SUMMARY
Translesion DNA synthesis (TLS) involves PCNA mono-ubiquitination and TLS DNA polymerases (pols). Recent evidence has shown that the mono-ubiquitination is induced not only by DNA damage but also by other factors that induce stalling of the DNA replication fork. We studied the effect of spontaneous DNA replication errors on PCNA mono-ubiquitination and TLS induction. In the pol1L868F strain, which expressed an error-prone pol α, PCNA was spontaneously mono-ubiquitinated. Pol α L868F had a rate-limiting step at the extension from mismatched primer termini. Electron microscopic observation showed the accumulation of a single-stranded region at the DNA replication fork in yeast cells. For pol α errors, pol ζ participated in a generation of +1 frameshifts. Furthermore, in the pol1L868F strain, UV-induced mutations were lower than in the wild-type and a pol δ mutant strain (pol3-5DV), and deletion of the RAD30 gene (pol η) suppressed this defect. These data suggest that nucleotide misincorporation by pol α induces exposure of single-stranded DNA, PCNA mono-ubiquitination, and activates TLS pols.
Keywords: DNA polymerase, translesion DNA synthesis, mutagensis, PCNA, ubiquitination
In organisms with a DNA genome, the genome is replicated by DNA polymerases (pol) and the duplicated genome is distributed to each daughter cell during cell division. When replicative pols are blocked by unrepaired DNA lesions, synthesis past DNA damage requires replacement of the stalled replicative pol by translesion synthesis (TLS) pols (1–3). Pol η has a unique property of being able to synthesize past DNA containing cis-syn cyclobutane thymine dimers (CPD) with efficiency similar to undamaged DNA by inserting the correct adenines most of the time. Pol ζ also belongs to the TLS pols, but unlike pol η it performs error-prone DNA synthesis on damaged DNA templates (4–6).
Recent studies have revealed that TLS pols are functionally and/or physically regulated by PCNA in promoting DNA synthesis. Hoege et al. showed that, following exposure of cells to a DNA damaging agent, PCNA becomes mono-ubiquitinated on Lys164 by Rad6 and Rad18 (7), and PCNA mono-ubiquitination mediates the switch to translesion synthesis (8). The mono-ubiquitination of PCNA increases its affinity for TLS pols through one or two copies of ubiquitin binding motif (UBM/Z, see (3) and references therein). Despite the fact that pol ζ belongs to the Rad6 epistasis group and requires PCNA as well as Rad6-Rad18-mediated ubiquitin conjugation at PCNA Lys164 (1,6,8,9), physical interaction between PCNA and pol ζ has not been demonstrated. Interestingly, Rev1 is stimulated by mono-ubiquitinated PCNA (10). It is able to interact with the other three Y-family polymerases as well as the Rev7 subunit of pol ζ (11–13), leaving the possibility that pol ζ activity is mediated by the ternary complex formation with Rev1.
Based on these results, it is considered that PCNA and its modified forms are the key regulator of TLS. PCNA modification was originally introduced by DNA damaging agents (7), although Northam et al. showed that it was also promoted by defects in the components of DNA replication machinery (14).
During DNA replication, pol α initiates DNA replication with the associated primase (15–17). Because pol α lacks proofreading activity, it performs DNA replication with a fidelity lower than the other replicative pols of δ and ε by 10-fold or more (2). Thus, pol α is estimated to generate 30,000 mutations each time a mammalian cell divides (18). More importantly, without the proofreading activity, it would leave more mismatched primers without farther extension (19). Some of the mismatches are removed by the mismatch repair system and the proofreading activity of pol δ (20,21). However, how cells deal with these replication intermediates remains to be understood.
In the present study, we used a mutator pol α strain of pol1L868F and L868F pol α enzyme to show that PCNA mono-ubiquitination occurred by pol α misincorporation. We presented evidence that this polymerase has a kinetic difficulty in extending the mismatched primer termini, and also showed populations of DNA replication forks contain long single-stranded DNA structures in yeast cells. Furthermore, TLS pols were activated in spontaneous and UV-induced mutagenesis. The results suggest that pol α may activate TLS through misincorporations during the DNA replication.
MATERIALS AND METHODS
Strains and plasmids
All mutants were isogenic with CG379 (MATα ade5 his7-2 leu2-3 leu2-112 trp1-289 ura3-52), and summarized in Supplementary Table 1. pol30K164R mutation was introduced using pCH1572-K164R (22), which was kindly provided by Dr. Shcherbakova (University of Nebraska Medical Center).
Detection of PCNA mono-ubiquitination
Yeast strains were grown to logarithmic phase in 3 ml of liquid YPAD medium. For analysis of MMS-treated cells, MMS was present in the medium at a concentration of 0.03% during the last hour of the culture growth. In some experiments, yeast cells were irradiated by UV with the indicated doses and cultured for 30 min at 27°C. The cells were collected by centrifugation, washed with 1 ml of ice-cold 20% trichloro acetic acid, and resuspended in 100 μl of the same solution. After cells were lysed by vigorous mixing with the same volume of acid-washed glass beads (300 microns; SIGMA), pellet protein was resuspended in 1 M Tris-HCl (pH 8.5), 0.1% SDS, and 100 mM β-mercaptoethanol. The extracts were loaded onto 12% polyacrylamide gel, subjected to electrophoresis and transferred to Immobilon P (Millipore). The blots were probed with a rabbit polyclonal antibody against yeast PCNA, which we raised using a PCNA fraction (23), and a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase. The bands were visualized using ECL kit (GE Healthcare).
Primer extension analysis
Primer extension analysis was performed using a template-primer as described previously (19,24). The reactions were performed at 37°C for 15 min with various amounts of wild-type or L868F pol α in 20 μl containing 2 mM MgCl2, 50 mM Tris-HCl, pH 8.0, 50 mM KCl, 2 mM DTT, 4 nM 32P-labeled primer-template DNA, and 100 μM dATP, dGTP, and dTTP, but not dCTP. Reactions were terminated by addition of an equal volume of a loading buffer containing 90% formamide, 20 mM EDTA, 0.05% xylene cyanol, and 0.05% bromophenol blue. Reaction products were analyzed by 14% polyacrylamide gel electrophoresis.
Electron microscopy
Logarithmic growing yeast cells were crosslinked by psoralen followed by extraction of genomic DNA according to the CTAB method (25). After the DNA was digested by BamHI, replication intermediate was enriched by BND-cellulose column (26). Glycerol was added to the eluate with a final concentration of 50%, and the mixtures were sprayed onto freshly cleaved mica surfaces using a painter’s airbrush. The protein-glycerol drops on mica were dried in a vacuum (1 × 106 Pa) for 8 min at −20°C in a freeze-etching device (FR-9000, Hitachi, Tokyo). Subsequently, specimens were rotary shadowed with platinum at an angle of 2.5° to the mica surface for 1 min, followed by carbon evaporation at an angle of 90° for 20 sec. The shadowed platinum films were removed from the mica by soaking in water and mounted on formvar membrane-coated copper grids. Images were taken with a Hitachi H-7600 electron microscope (access 100 KV) onto films at a magnification of ×20,000 and captured using an ES2000 scanner (EPSON). Images were further processed with Adobe PHOTOSHOP software (Adobe Systems, Mountain View, CA) (27). Single- and double-stranded DNAs, and size marker were determined by the control experiments using a restriction enzyme-digested plasmid DNA.
CAN1 forward and his7-2 reversion mutation assays
The fluctuation assays were performed as described previously with minor modifications (21,28). Essentially, strains were grown on YEPD plates at 27°C for 3–5 days. One or a few colonies were selected and spread on synthetic plates lacking histidine (his7-2 reversion assay) or containing 60 mg/liter of L-canavanine (CAN1 forward mutation assay). The rate was calculated on the median from 24 or more data with 95% confidence limits. For +1 frameshifts in CAN1 forward mutation assay, statistical significances were analyzed by Fisher’s exact t test with the two-sided significance level set.
Measurement of induced mutation frequency
For UV-induced mutagenesis, yeast strains were grown to logarithmic phase in liquid YEPD medium. They were appropriately diluted and plated onto YEPD plate for viability count, onto complete synthetic medium containing L-canavanine (60 mg/l) and lacking arginine for Canr mutant count. The cells were irradiated with UV light immediately after plating and incubated to allow visualization of colonies. The mutation frequency was calculated by dividing the Canr mutant count by the viable cell count. For MMS-induced mutagenesis, yeast strains were treated with the various concentrations of MMS during the last 10 min of the culture growth.
RESULTS
PCNA mono-ubiquitination status in wild-type and mutator pol strains
Upon DNA damages, a class of DNA polymerases is activated and employed for translesion DNA synthesis (TLS and TLS pols). Recent study has demonstrated that TLS is also induced in the absence of DNA damages, although the nature of the spontaneous pathways is yet to be studied. Previously, we isolated pol α mutants that frequently incorporate wrong nucleotides (21). Most of the reported pol mutants are associated with activity reduction. In contrast, mutant strains with amino acid substitutions at Leu868 or at the corresponding sites in other pols are active and leave pol errors (20,21,29–32).
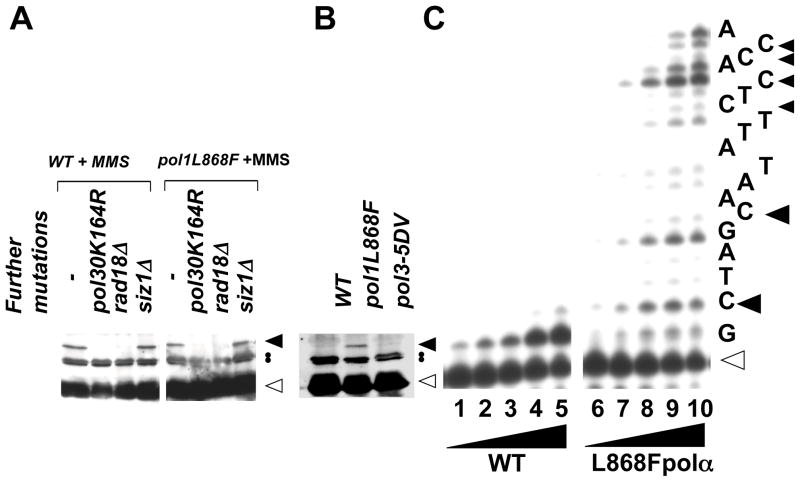
During the study of spontaneous mutagenesis in the pol1L868F allele, we found PCNA was constitutively mono-ubiquitinated (Fig. 1B for constitutive mono-ubiquitination. Fig 1A and C for control experiments). In Fig. 1A, we showed the reactivity of our antibody that detected PCNA and a modified form at ~37 kDa after cells were treated with MMS (closed arrowhead). Since the ~37 kDa PCNA was not seen in pol30K164R and rad18Δ strains, and it was not affected in siz1Δ strain (Fig. 1A), it corresponds to a mono-ubiquitinated form of PCNA. In the pol1L868F strain, PCNA mono-ubiquitination was also observed in the absence of any DNA damaging agent, but was not detected in the wild-type and a pol δ mutant strain carrying the pol3-5DV mutation inactivating the proofreading activity (33) (Fig. 1B, closed arrowhead).
Fig. 1.
Detection of PCNA mono-ubiquitination in strains with error-prone replicative pols (A–D). Whole cell extract of each strain was analyzed by Western blotting using an anti-PCNA polyclonal antibody. Positions of unmodified and mono-ubiquitinated PCNA were indicated by open and closed arrowheads, respectively. Closed circles indicate positions of non-specific bands. (A). After cells were treated with 0.03% MMS for 1 h, PCNA status was analyzed in wild-type, pol30K164R, rad18Δ, or siz1Δ, in the presence or absence of pol1L868F allele. (B). Spontaneous modification was analyzed in the presence of pol1L868F or pol3-5DV allele. (C). Polymerase stall was detected by a primer extension analysis. Wild-type or L868F pol α was incubated with a synthetic template-primer DNA in the presence of dA, dG, and dT, but not dCTP at 37°C for 15 min. Reaction mixtures included WT pol α at 17.5, 35, 70, 140, 280 nM in lanes 1 – 5, respectively, or L868F pol α at 1.8, 3.5, 7, 14, 28 nM in lanes 6 – 10, respectively. Nucleotides to be incorporated are shown at right. Open and closed arrowheads indicate positions for original primer size and for mismatched primers, respectively.
Primer extension analysis
To know what caused PCNA ubiquitination in pol1L868F cells, a primer extension assay was carried out to compare between wild-type and L868F pol α (Fig 1C). In order to generate misincorporations and extension during the DNA synthesis, we omitted one nucleotide of dCTP from the reaction. In this assay, elongation is limited by the rate of misincorporation at template dG, as well as the extension from the mispaired 3′-primer termini (24). Wild-type pol α (Fig. 1C, lanes 1–5) extended ~50% of the primer up to and opposite the first template dC at the highest polymerase concentration by incorporation of a single complementary nucleotide. Consistent with the previous result (21), L868F pol α showed infidelity DNA synthesis by extending the primer further downstream than the wild-type pol α (Fig. 1C, lanes 6–10). At the same time, L868F pol α left arrested products at the sites one-nucleotide shorter from the template dG (one base shorter than C positions, closed arrowheads) in Fig. 1C. These products corresponded to the ceased primers due to the kinetic difficulty in misincorporation. The ceased primers were also observed at the C positions which were with the mismatched primer termini (Fig. 1C, closed arrowheads), with intensities of the two major bands varying between the sites. These data suggest that L868F pol α, like wild type-pol α (19), has kinetic difficulties in incorporating the wrong nucleotides and in extension from the mismatched primers, although L868F pol α was more error-prone at both steps.
Electron microscopy analysis
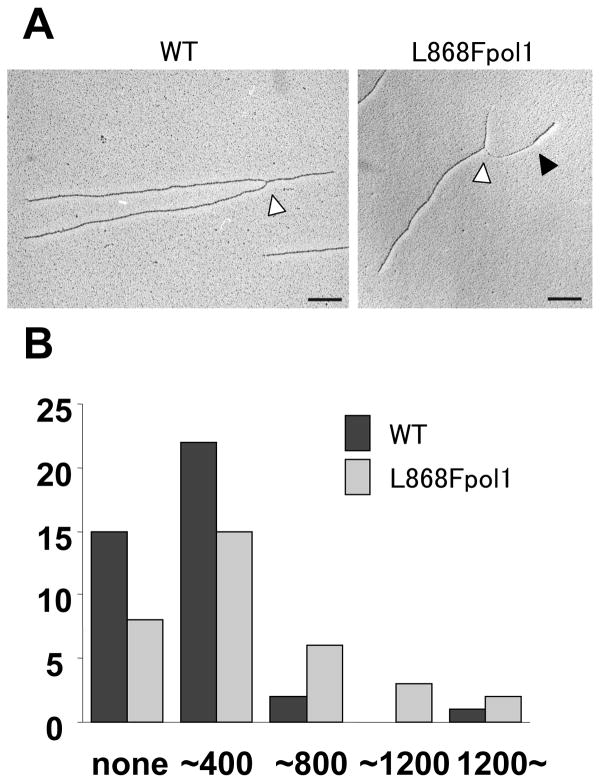
After the misincorporation takes place, L868F pol α, if not all, stalled at mismatched primer even in the presence of complementary dNTPs (Fig 1C). In order to know if such stalled primers leave aberrant DNA structures in living cells, we crosslinked and purified DNA from yeast cells, and observed replication fork structures by electron microscopy (Fig. 2A). In wild-type cells, most of the replication forks were associated with undetectable or a short stretch of thin DNA fiber at one of the nascent strands (Fig 2A, left), which was considered to be the lagging strand DNA with single-stranded (ss) DNA region (26,34). When length of the ssDNA regions of both wild-type and pol1L868F forks was measured and plotted on a graph (Fig. 2B), we found that replication forks in the pol1L868F strain were associated with the long stretches (sum of ~800, ~1200 and 1200~ bp, Fig 2A, right) of ssDNA regions more than wild-type. These data strongly suggest that pol α leaves the gapped DNA upon misincorporation in vivo, which caused PCNA mono-ubiquitination.
Fig. 2.
(A). Electron micrographs of representative forks isolated from yeast cells. The open arrowheads point to replication forks, the transition points from ds- to ssDNA are indicated by a black arrowhead. Size of standard insertion is 400 bp long. (B). Graphic representation of ssDNA length at replication forks isolated in the indicated conditions. Wild-type and pol1L868F were shown in black and gray columns, respectively. Total number of observed forks were 40 (wild-type) and 34 (L868F). Replication forks in the pol1L868F strain were associated with the long stretches (~800, ~1200 and 1200~ bp) of ssDNA regions more than wild-type (p = 0.008, Fischer’s exact t test).
Genetic interaction between pol α and pol ζ in spontaneous mutagenesis
PCNA mono-ubiquitination has been known to be the signal for recruitment of the TLS pols (1–3). The result prompted us to measure the spontaneous mutation rate using the pol1L868F, and double mutants with a point mutation in the POL30 gene that encodes PCNA (pol30K164R), or deletion of the REV3 gene encoding the catalytic subunit of the error-prone TLS pol of pol ζ (rev3Δ) (Table 1).
Table 1.
Spontaneous mutation rates of pol strains.
| Strain |
CAN1 forward mutation assay
|
his7 reversion assay
|
||
|---|---|---|---|---|
| MR (x 10−7) 1) | Fold change | MR (x 10−9) 1) | Fold change | |
| Wild-type | 2.4 (1.9–3.0) | 1 | 7.3 (5.3–9.1) | 1 |
| rev3 Δ | 1.5 (1.3–2.2) | 0.63 | 7.1 (4.3–1.1) | 0.97 |
| pol1L868F | 13 (10–15) | 5.3 | 89 (60–110) | 12 |
| pol1L868F rev3 Δ | 12 (8.9–14) | 5.1 | 42 (34–49) | 5.7 |
| pol1L868F pol30K164R | 15 (13–19) | 6.3 | ||
Spontaneous mutation rates (MR) of S. cerevisiae strains. Fluctuation tests were performed as described in MATERIALS AND METHODS. The rate was calculated on the median from 24 or more data with 95% confidence limits.
In accord with the previous observations (5), ~40% of the spontaneous mutation was dependent on pol ζ in wild-type (Table 1, Wild-type and rev3Δ). In contrast, against our expectation, the overall mutation rate was very similar with or without pol ζ in the pol1L868F strain (Table 1, pol1L868F and pol1L868F rev3Δ). It was also observed that the pol30K164R mutation did not change the mutation rate in the pol1L868F strain (Table 1, pol1L868F and pol1L868F pol30K164R). These results showed that L868F pol α did not depend on pol ζ for most of the DNA replication errors in vivo.
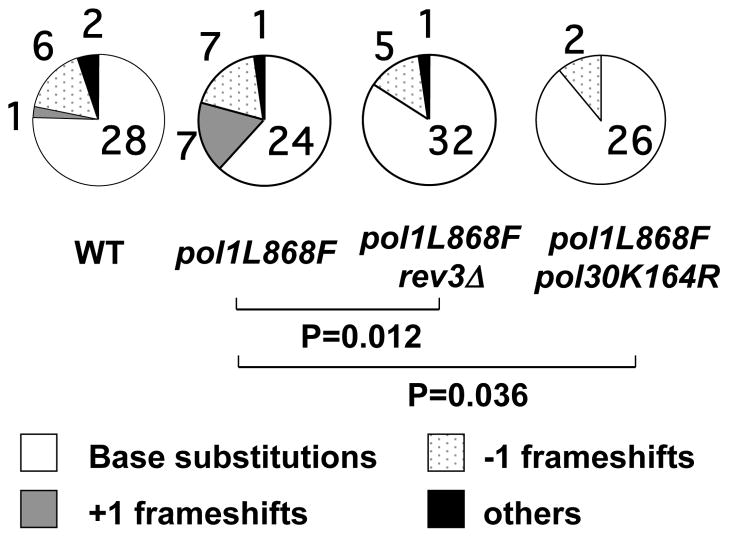
To know if pol ζ contributes to any particular types of mutations, we sequenced the can1 locus and compared the spectra between strains. We found the mutation spectra were similar in base substitutions and −1 frameshifts (Supplementary Table 2), but were different in +1 frameshifts (Fig. 3). In pol ζ-proficient pol1L868F cells, 7 out of 39 mutations (18%) were +1 frameshifts (Fig. 3, grey pie of pol1L868F). In contrast, in pol ζ-deficient pol1L868F and pol30K164R pol1L868F cells, there was no +1 frameshift (Fig. 3, pol1L868F rev3Δ and pol1L868F pol30K164R). The moderate but similar result was obtained by his7-2 reversion assay (Table 1, pol1L868F and pol1L868F rev3Δ). Therefore, in pol1L868F cells, the +1 frameshifts were fixed as mutations in combination with pol ζ.
Fig. 3.
Spontaneous mutation rates of S. cerevisiae strains. Sequencing result of can1 locus of canavanine-resistant colonies was shown. White, gray, dotted and black sectors indicate base substitutions, +1 frameshifts, −1 frameshifts and other mutations, respectively. p-values and the number of mutations of each type are indicated on each graph. Mutation rate of each strain is presented in Table 1. Sequences of the +1 frameshifts are presented in Supplementary Table 3.
UV-induced mutation frequencies in pol1L868F
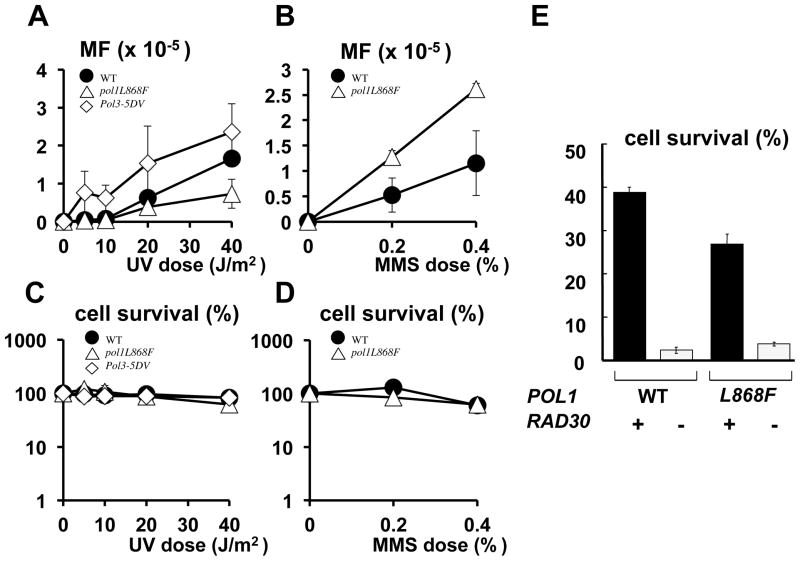
We also measured damage-induced mutation frequency. UV irradiation of the wild-type and the pol3-5DV strains resulted in a dose-dependent elevation of mutation frequency in a CAN1 forward mutation assay (Fig. 4A and Supplementary Fig. 1). On the other hand, the pol1L868F strain was moderately resistant to UV mutagenesis. When the UV dose was increased, mutations in the pol1L868F strain did not increase as sharply as in the wild-type and pol3-5DV strains, and at doses of 20 and 40 J/m2 the mutation frequencies were lower than those of the wild-type (Fig. 4A and Supplementary Fig. 1).
Fig. 4.
Effect of error-prone replicative pols on damage-induced mutation. UV-induced mutant frequency (MF) on canavanine plates (A) and % survival on YEPD plates (C) were measured in wild-type (filled circle), pol3-5DV (open rectangle) and pol1L868F (open triangle) strains. In (C) and (D), MMS-induced mutant frequency and % survival was shown, respectively. Data are averages of three independent experiments. Standard bar is shown when it exceeds the symbol size. In (A) and (B), spontaneous mutation frequencies are subtracted, i.e., 0.09, 0.62, 2.6 × 10−5 for wild-type, pol1L868F and pol3-5DV, respectively. The original data are provided in Supplementary Fig. 1A and B. (E). UV survival was measured at a dose of 80 J/m2. Genotypes of POL1 and RAD30 are indicated under the graph.
The observed difference in the UV mutagenesis effect was not explained by the altered UV-sensitivity of these strains, because their cell viability was similar (Fig. 4C). Furthermore, the suppressed mutagenesis was not observed in MMS-induced mutations (Fig. 4B and D).
L868F pol α has been characterized as having an increased ability to incorporate the correct dA over the 3′-T of CPD (21). This might suggest that, instead of pol η, L868F pol α was responsible for the TLS in the pol1L868F strain. However, this is unlikely because the TLS activity of L868F pol α over CPD, as determined by analysis of kinetics, is limited to as low as 1/60,000 of that of pol η (21). Furthermore, the pol1L868F mutation did not rescue the UV sensitivity phenotype of a mutant without pol η (Fig. 4E).
In strains with pol1L868F, pol η rather than pol ζ is favored to perform TLS
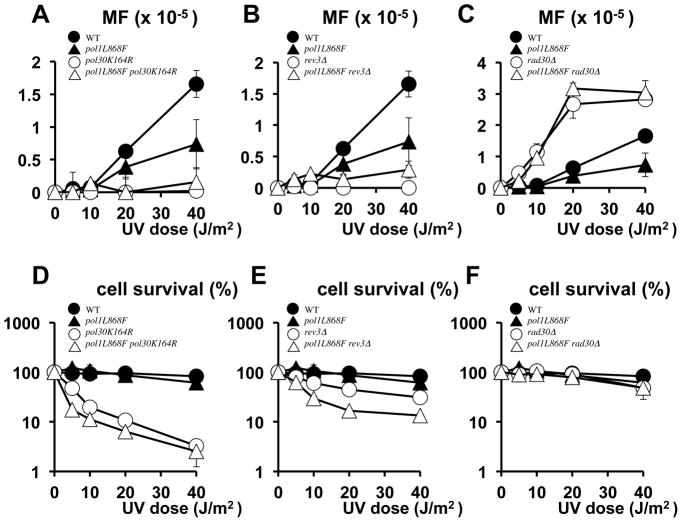
In order to know how significantly the TLS pols contribute to the above phenotype, we measured the mutation frequency under the depletion of TLS functions. Mutation frequency remained at a very low background level in the pol30K164R and pol1L868F pol30K164R double mutant (Fig. 5A), indicating that the wild-type and the pol1L868F strains rely on PCNA Lys164 for UV mutagenesis. It was also found that both the pol30K164R and pol1L868F pol30K164R strains were very UV-sensitive (Fig. 5D). These results were consistent with the previous observations that UV-induced mutations and cell viability were dependent on PCNA Lys164 (8).
Fig. 5.
Functions of PCNA Lys164 and translesion pols on UV mutation. UV-induced mutant frequency (MF) on canavanine plates (A, B, C) and % survival on YEPD plates (D, E, F) were measured. Data are averages of three independent experiments. Standard bar is shown when it exceeds the symbol size. (A) and (D). Wild-type (filled circle), pol30K164R (open circle), pol1L868F (filled triangle) and pol1L868F pol30K164R (open triangle). (B) and (E). Wild-type (filled circle), rev3Δ (open circle), pol1L868F (filled triangle) and pol1L868F rev3Δ (open triangle). (C) and (F). WT (filled circle), rad30Δ (open circle), pol1L868F (filled triangle) and pol1L868F rad30Δ (open triangle). In (A), (B) and (C), spontaneous mutation frequencies are subtracted. The original data are provided in Supplementary Fig. 1C, D and E, respectively.
We also performed UV-induced mutagenesis and UV-survival experiments using rev3Δ (Figure 5B and E) or rad30Δ strains (Figure 5C and F). The pol ζ-proficient POL1 (wild-type) strain showed mutation frequencies higher than those of the pol ζ-proficient pol1L868F strain (Figure 5B, closed circles and closed triangles). Like the pol30K164R mutation, the absence of pol ζ led to suppression of UV-induced mutagenesis in both wild-type and pol1L868F strains close to the background level (Fig. 5B, open circles and open triangles), suggesting that with the POL1 allele pol ζ participated in the mutations more than with the pol1L868F allele. In the absence of pol η (rad30Δ), mutation frequencies of the wild-type POL1 and the pol1L868F mutant increased to almost the same level (Fig. 5C, open circles and open triangles), suggesting that with the pol1L868F allele pol η suppressed the mutations more efficiently than with the POL1 allele.
DISCUSSION
PCNA mono-ubiquitination, mismatched primer termini and existence of ssDNA at DNA replication forks
In this manuscript, we reported that pol α errors induced PCNA mono-ubiquitination and activation of TLS pols. Because yeast cells were cultured in the absence of any stresses or DNA damaging agents, this mono-ubiquitination may not have been due to the DNA lesions. Instead, L868F pol α created numbers of mismatched primer termini that may be difficult to be extended. This might result in a situation similar to that reported by Northam et al., where PCNA was mono-ubiquitinated by DNA replication defects (14), although we observed the phenotypes distinct from those associated with the previously reported alleles (see discussion below).
Recently, Lopes et al. demonstrated that UV-irradiated S. cerevisiae cells uncoupled leading and lagging strand replication at UV lesions, and generated long ssDNA regions on one side of the fork (26). Despite the fact that our conditions were dependent on the spontaneous misincorporations, the fork structures and the gap sizes in the pol1L868F were strikingly similar to the UV-irradiated DNA (Fig. 2). Therefore, like the case in the UV-irradiation, we assume the mono-ubiquitination was invoked by this aberrant fork structure with a long stretch of ssDNA. In accord with this hypothesis, essential functions of the ssDNA and ssDNA-binding replication protein A (RPA) for PCNA ubiquitination have been proposed (35–37). These in vitro and in vivo data suggested that the mismatched primer termini induced PCNA mono-ubiquitination in S. cerevisiae via the aberrant DNA replication fork structure, although the precise molecular mechanisms between the ssDNA exposure and PCNA ubiquitination are yet to be studied.
In the UV-irradiation experiments, Lopes et al. also showed the existence of ss-gaps distant from the fork (26). Under our spontaneous conditions, the distant gaps were not frequently found (data not shown) and were not quantified. Because pol α is involved in the primer DNA synthesis, the gap formation may have been limited in the lagging strand and was not observed at the distant regions of both leading and lagging strands.
Genetic interaction between pol α and translesion pols for spontaneous mutations
We measured and compared spontaneous mutation rates between strains with or without pol ζ activity (Table 1 and Fig. 3). Our results showed that +1 frameshifts in the pol1L868F strain was pol ζ-dependent (Table 1 and Fig. 3), and suggest a possibility that pol ζ is intrinsically error-prone for +1 frameshifts, as observed in the rad1 strain (38). To our knowledge, our data provided the first evidence that pol α and ζ couple and generate mutations in vivo. In this context, it should be noted that +1 frameshifts are found in putative target genes in MMR-deficient cancer cells with microsatellite instability (39–43). Although +1 frameshifts are not the major mutations in this disease, it is feasible that this polymerase combination contributes to the mutations and carcinogenesis to some extent. Unexpectedly, we observed dispensable functions of pol ζ for base substitutions (Table 1, Fig. 2 and supplementary Table 2).
On the other hand, inspection of the mutation spectra showed that pol η was not involved in the generation of most spontaneous mutations, because in dT:dGMP incorporation (A -> C substitutions in Supplementary Table 2), a characteristic feature of pol η errors (44), mutation rates were similar between wild-type and pol1L868F strains. Similarly, we did not observe the increased rate of dCMP misincorporations (Supplementary Table 2), which are characteristic mutations by the dCMP transferase of REV1 (45). These results suggest that in the pol1L868F strain the mutant polymerase itself extended a fraction of the mismatched/misaligned primer termini, and was responsible for the generation of base substitutions and −1 frameshifts, as observed in an in vitro forward mutation assay (21). However, our data do not exclude the possibility that in the wild-type strain, PCNA mono-ubiquitination and TLS pols are involved in base substitutions and −1 frameshifts.
UV-induced mutagenesis
We observed moderate UV-induced mutations in the pol1L868F strain, while MMS treatment results in an increase of mutations in the same strain (Fig. 4). The in vitro translesion activities of L868F pol α over the abasic site (21) and other alkylated DNA lesions (data not shown) were far better than those over the UV-photoproducts. Therefore, the mutant pol α may have performed error-prone TLS over the alkylated DNA lesions, and increased the mutation frequency upon the increased MMS dose.
For UV-induced mutagenesis, further analysis showed that in pol1L868F cells pol η was responsible for the suppressed UV mutations more effectively than in wild-type cells (Fig. 5C). This phenotype was consistent with the Western blot result that PCNA was constitutively mono-ubiquitinated in the pol1L868F but not in wild-type (Fig. 1B), and with the results that the mutant pol α cannot efficiently bypass the UV-photoproducts ((21) and Fig. 4E). These results suggest that the mono-ubiquitination of PCNA, and the subsequent pol η activation may be a cause of the suppressed UV-induced mutations in the pol1L868F strain and that this process was promoted by the spontaneous PCNA mono-ubiquitination.
It is intriguing that we did not observe PCNA-mono ubiquitination or suppression of UV mutagenesis in the pol3-5DV strain, despite the fact that the pol3-5DV encodes error-prone pol δ and that it is associated with a spontaneous mutation frequency higher than the pol1L868F (Supplementary Fig. 1). Besides DNA replication, pol δ is also involved in nucleotide excision repair. pol3-5DV pol δ might leave mutations during the repair, and this would be a cause of the high UV-induced mutations.
It might be argued that pol α synthesizes DNA primers and may not have hit most of the UV damages. At present, several possible processes may explain why pol1L868F was resistant to UV-mutagenesis. For example, it has been known that RF-C-dependent PCNA loading onto the primer terminus takes place irrespective of the presence of matched or mismatched base pairs at the terminus (46). Therefore, after pol α dissociated prematurely from the mismatched primer termini, both PCNA loading and ubiquitination could occur. This PCNA, as a free form, might be able to move on the lagging strand (47) and facilitate pol η recruitment to a nearby CPD site. It has been also known that ubiquitination does not change pol δ-PCNA complex stability and DNA replication ability (10,48). Pol δ might be loaded, correct the mismatches, and continue DNA replication until it encountered the UV damages. During this process, the PCNA may interact with pol η via the putative PCNA interaction domains ((3) and references therein), thus providing the error-free TLS with an advantage. Third, since L868F pol α showed nucleotide incorporation ability higher than wild-type, we needed less L868F pol α than wild-type to achieve the same extent of primer extension in Fig. 1C. This intrinsic ability would have rendered high processivity to this mutant polymerase, and thus increased the possibility to hit the UV damages. However, further experiments are needed to clarify what processes took place upon pol α misincorporations.
In conclusion our data showed that pol α misincorporation resulted in exposure of ssDNA regions during DNA replication, and thereby induced PCNA mono-ubiquitination. This mono-ubiquitination facilitated two types of particular TLS activation, i.e., pol ζ to generate +1 frameshifts, and pol η to suppress UV-induced mutagenesis. The genome is replicated by various replicative pols and TLS pols. Understanding of these spontaneous processes may shed light on the genomic instability inherent in DNA replication and diseases.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan; and by the Japan Space Forum.
This work was supported by the National Institutes of Health (5R01CA129925-03 to Y.P., see J. Bioch, corrigendum J. Biochem. 2009; 146(1), 13–21; doi:10.1093/jb/mvp043).
We thank Drs Yasuo Kawasaki, Hiro-o Ishihara and Polina V Shcherbakova for critical discussion, and Haruo Ohmori for carefully reading the manuscript. We are also grateful to Tazuko Tomita, Naoaki Kano, Danuta Mizgalska, Rumiko Yagi, Yoshihiro Nakata and Azumi Yoshimura for sequencing can1 mutants and for EM analysis.
Abbreviations
- pol
DNA polymerase
- TLS
translesion DNA synthesis
References
- 1.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 2.Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 6.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 7.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 8.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence CW, Maher VM. Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein. Biochem Soc Trans. 2001;29:187–191. doi: 10.1042/0300-5127:0290187. [DOI] [PubMed] [Google Scholar]
- 10.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 13.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 14.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki M, Tamiya-Koizumi K, Takemura M, Furuta K, Izuta S, Savoysky E, Miura A, Yoshida S. Replication protein-A mediates the association of calf thymus DNA polymerase alpha-DNA primase complex with guanine-rich DNA sequence. J Biochem. 1996;120:766–772. doi: 10.1093/oxfordjournals.jbchem.a021477. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki M, Izuta S, Savoysky E, Sakurai T, Simbulan C, Tatebe M, Kojima K, Yoshida S. Deoxypyrimidine cluster mediates the priming by calf thymus DNA primase subunit. Biochem Mol Biol Int. 1993;29:645–652. [PubMed] [Google Scholar]
- 17.Suzuki M, Savoysky E, Izuta S, Tatebe M, Okajima T, Yoshida S. RNA priming coupled with DNA synthesis on natural template by calf thymus DNA polymerase alpha-primase. Biochemistry. 1993;32:12782–12792. doi: 10.1021/bi00210a030. [DOI] [PubMed] [Google Scholar]
- 18.Albertson TM, Preston BD. DNA replication fidelity: proofreading in trans. Curr Biol. 2006;16:R209–211. doi: 10.1016/j.cub.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 19.Limsirichaikul S, Ogawa M, Niimi A, Iwai S, Murate T, Yoshida S, Suzuki M. The Gly952 residue of saccharomyces cerevisiae DNA polymerase alpha is important in discriminating correct deoxyribonucleotides from incorrect ones. J Biol Chem. 2003;278:19079–19086. doi: 10.1074/jbc.M208604200. [DOI] [PubMed] [Google Scholar]
- 20.Pavlov YI, Frahm C, McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol Cell Biol. 2006;26:5509–5517. doi: 10.1128/MCB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki M, Avicola AK, Hood L, Loeb LA. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J Biol Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 25.Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- 26.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Shiomi Y, Usukura J, Masamura Y, Takeyasu K, Nakayama Y, Obuse C, Yoshikawa H, Tsurimoto T. ATP-dependent structural change of the eukaryotic clamp-loader protein, replication factor C. Proc Natl Acad Sci USA. 2000;97:14127–14132. doi: 10.1073/pnas.97.26.14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa M, Limsirichaikul S, Niimi A, Iwai S, Yoshida S, Suzuki M. Distinct function of conserved amino acids in the fingers of Saccharomyces cerevisiae DNA polymerase alpha. J Biol Chem. 2003;278:19071–19078. doi: 10.1074/jbc.M208605200. [DOI] [PubMed] [Google Scholar]
- 29.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair. 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J Biol Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 33.Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′->5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burhans WC, Vassilev LT, Wu J, Sogo JM, Nallaseth FS, DePamphilis ML. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, Hanaoka F, Yamamura K, Yamaizumi M, Tateishi S. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 37.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 38.Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 39.Sun X, Chen C, Vessella RL, Dong JT. Microsatellite instability and mismatch repair target gene mutations in cell lines and xenografts of prostate cancer. Prostate. 2006;66:660–666. doi: 10.1002/pros.20390. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi S, Takeuchi N, Fermin AC, Taguchi H, Koeffler HP. Frameshift mutations in caspase-5 and other target genes in leukemia and lymphoma cell lines having microsatellite instability. Leuk Res. 2003;27:359–361. doi: 10.1016/s0145-2126(02)00215-1. [DOI] [PubMed] [Google Scholar]
- 41.Souza RF, Appel R, Yin J, Wang S, Smolinski KN, Abraham JM, Zou TT, Shi YQ, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch PM, Frazier M, Powell SM, Harpaz N, Sugimura H, Young J, Meltzer SJ. Microsatellite instability in the insulin-like growth factor II receptor gene in gastrointestinal tumours. Nat Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 42.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 43.Chang HR, Cheng TL, Liu TZ, Hu HS, Hsu LS, Tseng WC, Chen CH, Tsao DA. Genetic and cellular characterizations of human TCF4 with microsatellite instability in colon cancer and leukemia cell lines. Cancer Lett. 2006;233:165–171. doi: 10.1016/j.canlet.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 45.Masuda Y, Takahashi M, Fukuda S, Sumii M, Kamiya K. Mechanisms of dCMP transferase reactions catalyzed by mouse Rev1 protein. J Biol Chem. 2002;277:3040–3046. doi: 10.1074/jbc.M110149200. [DOI] [PubMed] [Google Scholar]
- 46.Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 47.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 48.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.