×1.875×1.875 mm3) was reconstructed at every time frame. The user provided seeds on 7 sagittal slices only at time frame 13 (middle of 26 time frames), and the seeds were propagated to time frames 3, 10, 17, 24. For each slice, 26 time frames were stacked to form a 70×70×26 3D temporal stack volume, and it was segmented by RW using the seeds available at 5 time frames (3, 10, 13, 17, 24). For every time frame, corresponding 3D super-resolution volume was then segmented using the seeds generated from the temporal stack segmentations. Figure 4 shows two example segmented surfaces of subject 2 computed by RW at two time frames with user-provided (frame 13) and extracted (frame 20) seeds.
In order to evaluate the semi-automatic segmentation quality, a trained scientist manually segmented all 52 super-resolution volumes (1 volume/time frame × 26 time frames × 2 subjects). DSCs between the semi-automatic and the manual segmentations were 0.89 and 0.9 for the subject 1 and 2, respectively (Table 1). Since the tongue is known to be incompressible, i.e., the volume of the segmented tongue mask at every time frame should not vary [6, 9], we measured the volume variation of the segmented masks in the manual and semi-automatic methods. The volume changes are plotted in Fig. 5, and the mean and standard deviation of segmented volume sizes are summarized in Table 1, showing that the segmented volume size is more constant with the semi-automatic than manual segmentation.
Table 1.
Evaluation of the segmented volumes and the volume variability. The manual and semi-automatic segmentations are compared. DSC scores, and the mean and standard deviation of the sizes of the 26 segmented volumes for each subject are shown
| Subject | DSC | Segmented volume variability mean±std (cm3) |
|
|---|---|---|---|
| Semi-automatic | Manual | ||
| S1 | 0.89 | 116.5 ± 4.7 | 120.1 ± 6.4 |
| S2 | 0.90 | 102.9 ± 4.4 | 102.2 ± 5.2 |
Fig. 5.
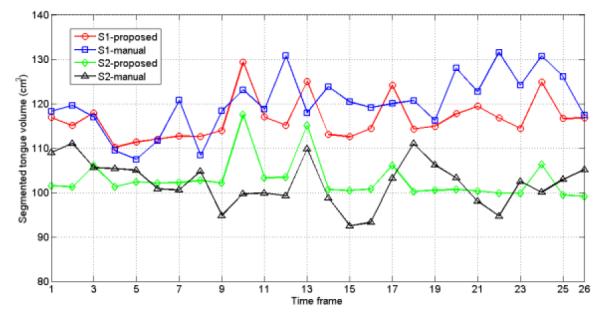
Segmented volume variability over time for two subjects (S1 and S2) with the manual and semi-automatic segmentations.
4. CONCLUSIONS
In this paper, we described a semi-automatic segmentation method for dynamic MR-based 3D motion analysis of the human tongue. The proposed semi-automatic segmentation method requires user-given seeds only on a few slice images and time frames and automatically propagates the seeds to the other frames by deformable registration. Furthermore, successive 2D temporal stack volume segmentation followed by the super-resolution volume segmentation by RW over all time frames enable segmentation of the time-varying volumes with minimal user interaction, thus significantly reducing the segmentation burden while keeping more consistent segmentation quality.
Acknowledgement
This work was supported by NIH/NCI grant 5R01CA133015.
5. REFERENCES
- [1].Kier WM, Smith KK. Tongues, tentacles and trunks: the biomechanics of movement in muscular-hydrostats. Zool. J. Linnean Soc. 1985;vol. 83:307–324. [Google Scholar]
- [2].Bokhari WA, Wang SJ. Tongue reconstruction: recent advances. Curr Opin Otolaryngol Head Neck Surg. 2007;vol. 15(no. 4):202–207. doi: 10.1097/MOO.0b013e3281fbd406. [DOI] [PubMed] [Google Scholar]
- [3].Dinkel J, Hintze C, Tetzlaff R, Huber PE, Herfarth K, Debus J, Kauczor HU, Thieke C C. 4D-MRI analysis of lung tumor motion in patients with hemidiaphragmatic paralysis. Radiother Oncol. 2009;vol. 91:449–454. doi: 10.1016/j.radonc.2009.03.021. [DOI] [PubMed] [Google Scholar]
- [4].Osman NF, Kerwin WS, McVeigh ER, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn. Reson. Med. 1999;vol. 42(no. 6):1048–1060. doi: 10.1002/(sici)1522-2594(199912)42:6<1048::aid-mrm9>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu X, Prince JL. Shortest path refinement for motion estimation from tagged MR images. IEEE Trans Med. Imag. 2010;vol. 29(no. 8):1560–1572. doi: 10.1109/TMI.2010.2045509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu X, Elmoniem K. Abd, Stone M, Murano EZ, Zhuo J, Gullapalli R, Prince JL. Incompressible deformation estimation algorithm (IDEA) from tagged MR images. IEEE Trans Med. Imag. 2011;vol. 31(issue 2):326–340. doi: 10.1109/TMI.2011.2168825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Human heart: tagging with MR imaging – a method for noninvasive assessment of myocardial motion. Radiology. 1988;vol. 169:59–63. doi: 10.1148/radiology.169.1.3420283. [DOI] [PubMed] [Google Scholar]
- [8].Parthasarathy V, Prince JL, Stone M, Murano E, Nessaiver M. Measuring tongue motion from tagged cine-MRI using harmonic phase (HARP) processing. J. Acoust. Soc. Am. 2007;vol. 121(no. 1):491–504. doi: 10.1121/1.2363926. [DOI] [PubMed] [Google Scholar]
- [9].Xing F, Lee J, Woo J, Murano E, Stone M, Prince JL. Estimating 3D tongue motion with MR images. Asilomar Conference on Signals, Systems, and Computers, Monterey, CA. 2012 Nov. [Google Scholar]
- [10].Balfar MA, Ramli AR, Saripan MI, Mashohor S. Review of brain MRI image segmentation methods. Artif Intell Rev. 2010;vol. 33:261–274. [Google Scholar]
- [11].Woo J, Murano EZ, Stone M, Prince JL. Reconstruction of high-resolution tongue volumes from MRI. accepted to IEEE Trans Biomed. Eng. 2012 doi: 10.1109/TBME.2012.2218246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Grady L. Random walks for image segmentation. IEEE Trans Pattern Anal. Mach. Intell. 2006;vol.28(no.1):1768–1783. doi: 10.1109/TPAMI.2006.233. [DOI] [PubMed] [Google Scholar]


