ABSTRACT
Recent studies have shown that individuals with colorectal cancer have an altered gut microbiome compared to healthy controls. It remains unclear whether these differences are a response to tumorigenesis or actively drive tumorigenesis. To determine the role of the gut microbiome in the development of colorectal cancer, we characterized the gut microbiome in a murine model of inflammation-associated colorectal cancer that mirrors what is seen in humans. We followed the development of an abnormal microbial community structure associated with inflammation and tumorigenesis in the colon. Tumor-bearing mice showed enrichment in operational taxonomic units (OTUs) affiliated with members of the Bacteroides, Odoribacter, and Akkermansia genera and decreases in OTUs affiliated with members of the Prevotellaceae and Porphyromonadaceae families. Conventionalization of germfree mice with microbiota from tumor-bearing mice significantly increased tumorigenesis in the colon compared to that for animals colonized with a healthy gut microbiome from untreated mice. Furthermore, at the end of the model, germfree mice colonized with microbiota from tumor-bearing mice harbored a higher relative abundance of populations associated with tumor formation in conventional animals. Manipulation of the gut microbiome with antibiotics resulted in a dramatic decrease in both the number and size of tumors. Our results demonstrate that changes in the gut microbiome associated with inflammation and tumorigenesis directly contribute to tumorigenesis and suggest that interventions affecting the composition of the microbiome may be a strategy to prevent the development of colon cancer.
IMPORTANCE
The trillions of bacteria that live in the gut, known collectively as the gut microbiome, are important for normal functioning of the intestine. There is now growing evidence that disruptive changes in the gut microbiome are strongly associated with the development colorectal cancer. However, how the gut microbiome changes with time during tumorigenesis and whether these changes directly contribute to disease have not been determined. We demonstrate using a mouse model of inflammation-driven colon cancer that there are dramatic, continual alterations in the microbiome during the development of tumors, which are directly responsible for tumor development. Our results suggest that interventions that target these changes in the microbiome may be an effective strategy for preventing the development of colorectal cancer.
INTRODUCTION
Colorectal cancer (CRC) is one of the most commonly diagnosed malignancies worldwide, resulting in over a half-million deaths annually (1). Significant risk factors for CRC include diets rich in red and processed meat, alcohol consumption, and chronic inflammation of the gastrointestinal tract (2–5). Each of these factors is closely associated with changes in composition and function of the complex community of microorganisms that inhabits our gastrointestinal tract. This community, known as the gut microbiome, promotes various physiological functions that are associated with cancer, including cell proliferation, angiogenesis, and apoptosis (6–9). Therefore, we hypothesized that the composition, structure, and functional capacity of the gut microbiome all directly affect tumor development in the colon.
Several recent studies have addressed this hypothesis by characterizing the composition of the gut microbiome associated with patients with CRC (10–16). Using culture-independent approaches, each of these studies observed a significant shift in the composition of the gut microbiome in patients with CRC compared to that in healthy controls. This phenomenon, referred to as dysbiosis, can be observed in both the luminal microbiome from feces and the mucosa-associated microbiome from tumor biopsy specimens. Interestingly, each of these studies obtained conflicting results regarding the composition and structure of the CRC-associated microbial community. Furthermore, there are no bacterial populations that have consistently been identified across each study that can be attributed to the development or presence of CRC. These data clearly show an association between abnormalities in the gut microbiome and CRC; however, the conflicting results point out the need for a mechanistic understanding of the role of the gut microbiome in this process.
The combination of factors that could lead to dysbiosis is complex and not well understood. In addition, the effect of the development of this abnormal community on colon tumorigenesis remains unclear. Recent evidence suggests that certain strains of Bacteroides fragilis and Escherichia coli can directly affect tumor development in the colon through the production of virulence factors (e.g., toxins and gene products) (17, 18). Furthermore, bacterial populations that produce the short-chain fatty acid butyrate have antitumor effects in the colon by promoting apoptosis of colonic cancer cells (19, 20). We reason that dysbiosis of the gut microbiome leads to both enrichment of cancer-promoting bacterial populations and loss of protective populations. Thus, understanding the dynamics changes in the gut microbiome on a community-wide scale will be essential for understanding colon tumor development.
The gut microbiome is also likely to contribute to CRC through the initiation of inflammation. The link between inflammation and cancer is well established, and patients with inflammatory bowel diseases, such as ulcerative colitis, are at a greater risk of developing CRC in their lifetime. In the case of ulcerative colitis, the risk for cancer is related to both the duration and severity of inflammation, with an increasing rate of 0.5 to 1% per year after the first decade (2, 21, 22). Chronic inflammation of the colon leads to the production of various inflammatory cytokines and reactive oxygen species that work in concert to generate a tumor microenvironment that promotes carcinogenesis (21, 23, 24). It has been suggested that this process is microbe driven, but it is unclear how the normally beneficial gut microbiome becomes inflammatory.
To determine the role of the gut microbiome in inflammation and colon tumorigenesis, we used a well-established model of colitis-associated CRC that recapitulates the progression from chronic inflammation to dysplasia and adenocarcinoma in humans (25). We characterized the dynamics of the gut microbiome in this model and demonstrated that community-wide changes promote tumorigenesis in the colon. Our data support a model in which epithelial cell mutation and inflammatory perturbations of the gut microbiome lead to the development of an abnormal microbial community with enhanced tumor-promoting activity.
RESULTS
Inflammation-associated colon tumorigenesis.
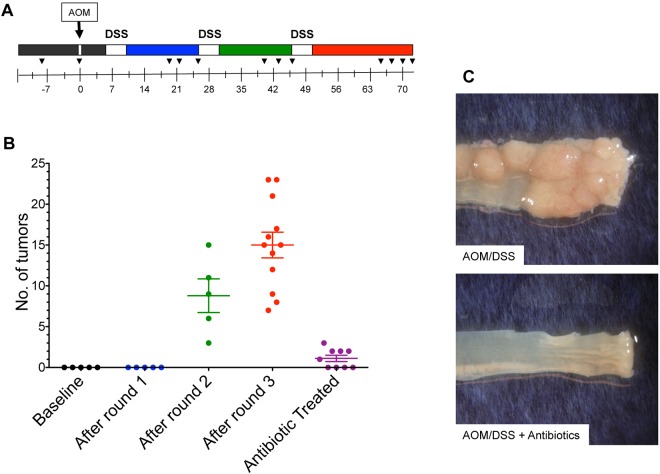
We were able to replicate an inflammation-based murine model of tumorigenesis in specific-pathogen-free (SPF) C57BL/6 mice (n = 12) using an intraperitoneal injection of the chemical carcinogen azoxymethane (AOM) followed by three subsequent rounds of water-administered 2% dextran sodium sulfate (DSS) treatment (26, 27) (Fig. 1A). The mice showed a consistent pattern of weight loss following each round of DSS treatment, with the most pronounced change occurring after the first round of DSS (see Fig. S1A in the supplemental material). We did not observe macroscopic tumors following the first round of DSS administration; however, we did observe increased infiltration by immune cells, lytter eff significant epithelial damage, and submucosal edema (see Fig. S1B). In addition, we observed a significant increase in the proinflammatory mediators macrophage inflammatory protein 2 (MIP-2), gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), and IL-1β (see Fig. S1A). Macroscopic tumors and epithelial hyperplasia were apparent following the second round of DSS (Fig. 1B; see also Fig. S1B). At the end of the model, the cohort had a median of 14.5 tumors per mouse (n = 12), the majority of which were greater than 1 mm in diameter and located in the distal colon and rectum (see Fig. S1B). These results demonstrate that our cohort of AOM/DSS-treated mice developed a significant number of colonic tumors with complete penetrance that could be detected as early as 7 weeks after AOM injection.
FIG 1 .
Inflammation-induced tumorigenesis is commensal dependent. (A) Mice were injected with azoxymethane (AOM) on day 1, followed by 3 subsequent rounds of water-administered 2% DSS. Colons were harvested 73 days after AOM, and tumors were grossly counted. Black wedges indicate fecal samples used for gut microbiome analysis (n = 12). (B) Representative mice were euthanized following each round of DSS to identify macroscopic tumors (n = 5 for each time point). An antibiotic cocktail of metronidazole, streptomycin, and vancomycin was administered in the drinking water of a separate cohort of mice for the duration of the model (n = 9). Statistical analysis was performed using a two-tailed Student’s t test. *, P < 0.01. (C) Representative images of tumors in the distal colon of conventional mice treated with AOM/DSS (n = 12) and mice treated with an antibiotic cocktail and AOM/DSS (n = 9). Error bars represent ±SEM.
To determine whether tumor incidence and penetrance were dependent on the gut microbiome, we treated mice (n = 9) with an antibiotic cocktail of metronidazole, vancomycin, and streptomycin ad libitum for 2 weeks prior to AOM and then throughout the model, including the days of AOM injection and throughout the DSS treatment and recovery periods. Antibiotic-treated mice had significantly fewer tumors in the colon than untreated mice (Fisher exact test, P < 0.001) (Fig. 1B). Tumors that were present in antibiotic-treated mice were also significantly smaller than those observed in untreated mice (Student’s t test, P = 0.002) (Fig. 1C; see also Fig. S5A in the supplemental material). These results suggest that specific populations within the microbiome were essential for tumorigenesis. To determine whether the relative change in bacterial density following antibiotic treatment was due to a change in the bacterial load, we performed quantitative PCR (qPCR) with the 16S rRNA gene from stool samples of antibiotic-treated mice. The number of 16S rRNA gene copies per mg of feces was not significantly different from that for untreated stool samples (P = 0.21) (see Fig. S2). Combined, these results indicate that changes to the structure of the community rather than total bacterial numbers affected tumorigenesis.
Significant shifts in the microbiome are associated with colon tumorigenesis.
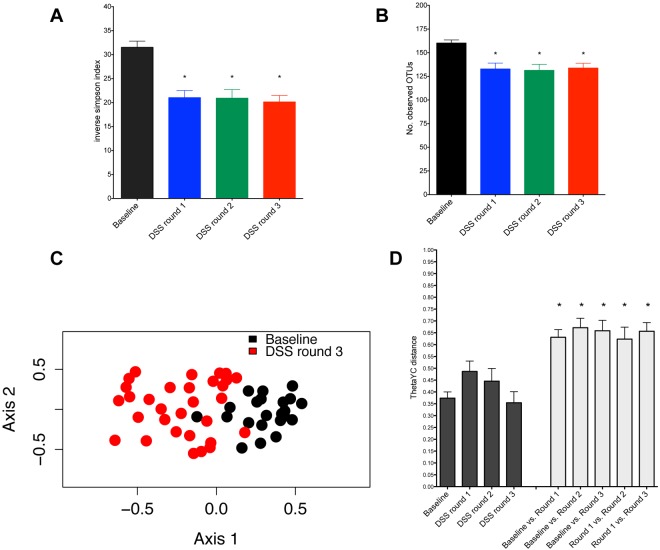
To further test the hypothesis that specific changes in the microbial community structure were associated with inflammation and tumorigenesis, we examined the dynamics of the gut microbiome throughout the model using stool samples from a subset of the original cohort of conventional mice treated with AOM/DSS for Fig. 1 (n = 10). We used the fecal samples taken prior to AOM injection as a baseline control for each mouse and then took samples following each subsequent round of DSS administration (Fig. 1A). Mice showed a significant decrease in microbial diversity in the gut microbiome following the first round of DSS administration through tumor development (P < 0.001) (Fig. 2A and B). Ordination of the distances between fecal samples showed that at the time of euthanization, tumor-bearing mice developed a significantly altered microbiome that clustered separately from that in baseline samples taken prior to the first round of DSS (Fig. 2C). Further examination of fecal samples collected at various time points during the AOM/DSS tumor induction protocol revealed that significant alterations in the microbiome could be observed as early as the first round of DSS administration in 7 of the 10 mice. Each round of DSS treatment resulted in a significant change in the structure of the microbiome (Fig. 2D). Fecal samples taken from tumor-bearing mice after the third round of DSS until the time of euthanization also clustered separately from earlier samples. The distances between clusters were significantly higher than the distances within clusters (Fig. 2D). These clusters were observed using operation taxonomic unit (OTU) and phylogenetics-based metrics of β-diversity (i.e., θYC and unweighted or weighted UniFrac) and could be distinguished from one another using the Random Forest machine learning algorithm (accuracy for each group: baseline, 100%; DSS round 1, 72.4%; DSS round 2, 71.9%; DSS round 3, 80.6%). These results highlighted the association between a dramatically altered microbiome structure and the presence of tumors.
FIG 2 .
Development of a dysbiotic gut microbiome during colon tumorigenesis. Microbiome analysis was performed with fecal samples from 10 representative mice; color coding is as indicated in Fig. 1A. (A) Inverse Simpson’s diversity index. (B) Observed community richness estimate. Statistical analysis was performed using repeated-measures paired group analysis of variance. (C) Nonmetric multidimensional scaling (NMDS) ordination based on θyc distances for all 10 mice during the AOM/DSS model. (D) Average θYC distance within (black) and between (gray) phases of the model. Error bars represent ±SEM.
To determine the effect of inflammation on the microbial community independent of tumorigenesis, we treated mice with three rounds of DSS without the AOM injection (n = 5). There was an initial community shift following the first round of DSS, but the subsequent stepwise shifts that occurred in AOM/DSS-treated mice were not observed in mice treated with DSS only (see Fig. S3 in the supplemental material). Furthermore, we did not observe the sustained drop in microbial diversity that was observed in AOM/DSS-treated animals (see Fig. S3A). These results suggest that inflammation alone is not sufficient to cause microbial community changes. Rather, the synergistic effects of the AOM/DSS model are necessary for the development of the altered microbiome structure and tumorigenesis.
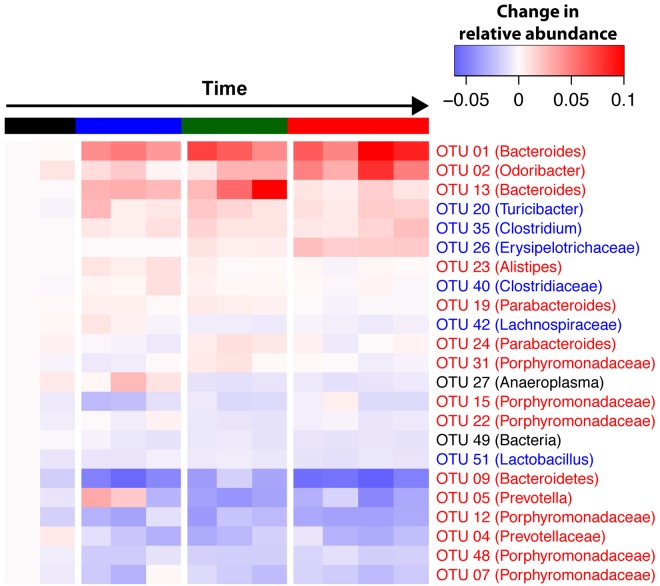
We next identified which OTUs were responsible for the dramatic shifts in the microbial community structure during inflammation and tumorigenesis (Fig. 3). Consistent with our communitywide β-diversity analyses, we observed changes in 37 bacterial populations (after excluding OTUs representing <0.5% of the community) during the time course of the model relative to those in baseline samples prior to treatment. Fecal samples taken after the first round of DSS were enriched in the relative abundance of OTUs affiliated with members of the genus Bacteroides (OTUs 1 and 13). We also observed a significant decrease in the relative abundances of OTUs associated with members of the genus Prevotella and unclassified genera within the family Porphyromonadaceae. Following the second round of DSS, we observed a further loss of the same Prevotella (OTUs 4 and 5) and Porphyromonadaceae (OTUs 7, 12, 15, 22, 31, and 48) and the continued enrichment of Bacteroides (OTUs 1 and 13). Samples taken from mice following the third round of DSS showed significant differences compared to those taken following the first round of DSS and from healthy baseline mice (Fig. 3) (all P values were <0.001 as determined by analysis of molecular variance [AMOVA]). Tumor-bearing mice showed enrichment in OTUs affiliated with Bacteroides (OTU 1), Odoribacter (OTU 3), and Turicibacter (OTU 20). Additionally, we detected a marked bloom of a member of the Erysipelotrichaceae family (OTU 26), which was undetectable in all of the mice prior to the second round of DSS, when tumors are not evident. Simultaneous with the blooming of several bacterial populations, there was a significant decrease in the relative abundance of OTUs associated with members of the genus Prevotella (OTUs 4 and 5) and the family Porphyromonadaceae (OTUs 7, 12, 15, 22, 31, and 48). An OTU associated with the Bacteroides genus (OTU 13), which bloomed during the onset of inflammation, decreased significantly following the third round of DSS. These results strongly suggest that both inflammation and tumorigenesis promote gut microbiome dysbiosis, as highlighted by major shifts in bacterial populations from a wide range of taxonomic groups.
FIG 3 .
Heat map of OTUs with relative abundances that are significantly different from their relative abundances at the time of AOM administration. The average OTU abundance between mice for each OTU was calculated for each time point. The timeline is colored for the following groups: baseline samples (prior to AOM), black; following the first round of DSS, blue; following the second round of DSS, green; following the third round of DSS, red. The OTU number and taxonomic group based on RDP classification are represented for each row. Repeated-measures paired group analysis of variance was used to identify significantly altered OTUs.
We hypothesized that the variability in tumor burden among AOM/DSS-treated mice was associated with variability in the gut microbiome between mice (coefficient of variation for tumor burden = 37.9) (Fig. 1C). We identified an OTU related to an unclassified genus within the family Porphyromonadaceae (OTU 12) that was negatively correlated with tumor burden (Spearman correlation = −0.73; P < 0.05). The relative abundance of this bacterial population decreased with each round of DSS, and this drop in abundance was more pronounced in mice with higher tumor burdens. These results suggest that alterations in the relative abundances of specific bacterial populations were associated not only with the incidence of tumors but also with their prevalence.
Tumor-associated alterations in the microbiome increase tumorigenesis in germfree mice.
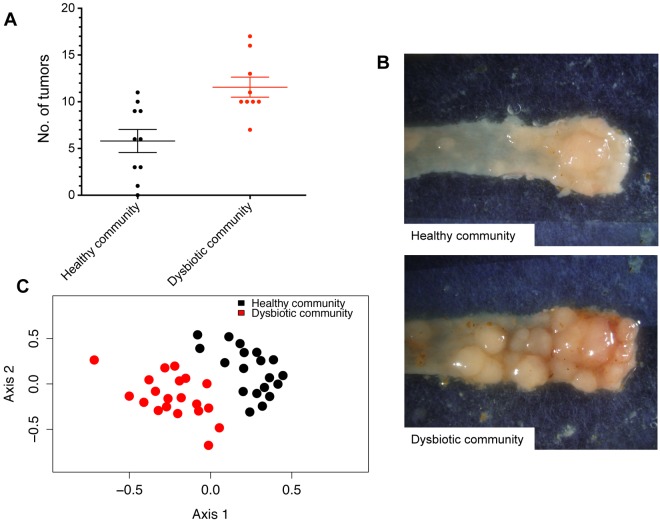
To determine whether the community-wide microbiome changes directly contributed to tumor incidence in the colon, we conventionalized germfree mice with either the healthy microbiome of untreated mice or the microbiome of tumor-bearing mice analyzed in Fig. 1. To ensure that mice were repeatedly inoculated and stably colonized, we transferred fresh feces and bedding to two groups of germfree mice (n = 10/group). One group was housed with the bedding from healthy, untreated SPF mice, and a second group was housed with bedding from tumor-bearing AOM/DSS-treated mice. To minimize litter effects, each group was comprised of two cages of 5 mice collected from separate litters that were randomly assigned to each of the cages. Following conventionalization, mice were treated with AOM/DSS under germfree conditions, as described above (Fig. 1). All bacterial phyla and 90% (62 of 69) of genus-level taxa detected in donor samples were detected within the recipient germfree mice (see Table S3 in the supplemental material), which is higher than has been previously reported (28, 29). Furthermore, 81% of the sequences we obtained from the donor mice belonged to OTUs that were found in the recipient germfree mice. Mice conventionalized with the microbiome of tumor-bearing mice had a 2-fold increase in tumor burden (P = 0.002) relative to that of mice conventionalized with a healthy microbiome (Fig. 4A). Additionally, tumors from these mice were significantly larger than those observed in recipients of a healthy microbiome (P = 0.002) (see Fig. S5B). Similar to our results with SPF mice, germfree mice conventionalized with the community of tumor-bearing mice had a significantly less diverse gut microbiome (P < 0.001). Using community-wide β-diversity analyses, we determined that conventionalization with these two treatments of bedding resulted in two distinct microbial community structures (AMOVA. P < 0.001) (Fig. 4C). Germfree mice conventionalized with the microbiome of tumor-bearing mice showed significant enrichment in the relative abundance of OTUs affiliated with the genera Bacteroides (OTU 1) and the family Erysipelotrichaceae (OTU 26). Additionally, these germfree mice had significantly fewer members of the Porphyromonadaceae (OTU 12) compared to germfree mice conventionalized with bedding from healthy mice. Finally, germfree mice conventionalized with a healthy microbiome successfully recapitulated the community dynamics seen in conventional mice during tumorigenesis. We observed significant changes in 34 OTUs following the AOM/DSS model. Similar to tumor-bearing conventional mice, germfree tumor-bearing mice showed enrichment in OTUs affiliated with members of the Bacteroides (OTU 1), Odoribacter (OTU 3), Turicibacter (OTU 20), and a bloom in the Erysipelotrichaceae (OTU 26), which was undetectable before AOM administration. There was also a significant decrease in the relative abundance of OTUs associated with members of the genus Prevotella (OTUs 4 and 5) and the family Porphyromonadaceae (OTUs 6, 7, and 12). These results demonstrate that alterations to the gut microbiome that were associated with chronic inflammation and tumorigenesis in SPF mice were transmitted to germfree mice and can exacerbate colon tumorigenesis.
FIG 4 .
Tumor-associated gut microbiome alterations exacerbate tumorigenesis in germfree mice. (A) Number of tumors observed at the end of the model when germfree mice were colonized using bedding from healthy mice (Healthy community) or mice with tumors (Dysbiotic community). (B) Representative images of tumors in the distal colon of mice conventionalized with a healthy microbiome (n = 10) or the microbiome of tumor-bearing mice (n = 9). (C) NMDS ordination based on θyc distances for all 19 mice following conventionalization with a healthy microbiome (Healthy community) or the microbiome of tumor-bearing mice (Dysbiotic community). Error bars represent ±SEM.
DISCUSSION
In the present study, we established a causal role for the gut microbiome in exacerbating tumor formation in an inflammation-based model of tumorigenesis. Manipulation of the microbiome using antibiotics reduced tumor formation, which highlighted the importance of bacterium-driven factors in tumorigenesis. We demonstrated dynamic changes in the microbial community structure associated with dysbiosis, which occurs prior to the first signs of macroscopic tumor formation. We established the synergistic effect of AOM- and DSS-induced inflammation and tumorigenesis in driving microbial community changes that occur in a stepwise fashion. Finally, transfer of microbiota from tumor-bearing mice into germfree mice significantly increased the number and size of tumors compared to those in germfree mice inoculated with healthy microbiota. Our experiments also demonstrated dramatic shifts in the relative abundances of bacterial populations, including those related to the genus Bacteroides, that were associated with increased tumorigenesis.
Several recent studies have compared the gut microbiome of patients with CRC to that of healthy controls (10–16). These studies have consistently demonstrated significant differences in the microbial community structure of patients with CRC, but each study has disagreed in terms of the specific gut microbiome composition and profile associated with CRC. The inability to identify a consensus community profile or etiological agent is likely due to the large variation in the structure of the microbiome across individuals and the improbability of there being one community profile or bacterial population that is associated with all CRCs. We were able to reduce the interindividual variation and the diversity of cancer types using a murine model of inflammation-induced CRC. Unlike the human cross-sectional studies, we demonstrated dynamic changes in the microbiome during the development of inflammation and tumorigenesis and that these changes directly cause disease.
Based on this study, the gut microbiome complements the activity of AOM and DSS to cause tumorigenesis, but the underlying mechanisms driving microbially mediated tumorigenesis observed remain to be elucidated. Although a number of bacterial populations have altered relative abundances throughout the model, it is as yet unclear whether there is an increase in bacterial populations that induce inflammation or a loss of populations that produce anti-inflammatory signals and help maintain immune homeostasis in the gut. Regardless, an increasingly inflammatory environment would generate a self-reinforcing pathogenic cascade between the gut microbiome and the host, fostering the development of cancer through the development of, for example, genotoxic reactive oxygen species and protumor inflammatory mediators (e.g., TNF-α, IL-6, IL-1β, and IL-23). In addition to the role of the gut microbiome in inflammation, changes mediated by chronic inflammation and tumorigenesis could lead to the enrichment of bacterial populations (30) that have a direct role in tumor development through the production of metabolites, antigens, virulence factors, and other potential tumor-promoting gene products. A recent study by Arthur et al. (17) demonstrated that colonic inflammation in the IL-10-deficient mouse impacts the composition of the gut microbiome, leading to an enrichment of tumor-promoting E. coli strains. Although we did not detect any significant changes in populations related to the genus Escherichia, it is likely that the microbial community alterations we observed in our tumor model are enriched with populations that fill a similar role. Specifically, marked increases in Bacteroides species in our study may contribute to tumorigenesis. Human commensals belonging to the genus Bacteroides, specifically enterotoxigenic B. fragilis (ETBF), have been associated with inflammation and CRC (18, 31). ETBF has been shown to strongly induce colonic tumors in multiple-intestinal-neoplasia mice through secretion of a metalloprotease toxin, and certain strains are thought to contribute to CRC risk in humans. We did not detect ETBF in the murine gut microbiome (data not shown), but it is possible that similar processes are occurring during tumorigenesis in mice.
Chronic inflammation and tumorigenesis are also likely to lead to the loss of members of the gut microbiome that are important for maintaining epithelial health and immune homeostasis. In this study, we observed a dramatic decrease in OTUs from unclassified genera within the family Porphyromonadaceae. We hypothesize that these bacterial populations serve a protective role and are important mediators of gut health in the murine gut microbiome. One mechanism of protection could be through the fermentation of complex carbohydrates (e.g., fiber) into short-chain fatty acids (SCFA), such as butyrate. Butyrate reduces inflammation (32) and inhibits growth and induces apoptosis in cancer cells (19, 20). Therefore, loss of butyrate-producing populations in the gut could increase both inflammation and tumorigenesis. This is supported by extensive epidemiological data that demonstrate a link between diets high in fiber and a decreased CRC risk (33). Furthermore, recent studies have shown that individuals who consume low-fiber diets or are diagnosed with CRC have a lower level of SCFAs in their feces (34). It is also possible that members of the family Porphyromonadaceae are important mediators of anti-inflammatory signals in the gut. A loss of such anti-inflammatory populations would lead to a dramatic intensification of inflammation in the gut during DSS-induced colitis and a marked increase in tumor-promoting signals.
It is important to note that the gut microbiome is an extremely complex and diverse community, and therefore it is unlikely that a single bacterial population is responsible for driving tumorigenesis or that one CRC-associated microbiome can be found in all CRC patients. Rather, as our data suggest, a community-wide effect involving the gain and loss of bacterial populations and general metabolic functions likely plays a critical role in CRC development. As we demonstrated in this study, changes in the entire gut microbiome can dramatically alter the tumor burden, and identifying the mechanisms behind this phenomenon will be critical for addressing how the microbiome can be altered therapeutically to reduce colon tumorigenesis.
MATERIALS and METHODS
Animals and animal care.
Studies were conducted on adult (8 to 12 weeks old) age-matched male C57BL/6 mice that were bred and maintained under SPF or germfree conditions as specified above. Both SPF and germfree mice were fed the same autoclaved chow diet. All animal experiments were approved by the University Committee on Use and Care of Animals at the University of Michigan.
Inflammation-induced colon tumorigenesis.
Eight- to twelve-week-old mice received a single intraperitoneal (i.p.) injection of azoxymethane (10 mg/kg of body weight). Water containing 2% DSS was administered to mice beginning on day 5 for 5 days, followed by 16 days of water. This was repeated twice for a total of 3 rounds of DSS (26, 27). Mice were euthanized on days 14, 24, 38, and 45 for intermediate time point analysis. The remaining mice were euthanized 3 weeks after the third round of DSS administration for tumor counting.
Histological analysis.
At necropsy, all colons were harvested, flushed of luminal contents, and cut open longitudinally to count and measure tumors. The largest dimension of each tumor was measured with calipers. Tumors were categorized based on size (<1 mm, 1 to 2 mm, or >2 mm). Colons were then “jelly rolled,” fixed in Carnoy’s solution, and embedded in paraffin. Five-micrometer sections were used for hematoxylin-and-eosin (H&E) staining, and slides were analyzed under magnification ×100.
RNA isolation and cytokine analysis.
Distal colon tissue was homogenized and total RNA was isolated using the Nucleospin RNA kit (Macherey-Nagel). cDNA was synthesized using an iScript kit (Bio-Rad). and the cDNA was then used for quantitative PCR (qPCR) using a SYBR green expression assay (Applied Biosystems).
Antibiotic treatment.
Mice were treated with an antibiotic cocktail of metronidazole (0.75 g/liter), vancomycin (0.5 g/liter), and streptomycin (2 g/liter) in their drinking water for 2 weeks prior to and throughout the duration of AOM/DSS administration.
Germfree conventionalization.
Eight-weeks-old C57BL/6 germfree male mice were used. Fresh feces and bedding were collected from untreated and AOM/DSS-treated tumor-bearing mice and immediately transferred to cages of germfree mice 2 weeks prior to AOM injection to allow stable colonization. Germfree mice were divided into two treatment groups, one group receiving bedding from untreated, healthy mice and the other group receiving bedding from AOM/DSS-treated tumor-bearing mice. To ensure that there were no cage effects, each treatment group was comprised of two cages of mice. The mice were obtained from separate litters and randomly assigned to the four cages. Mice were then treated with AOM/DSS to induce tumors as described above. Three weeks after the last round of DSS, mice were euthanized and colons were harvested as described above.
DNA extraction.
Fecal samples were collected daily from the mice throughout the AOM/DSS protocol and immediately frozen for storage at −20° C. We selected 12 fecal samples distributed over the 73-day timeline of the AOM/DSS model for 10 representative mice. Microbial genomic DNA was extracted using the PowerSoil-htp 96 Well Soil DNA isolation kit (Mo Bio laboratories) using an EpMotion 5075 pipetting system.
16S rRNA gene sequencing and curation.
The V35 region of the 16S rRNA gene from each sample was amplified and sequenced using the 454 Titanium sequencing platform at the Baylor College of Medicine Human Genome Sequencing Center as described elsewhere (http://www.mothur.org/wiki/454_SOP). We curated our sequences as described previously using the mothur software package (35–37). Briefly, we denoised sequences using the PyroNoise algorithm after trimming each flowgram to 450 flows (38), aligned the resulting sequences to a reference alignment derived from the SILVA 16S rRNA sequence database (39), and removed sequences that were flagged as possible chimeras by UCHIME (50) or that did not align to the V35 region. After curating the sequence data, we obtained between 6 and 10,742 sequences (median = 5,681), with a median length of 253 bp. To minimize biased effects of uneven sampling, we rarefied to 1,800 sequences per sample. Seven samples either did not pass through sequence curation or had less than 1,800 sequences and were therefore not used for further analysis. Parallel sequencing of a mock community allowed us to measure a median error rate of 0.06%.
Analysis of the microbiome.
Sequences were clustered into OTUs based on a 3% distance cutoff using the average-neighbor algorithm. All sequences were classified using the RDP training set, version 9 (http://sourceforge.net/projects/rdp-classifier/), and OTUs were assigned a classification based on which taxonomy had the majority consensus of sequences within a given OTU using a naive Bayesian classifier (40). Microbial diversity was calculated using the inverse Simpson index (41) and the observed number of OTUs. To calculate β-diversity, we used the θYC distance metric with OTU frequency data (42), and we calculated UniFrac statistics (43) using neighbor-joining phylogenetic trees generated using the nonheuristic neighbor-joining algorithm implemented in the software program Clearcut (44). Analysis of molecular variance (AMOVA) was performed to determine significance between the community structures of different groups of samples based on θYC and UniFrac distance matrices (45). To identify OTUs important for driving differences between groups (baseline, after DSS round 1, after DSS round 2, and after DSS round 3), we used a repeated-measure paired treatment analysis of variance for each OTU and corrected for multiple comparisons using an experiment-wise error rate of 0.01 (46). Additionally, we identified features (OTUs) important for each group using the machine learning algorithm random forest as implemented in the software environment R (http://CRAN.R-project.org) (47). The abundance-based Jaccard dissimilarity index indicates the fraction of all sequences that affiliate with OTUs that are shared between two communities and was used to calculate the fraction of OTUs that were shared between donor samples and germfree recipient samples (48). All sff files and the MIMARKS spreadsheet are available at http://www.mothur.org/aomdss_dynamics/.
16S rRNA qPCR analysis.
Relative bacterial loads in stool samples were quantified by qPCR analysis of bacterial genomic DNA using Kapa SYBR-fast master mix (Kapa biosciences) and universal 16S rRNA gene primers (F, ACTCCTACGGGAGGCAGCAGT; R, ATTACCGCGGCTGCTGGC) (49). Samples were normalized to fecal mass, and the relative fold change was determined using untreated stool samples for each replicate mouse (n = 5). Note that qPCR measures the relative fold change of the 16S gene copy number, not actual bacterial numbers.
SUPPLEMENTAL MATERIAL
Colonic inflammation during tumorigenesis. (A) Relative cytokine expression levels in the colon. Statistical analysis was performed using a two-tailed Student’s t test. *, P < 0.01. (B) Representative H&E-stained histological sections from colons of mice during AOM/DSS tumor induction protocol. Single arrows indicate mucosal ulceration, double arrows indicate submucosa edema, and dashed arrows indicate adenoma. Error bars represent SD. Download
16S rRNA quantitative qPCR. Relative fold decrease in 16S rRNA copy number during antibiotic treatment compared to findings for baseline untreated samples. All time points showed no significant decrease compared to data for baseline untreated samples (P = 0.21). Error bars represent SEM. Download
Microbiome alterations associated with DSS-induced inflammation. (A) Inverse Simpson’s diversity index. (B) Average θYC distance between phases of the model. Error bars represent SEM. Download
Numbers of tumors observed at the end of the model when germfree mice were colonized using bedding from healthy mice (Healthy community) and mice with tumors (Dysbiotic community). Each treatment had 2 replicate cages. The mice were obtained from separate litters that were randomly assigned to each of four cages. Error bars represent SD. Download
Measurement of largest dimension of each tumor (mm) was performed using calipers for each mouse following AOM/DSS administration. (A) Antibiotic-treated mice (n = 10) versus conventional mice (n = 12). P = 0.002. (B) Germfree mice conventionalized with a healthy community (n = 10) versus a dysbiotic community (n = 10). P = 0.0019. Statistical analysis of tumor count data was performed using Fisher’s exact test. Download
Average relative abundances of significantly altered OTUs during AOM/DSS tumor induction; OTUs included are significantly altered; significance was determined using repeated-measure paired treatment analysis of variance and correcting for multiple comparisons using an experiment-wise error rate of 0.01; taxonomic group based on RDP classification is represented for each OTU; “*” indicates significant alterations relative to DSS round 1
Average relative abundances of OTUs for baseline and DSS round 3 in germfree mice conventionalized with healthy microbiota; OTUs included are significantly altered from baseline over time in mice conventionalized with bedding from healthy mice; significance was determined using repeated-measure paired treatment analysis of variance and correcting for multiple comparisons using an experiment-wise error rate of 0.01; taxonomic group based on RDP classification is represented for each OTU
Average relative abundances of genus-level bacterial taxa are represented for conventional donor mice (n = 12) and recipient germfree mice (n = 10 for each treatment); RDP classification was used for phylotype analysis
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes for Health to P.D.S. (R01GM095356, R01HG005975, P30DK034933, and University of Michigan GI SPORE) and G.Y.C. (University of Michigan GI SPORE and ARRA supplement P30CA4659-22S3).
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. 2013. The gut microbiome modulates colon tumorigenesis. mBio 4(6):e00692-13. doi:10.1128/mBio.00692-13.
REFERENCES
- 1. Parkin DM, Bray F, Ferlay J, Pisani P. 2005. Global cancer statistics, 2002. CA Cancer J. Clin. 55:74–108 [DOI] [PubMed] [Google Scholar]
- 2. Chambers WM, Warren BF, Jewell DP, Mortensen NJ. 2005. Cancer surveillance in ulcerative colitis. Br. J. Surg. 92:928–936 [DOI] [PubMed] [Google Scholar]
- 3. Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M. 2009. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int. J. Cancer 125:171–180 [DOI] [PubMed] [Google Scholar]
- 4. Larsson SC, Rafter J, Holmberg L, Bergkvist L, Wolk A. 2005. Red meat consumption and risk of cancers of the proximal colon, distal colon and rectum: the Swedish Mammography Cohort. Int. J. Cancer 113:829–834 [DOI] [PubMed] [Google Scholar]
- 5. Slattery ML. 2000. Diet, lifestyle, and colon cancer. Semin. Gastrointest. Dis. 11:142–146 [PubMed] [Google Scholar]
- 6. Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. 2011. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci. U. S. A. 108:4570–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dolara P, Caderni G, Salvadori M, Morozzi G, Fabiani R, Cresci A, Orpianesi C, Trallori G, Russo A, Palli D. 2002. Fecal levels of short-chain fatty acids and bile acids as determinants of colonic mucosal cell proliferation in humans. Nutr. Cancer 42:186–190 [DOI] [PubMed] [Google Scholar]
- 8. Stappenbeck TS, Hooper LV, Gordon JI. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 99:15451–15455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rakoff-Nahoum S, Medzhitov R. 2007. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science 317:124–127 [DOI] [PubMed] [Google Scholar]
- 10. Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, Yang L, Liu ZJ, Yuan YZ, Liu F, Wu JX, Zhong L, Fang DC, Zou W, Fang JY. 2013. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 97:1044–1052 [DOI] [PubMed] [Google Scholar]
- 11. Chen W, Liu F, Ling Z, Tong X, Xiang C. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7:e39743. 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geng J, Fan H, Tang X, Zhai H, Zhang Z. 2013. Diversified pattern of the human colorectal cancer microbiome. Gut Pathog. 5:2. 10.1186/1757-4749-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. 2010. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1:138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. 2011. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6:e16393. 10.1371/journal.pone.0016393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. 2012. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6:320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hague A, Elder DJ, Hicks DJ, Paraskeva C. 1995. Apoptosis in colorectal tumor-cells—induction by the short-chain fatty-acids butyrate, propionate and acetate and by the bile-salt deoxycholate. Int. J. Cancer 60:400–406 [DOI] [PubMed] [Google Scholar]
- 20. Ruemmele FM, Schwartz S, Seidman EG, Dionne S, Levy E, Lentze MJ. 2003. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut 52:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ullman TA, Itzkowitz SH. 2011. Intestinal inflammation and cancer. Gastroenterology 140:1807–1816 [DOI] [PubMed] [Google Scholar]
- 22. Eaden J, Abrams K, Ekbom A, Jackson E, Mayberry J. 2000. Colorectal cancer prevention in ulcerative colitis: a case-control study. Aliment. Pharmacol. Ther. 14:145–153 [DOI] [PubMed] [Google Scholar]
- 23. Arthur JC, Jobin C. 2013. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes 4:253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin WW, Karin M. 2007. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 117:1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, Signori E, Fazio VM. 2011. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J. Carcinog. 10:9. 10.4103/1477-3163.78279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. 2003. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 94:965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen GY, Shaw MH, Redondo G, Núñez G. 2008. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 68:10060–10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. 2013. Gut microbiomes of Malawian twin pairs discordant for Kwashiorkor. Science 339:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Nùñez G, Chen G, Rosenstiel P, Chamaillard M. 2013. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123:700–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sears CL, Islam S, Saha A, Arjumand M, Alam NH, Faruque AS, Salam MA, Shin J, Hecht D, Weintraub A, Sack RB, Qadri F. 2008. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 47:797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segain JP, de la Bletiere DR, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottière HM, Galmiche JP. 2000. Butyrate inhibits inflammatory responses through NF kappaB inhibition: implications for Crohn’s disease. Gut 47:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aune D, Chan DS, Lau R, Vieira R, Greenwood DC, Kampman E, Norat T. 2011. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ 343:d6617. 10.1136/bmj.d6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O’Keefe SJ, Ou J, Aufreiter S, O’Connor D, Sharma S, Sepulveda J, Fukuwatari T, Shibata K, Mawhinney T. 2009. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J. Nutr. 139:2044–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. 2012. Stabilization of the murine gut microbiome following weaning. Gut Microbes 3:383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641 [DOI] [PubMed] [Google Scholar]
- 39. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Magurran AE. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ. [Google Scholar]
- 42. Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Statist. Theory Methods 34:2123–2131 [Google Scholar]
- 43. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheneman L, Evans J, Foster JA. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823–2824 [DOI] [PubMed] [Google Scholar]
- 45. Martin AP. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57:289–300 [Google Scholar]
- 47. Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22 [Google Scholar]
- 48. Chao A, Chazdon RL, Colwell RK, Shen TJ. 2006. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics 62:361–371 [DOI] [PubMed] [Google Scholar]
- 49. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colonic inflammation during tumorigenesis. (A) Relative cytokine expression levels in the colon. Statistical analysis was performed using a two-tailed Student’s t test. *, P < 0.01. (B) Representative H&E-stained histological sections from colons of mice during AOM/DSS tumor induction protocol. Single arrows indicate mucosal ulceration, double arrows indicate submucosa edema, and dashed arrows indicate adenoma. Error bars represent SD. Download
16S rRNA quantitative qPCR. Relative fold decrease in 16S rRNA copy number during antibiotic treatment compared to findings for baseline untreated samples. All time points showed no significant decrease compared to data for baseline untreated samples (P = 0.21). Error bars represent SEM. Download
Microbiome alterations associated with DSS-induced inflammation. (A) Inverse Simpson’s diversity index. (B) Average θYC distance between phases of the model. Error bars represent SEM. Download
Numbers of tumors observed at the end of the model when germfree mice were colonized using bedding from healthy mice (Healthy community) and mice with tumors (Dysbiotic community). Each treatment had 2 replicate cages. The mice were obtained from separate litters that were randomly assigned to each of four cages. Error bars represent SD. Download
Measurement of largest dimension of each tumor (mm) was performed using calipers for each mouse following AOM/DSS administration. (A) Antibiotic-treated mice (n = 10) versus conventional mice (n = 12). P = 0.002. (B) Germfree mice conventionalized with a healthy community (n = 10) versus a dysbiotic community (n = 10). P = 0.0019. Statistical analysis of tumor count data was performed using Fisher’s exact test. Download
Average relative abundances of significantly altered OTUs during AOM/DSS tumor induction; OTUs included are significantly altered; significance was determined using repeated-measure paired treatment analysis of variance and correcting for multiple comparisons using an experiment-wise error rate of 0.01; taxonomic group based on RDP classification is represented for each OTU; “*” indicates significant alterations relative to DSS round 1
Average relative abundances of OTUs for baseline and DSS round 3 in germfree mice conventionalized with healthy microbiota; OTUs included are significantly altered from baseline over time in mice conventionalized with bedding from healthy mice; significance was determined using repeated-measure paired treatment analysis of variance and correcting for multiple comparisons using an experiment-wise error rate of 0.01; taxonomic group based on RDP classification is represented for each OTU
Average relative abundances of genus-level bacterial taxa are represented for conventional donor mice (n = 12) and recipient germfree mice (n = 10 for each treatment); RDP classification was used for phylotype analysis