ABSTRACT
Two-component systems (TCS) comprise histidine kinases and their cognate response regulators and allow bacteria to sense and respond to a wide variety of signals. Histidine kinases (HKs) phosphorylate and dephosphorylate their cognate response regulators (RRs) in response to stimuli. In general, these reactions appear to be highly specific and require an appropriate association between the HK and RR proteins. The Myxococcus xanthus genome encodes one of the largest repertoires of signaling proteins in bacteria (685 open reading frames [ORFs]), including at least 127 HKs and at least 143 RRs. Of these, 27 are bona fide NtrC-family response regulators, 21 of which are encoded adjacent to their predicted cognate kinases. Using system-wide profiling methods, we determined that the HK-NtrC RR pairs display a kinetic preference during both phosphotransfer and phosphatase functions, thereby defining cognate signaling systems in M. xanthus. Isothermal titration calorimetry measurements indicated that cognate HK-RR pairs interact with dissociation constants (Kd) of approximately 1 µM, while noncognate pairs had no measurable binding. Lastly, a chimera generated between the histidine kinase, CrdS, and HK1190 revealed that residues conferring phosphotransfer and phosphatase specificity dictate binding affinity, thereby establishing discrete protein-protein interactions which prevent cross talk. The data indicate that binding affinity is a critical parameter governing system-wide signaling fidelity for bacterial signal transduction proteins.
IMPORTANCE
Using in vitro phosphotransfer and phosphatase profiling assays and isothermal titration calorimetry, we have taken a system-wide approach to demonstrate specificity for a family of two-component signaling proteins in Myxococcus xanthus. Our results demonstrate that previously identified specificity residues dictate binding affinity and that phosphatase specificity follows phosphotransfer specificity for cognate HK-RR pairs. The data indicate that preferential binding affinity is the basis for signaling fidelity in bacterial two-component systems.
INTRODUCTION
In response to nutrient limitation, Myxococcus xanthus undergoes a complex developmental process culminating in the formation of multicellular structures termed fruiting bodies. As the fruiting body matures, individual cells differentiate into metabolically dormant and stress-resistant spores. The developmental process is characterized by significant changes in gene expression (1). Many developmentally regulated genes are σ54 dependent, thus requiring the alternative sigma factor RpoN (2). Transcription from σ54-dependent promoters requires an upstream activator homologous to bacterial NtrC proteins. Mutations inactivating most of the 27 NtrC homologs in M. xanthus produce defects in development, leading to the hypothesis that NtrC-like activators (NLAs) are involved in complex signaling networks (2–12). NtrC homologs are response regulators (RRs) for two-component signal transduction systems (TCS), which require activation by a histidine kinase (HK). While NtrC homologs are regulated in M. xanthus (2), there is a significant lack of data regarding the associated HKs. Indeed, M. xanthus NLAs regulate development through a complex cascade of gene expression (2) and may interact biochemically in functionally complex signaling pathways.
Many organisms utilize complex signal transduction cascades to regulate critical cellular processes. Complex regulatory pathways allow integration of multiple signals and checkpoints for the control of important cellular decisions. For instance, Bacillus subtilis employs multiple kinases within a complex signaling cascade to regulate phosphorylation of Spo0A to control sporulation (13). Alternatively, Escherichia coli CheA regulates the activity of both CheY and the negative feedback controller, CheB, to control chemotaxis (14, 15). In M. xanthus, the CrdS-CrdA TCS interacts with the Che3 chemosensory system to regulate timing of aggregation and fruiting body development (5, 6, 16). These examples highlight the variability in signal transduction system architecture and suggest that further analysis of M. xanthus two-component systems may provide additional insights into the regulation of complex signaling pathways.
Upon sensing an appropriate ligand, a TCS HK autophosphorylates on a conserved histidine. The phosphoryl group is then transferred to a conserved aspartate within the RR, thus regulating activity of the system. Many HKs are bifunctional enzymes possessing the ability to both phosphorylate and subsequently dephosphorylate their cognate RRs. Dephosphorylation of a phosphorylated RR provides a mechanism to eliminate cross talk from other sources which would otherwise lead to fitness disadvantages. Specific residues within the HK are known to be required for proper phosphatase activity (16–32), and specificity residues are known to prevent cross talk (27, 33–36). One aspect of signaling fidelity that has yet to be determined conclusively is whether specificity residues dictate binding affinity to ensure cognate HK-RR interactions. Some examples have implicated binding or competition as a factor in the prevention of cross talk (27, 30, 36–43), but information regarding a direct link between specificity residues and binding affinity is limited.
Studies with E. coli and Caulobacter crescentus established the basis for signaling fidelity regarding TCS phosphotransfer (44, 45). However, by comparison to M. xanthus, those organisms have fewer and relatively distant TCS homologs. M. xanthus encodes ~127 TCS, including those utilizing NtrC homologs which likely arose through duplication (46), suggesting that signaling fidelity and specificity may be more complex in this case (43). To investigate fidelity and binding affinity, we performed phosphotransfer profiling with each of the 27 predicted NtrC RR homologs in combination with a subset of corresponding HKs. We also profiled phosphatase activity for ten putative HK-RR systems and extended our analysis to binding affinity for a subset of these proteins. Lastly we generated a chimera that switched its binding affinity as well as its preferred target for phosphotransfer and phosphatase specificity to its predicted new RR partner. Our results demonstrate a clear kinetic preference for phosphotransfer and phosphatase activities and correspond directly with binding affinity. The results support the model that binding affinity is controlled by specificity residues which thereby dictate kinetic preference.
RESULTS
Bioinformatic analysis of signaling systems utilizing NtrC homologs in M. xanthus.
The M. xanthus genome encodes a minimum of 127 HKs and 143 RRs, 27 of which have domain topology identical to that of the E. coli protein NtrC. These proteins were identified using the Microbial Signal Transduction Database by searching for proteins with NtrC-like domain topology: each homolog possesses an amino-terminal receiver domain (REC), a central σ54 activation domain, and a carboxyl-terminal helix-turn-helix DNA-binding domain (47). In addition, all of the associated REC domains have conserved residues required for phosphorylation by a histidine kinase, indicating that these proteins are likely involved in signaling (48). The 27 NtrC homologs are distributed throughout the M. xanthus chromosome and display high sequence similarity, which is indicative of recent duplication events rather than acquisition via horizontal transfer (46). A subset of NtrC-like activators have also been annotated and studied previously (12). Many NtrC homologs have been studied in M. xanthus, and most are reported to affect development (2–12) (Table 1). Based on these studies, it has been suggested that NtrC regulators may be involved in cross talk or cross-regulated systems (2, 6).
TABLE 1 .
HK-NtrC pairs in M. xanthusa
| HK (Mxan no.) |
HK designation | NtrC (Mxan no.) | NtrC designation |
ntrC (nla) mutant phenotype |
Reference for HK phosphorylation or phosphotransfer |
|---|---|---|---|---|---|
| 0172b | NtrC0172 | Development (3) | |||
| 0938 | HK0938 | 0937 | Nla7 | (12) | This study |
| 1077 | 1078 | Nla19 | Development (12) | ||
| 1129 | FrgB | 1128 | FrgC | Development (49) | This study |
| 1166 | Nla28S | 1167 | Nla28 | Development (12) | 52 |
| 1190 | HK1190 | 1189 | NtrC1189 | This study; 6, 16 | |
| 1249 | SasS | 1245 | SasR | Development (7) | This study |
| 2516b | Nla4 | Development (12) | |||
| 3214b | ActB | Development (8) | |||
| 3419 | 3418 | NtrC3418 | |||
| 3555b | NtrC3555 | (3) | |||
| 3812 | HK3812 | 3811 | NtrC3811 | This study | |
| 4043 | Nla6S | 4042 | Nla6 | Development (12) | 51 |
| 4197 | 4196 | NtrC4196 | Development (3) | ||
| 4240b | NtrC4240 | ||||
| 4251 | 4252c | ||||
| 4262 | 4261 | NtrC4261 | Development (3) | 6, 16 | |
| 4579 | 4580c | ||||
| 4786 | HK4786 | 4785 | NtrC4785 | This study | |
| 4977b | NtrC4977 | (3) | |||
| 5123 | MrpA | 5124 | MrpB | Development (50) | |
| 5184 | CrdS | 5153 | CrdA | Development (5, 6) | This study; 6, 16 |
| 5365 | HsfB | 5364 | HsfA | Heat shock (10) | This study; 10 |
| 5778 | HK5778 | 5777 | Nla23 | Development (12) | This study |
| 5785 | PilS | 5784 | PilR | Motility (11) | |
| 7142 | 7143 | NtrC7143 | Development (3) | ||
| 7439 | HK7439 | 7440 | Nla24 | Development (12) | This study |
Shown is the list of M. xanthus NtrC homologs and their predicted cognate partner histidine kinases (HK). The locus tags for each HK or NtrC RR homolog are shown. Alternative names for some of the homologs and phenotypes for mutants are also listed. Biochemical determination of in vitro phosphorylation of the HK is listed in the last column. Mxan, the locus identifier for each M. xanthus gene in the NCBI database.
Orphan ntrC gene (lacking a corresponding kinase).
The NtrC homolog was not tested.
To determine if signaling fidelity is maintained for this set of TCS RR proteins, as described previously (33, 36), or if there might be some level of cross-regulation for these systems, we assessed the phosphorylation kinetics for several HK-RR systems in vitro. First, we sought to identify cognate kinases for each NtrC homolog, as those proteins have not been extensively characterized. The gene neighborhood for each RR was analyzed for instances where an HK was encoded within a putative operon. Of the 27 M. xanthus NtrC homologs that met our criteria, we were able to identify 19 HKs encoded adjacent to or within the same putative operon or gene cluster (Table 1), 6 of which have been studied previously: FrgBC (49), HsfBA (10), MrpAB (50), PilSR (11), Nla6S (HK)-Nla6 (RR) (12, 51), and Nla28S (HK)-Nla28 (RR) (12, 52). The NtrC homologs CrdA and SasR are orphans but have experimentally determined cognate HKs, CrdS and SasS, respectively (6, 7). Six additional NtrC homologs are orphans (NtrC0172, NtrC3555, NtrC4240, NtrC4977, ActB, and Nla4 [3, 8, 12]) and might be secondary targets for prototypical TCS or targets of orphan HKs. Genetic analyses were performed for 7 additional RRs (NtrC4196, NtrC4261, NtrC7143, Nla7, Nla19, Nla23, and Nla24 [3, 12]) but did not assess a role for the associated putative HK. The 6 remaining RRs and their putative associated HKs have not been assayed for function in vivo. Moreover, none of the aforementioned kinases, regulators, or predicted cognate pairs have been tested for specificity via phosphotransfer profiling. The predicted or experimentally determined cognate HK and RR homolog and known phenotypes for corresponding mutants are listed in Table 1.
NtrC homologs are preferentially phosphorylated by cognate histidine kinases.
Previous studies demonstrated that specificity during in vitro phosphotransfer profiling reflects in vivo phosphotransfer preference for a given HK-RR signaling system (44, 53). To characterize the phosphotransfer preference for a subset of M. xanthus HK-RR pairs, we first generated constructs to express the signaling domains of the 27 NtrC RRs and the 21 associated HKs. Each construct was designed to express a soluble REC domain for each RR or a truncated form of the kinase (including the dimerization and histidine phosphotransfer [DHp] and the catalytic and ATP-binding [CA] domains [54]) where the input domain was replaced by a 6-His affinity tag. Using this method, we were able to purify 25 of 27 NtrC REC domains and soluble, active forms of 10 kinases (Fig. 1): HK0938, HK1190, HK3812, HK4786, HK5778, HK7439, FrgB, HsfB, CrdS, and SasS. Activity for each of these proteins was first determined by in vitro autophosphorylation. For REC domains, the ability to incorporate radioactive phosphoryl groups was determined after incubation with [32P]acetyl-phosphate (AcP) (see below). The activity for each HK was determined by its ability to incorporate radioactive phosphoryl groups when incubated with [γ-32P]ATP. The 10 HK proteins displayed variation in both the amount and rate of autophosphorylation (Fig. 1; also, see Fig. S1 in the supplemental material). For instance, HsfB reached maximal phosphorylation in approximately 5 min, while HK4786 did not attain peak levels even after 60 min. Additionally, the apparent maxima varied significantly between proteins: HK3812 was able to generate only 0.38 pmol of phosphorylated protein, while FrgB generated 2.4 pmol at 60 min. A similar level of variability has been documented for HKs purified from E. coli, where both the rate and level of phosphorylation varied significantly (55), and this may reflect variation in the fraction of active or properly folded proteins.
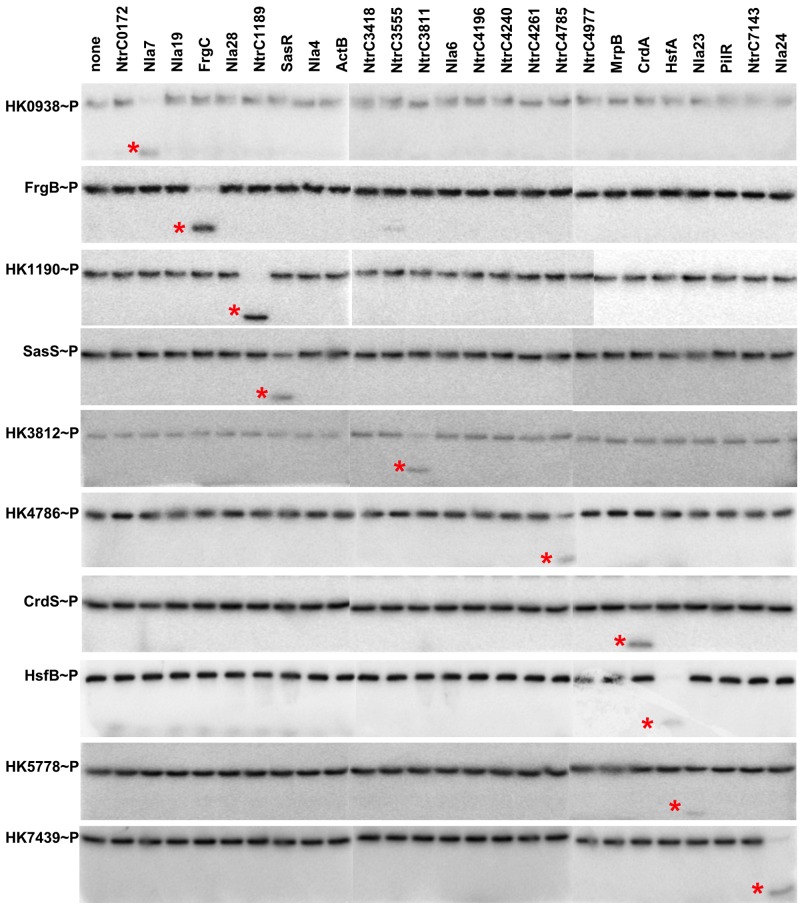
FIG 1 .
Phosphotransfer profiling indicates specificity for cognate TCS. Each HK was allowed to autophosphorylate to maximum levels before incubation with an equimolar amount of each individual NtrC RR homolog. All phosphotransfer reactions were stopped after 10 s, and products were resolved using SDS-PAGE and analyzed on a phosphorimager as described in Materials and Methods. Each phosphorylated NtrC homolog is indicated by an asterisk. Three separate gels were run and processed simultaneously for each HK to enable profiling for all 25 RRs. The resulting images were assembled into rows as shown.
To test kinetic preference for each HK, we performed a series of phosphotransfer profiling experiments similar to those described previously (44). In these experiments, each individual HK was autophosphorylated prior to incubation with each individual RR. As a baseline, we performed phosphotransfer profiling with CrdS, for which both genetic and biochemical evidence indicates that CrdA is the cognate RR in vivo (6, 16). When we allowed the experiment to proceed for 10 s, we saw phosphotransfer from CrdS to CrdA (Fig. 1). However, when we allowed the same experiment to proceed for an hour, CrdS was able to phosphorylate three other NtrC homologs, NtrC0172, NtrC1189, and the orphan NtrC3555 (see Fig. S2 in the supplemental material). It is likely that CrdS phosphorylates these alternative targets only under the conditions of this assay and is not physiologically relevant. This result is similar to other phosphotransfer profiling experiments where the E. coli kinases EnvZ, CheA, and CpxA phosphorylate their cognates at short time points but allow transfer to alternative targets over longer time courses (44). Because longer time points are generally permissive for low-level phosphorylation of nonspecific targets, we performed all subsequent phosphotransfer profiling experiments using a 10-s time point.
In addition to CrdS, we performed phosphotransfer profiling experiments using the M. xanthus kinases HK0938, HK1190, HK3812, HK4786, HK5778, HK7439, FrgB, HsfB, and SasS. The results for the phosphotransfer profiles were similar to those obtained with CrdS. Each HK phosphorylated its predicted cognate RR within 10 s (Fig. 1), although variability was apparent in this experiment. For instance, there was no observable loss of phosphorylated HK5778 (HK5778~P) which corresponded with only modest production of the phosphorylated target, Nla23~P. In stark contrast, HK1190 exhibited significantly higher levels of phosphotransfer to NtrC1189, as indicated by the complete turnover of HK1190~P which corresponded with production of high levels of phosphorylated target protein, NtrC1189~P. Collectively, the results show that each HK displays a kinetic preference for phosphotransfer to its predicted cognate RR and that these systems comprise discrete signaling pathways: HK0938-Nla7, HK1190-NtrC1189, HK3812-NtrC3811, HK4786-NtrC4785, HK5778-Nla23, HK7439-Nla24, CrdSA, FrgBC, HsfBA, and SasSR.
Phosphatase profiling demonstrates specificity for bifunctional histidine kinases.
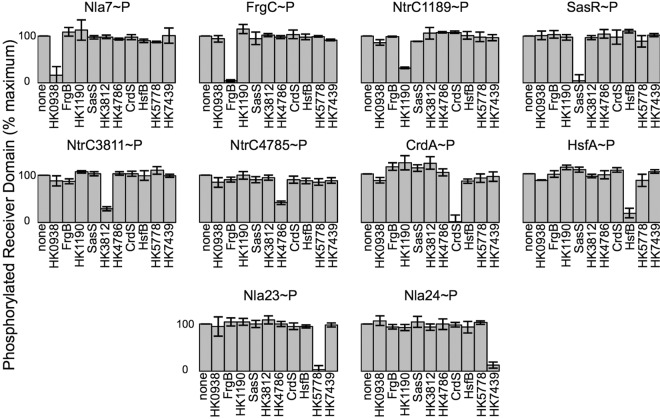
Many HKs are known to be bifunctional enzymes capable of both phosphorylation and dephosphorylation of the cognate RR (17–20, 23–26, 30–32). Phosphatase activity is critical to signaling fidelity, as it provides a mechanism to negate cross talk or aberrant phosphorylation by other sources (27, 33–36, 43). Thus, we sought to test the fidelity of HK phosphatase activity by conducting a corollary assay termed phosphatase profiling. In this assay, we generated RR~P using the high energy phosphodonor acetyl-phosphate (AcP). We selected the 10 NtrC homologs that were specifically phosphorylated by their cognate HK in our phosphotransfer profiling assays (Fig. 1): NtrC1189, NtrC3811, NtrC4785, Nla7, Nla23, Nla24, CrdA, FrgC, HsfA, and SasR. Each RR was labeled using [32P]AcP as described previously (6, 36, 56). Each RR~P incubated alone in buffer was used to normalize the data for subsequent reactions (Fig. 2). Each RR~P was incubated with an equimolar amount of each of the ten purified kinases. A 5-min incubation was chosen because the assay requires measuring the disappearance of label, and that time point had been used previously to demonstrate phosphatase activity for several other kinases (16).
FIG 2 .
Phosphatase profiling reveals specificity within cognate TCS. Phosphatase assays were performed using acetyl-phosphate (AcP)-labeled response regulators as described in Materials and Methods. The amount of phosphorylated RR incubated in buffer alone was set to 100%. A decrease in the amount of phosphorylated RR is indicative of phosphatase activity.
Initially, we performed the phosphatase profiling experiment using the previously described NtrC homolog, CrdA. In this experiment, CrdA was dephosphorylated only by its cognate kinase, CrdS. In the presence of CrdS, CrdA~P levels were reduced to 1.6% relative to the buffer control (Fig. 2). When CrdA~P was incubated with each of the other HKs, there was no statistically significant diminution of CrdA~P, indicating that CrdS phosphatase activity is specific for CrdA~P. Using this phosphatase profiling assay, we observed a similar trend for each of the other nine NtrC homologs. Each RR~P was dephosphorylated only when incubated with its predicted cognate HK (statistical significance determined by Student’s t test; P < 0.05). Four HKs (HK5778, CrdS, FrgB, and SasS) were able to dephosphorylate their cognate RR~P by 90%. The remaining 6 HKs (HK0938, HK1190, HK3812, HK4786, HK7439, and HsfB) dephosphorylated their targets by ~60 to 90% (Fig. 2). The variability for phosphatase activity observed here is similar to that observed for phosphotransfer activity exhibited by each HK. Importantly, each HK displayed specificity for the same RR homolog for both phosphatase and phosphotransfer activities (Fig. 1 and 2). Together, these data allow us to conclude that phosphatase activity, like phosphotransfer activity, is highly specific and can be used to define cognate signaling systems.
A combined assay illustrates differences in phosphorylation kinetics for TCS.
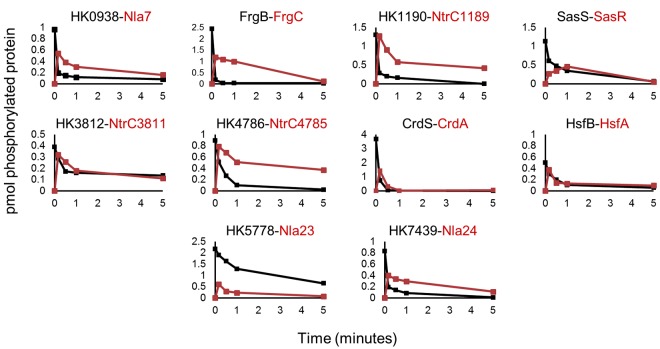
After determining the phosphotransfer and phosphatase specificity independently for each HK via profiling, we then analyzed the combined phosphotransfer and phosphatase functions over time in one assay. In essence, the assay measures the flux of phosphoryl groups through the HK-RR system. We tested the 10 HK-RR pairings identified above in the profiling experiments. CrdS~P displayed rapid phosphotransfer to CrdA. Subsequently, CrdS dephosphorylated CrdA~P (Fig. 3), resulting in a complete loss of radiolabel from the reaction by 1 min in this assay. Because the half-life of CrdA~P is about 1 h (6), the disappearance of both CrdS~P and CrdA~P indicates fast phosphotransfer and subsequent dephosphorylation by CrdS. HK0938, HK1190, HK4786, HK7439, FrgB, and HsfB demonstrated kinetics similar to that of CrdS, where <10% of the HK~P remained at the 1-min time point. Additionally, these HKs were able to dephosphorylate their cognate RRs to various degrees over the 5-min time course. In contrast, HK3812, HK5778, and SasS displayed lower phosphotransfer rates, with more than 25% of HK~P being present at the 1-min time point. HK5778 displayed the slowest turnover, with more than 30% of HK5778~P remaining after 5 min. Overall, the results are consistent with the phosphotransfer and phosphatase profiling data yet reveal differences in rates that may reflect physiologically relevant aspects of TCS control in M. xanthus.
FIG 3 .
Time course assays for phosphotransfer activity. Cognate HK and RR pairs identified by the above profiling experiments were assayed in a time course for phosphotransfer activity. Each HK protein (black) was allowed to reach maximal phosphorylation and then mixed with equimolar amounts of cognate NtrC RR protein (red). Samples were collected at 0, 0.25 0.5, 1 and 5 min, resolved by electrophoresis, imaged, and quantified as described in Materials and Methods. The quantified results are presented as pmol of phosphorylated proteins.
Cognate systems display preferential binding as measured by isothermal titration calorimetry.
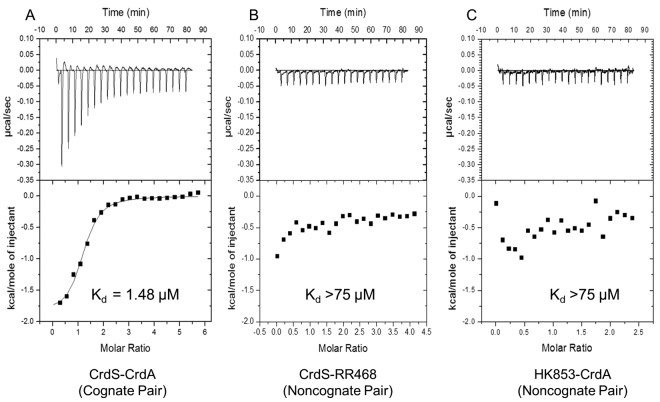
Previous work led to the hypothesis that binding affinity facilitates preferential interactions between TCS proteins to maintain signaling fidelity (27, 30, 36–43). In this study, we used isothermal titration calorimetry (ITC) to determine binding affinities of four pairs of TCS proteins and tested all cognate and noncognate pairwise interactions. We chose M. xanthus HK1190-NtrC1189, CrdS-CrdA, FrgB-FrgC, and T. maritima HK853-RR468. We chose HK1190 because it is most similar to CrdS within the α1 helix of the DHp domain (see below). Additionally, RR NtrC1189 was phosphorylated, albeit inefficiently, by CrdS at the 1-h time point in the phosphotransfer profiling experiment (see Fig. S2 in the supplemental material), suggesting the possibility of cross talk between these two systems. We chose FrgBC because genetic analyses confirmed its likely function as a cognate TCS affecting motility in M. xanthus (49). Lastly, we chose the well-studied yet evolutionarily distant HK853-RR468 TCS from T. maritima to serve as a control, and we determined that HK853 exhibited no capacity to phosphorylate any of the 25 M. xanthus NtrC RR homologs in this study (see Fig. S3 in the supplemental material).
Background-corrected, integrated thermograms for three representative binding reactions are shown in Fig. 4. Calorimetric measurements and subsequent fitting of binding isotherms allowed us to determine the affinity for each pairwise protein-protein interaction. For the cognate pair, CrdS-CrdA, the reaction curve clearly indicates an interaction with an estimated dissociation constant (Kd) of 1.4 ± 0.3 µM. Similar results were obtained for each pairwise interaction of cognate partner proteins, where the resulting Kd was estimated to be about 1 µM (Table 2). It is worth noting that, under the conditions of the assay, the proteins are unlikely to be phosphorylated. Nevertheless, experiments performed using noncognate proteins pairs, CrdS-RR468 and HK853-CrdA, exhibited Kds above the limit of detection (greater than 75 µM), indicating no detectable binding (Fig. 4B and C). Similar results were obtained for each pairwise interaction between noncognate partners where the Kd was above the limit of detection (Table 2). One caveat for this assay is that interactions can occur where ΔH is near 0 at a given temperature and would therefore be undetectable via ITC under the conditions tested. Thus, we performed additional ITC experiments at a different temperature (10°C). At this lower temperature, there was still no evidence for binding between noncognate proteins (data not shown). To provide further support for the above results, we measured changes in the intrinsic fluorescence in CrdS upon binding to CrdA. CrdS contains a native tryptophan residue near the predicted binding site for CrdA and thus could be utilized in fluorescence binding assays. Fluorescence binding indicated that CrdS and CrdA interact with an apparent Kd of 2.3 µM, validating the ITC data (see Fig. S4 in the supplemental material). Together, the results are consistent with the hypothesis that binding affinity corresponds directly to kinetic preference for both phosphotransfer and phosphatase function.
FIG 4 .
Isothermal titration calorimetry analysis of HK-RR binding affinity. Thermograms and binding isotherms for the cognate CrdS-CrdA TCS pair (A), the noncognate CrdS-RR468 pair (B), and the noncognate HK853-CrdA pair (C) are shown. Enthalpy and Kd for binding were determined using isothermal titration calorimetry in a VP-ITC microcalorimeter as described in Materials and Methods. The top portion of each panel shows baseline corrected thermograms. The bottom portion shows the corresponding binding isotherms generated using nonlinear binding models.
TABLE 2 .
Dissociation constants determined by isothermal titration calorimetrya
| Protein target | Ligand | Kd (µM)b | Phosphotransfer and phosphatase specificity |
|---|---|---|---|
| CrdS | CrdA | 1.41 ± 0.33 | Yes |
| CrdS | FrgC | >75 | |
| CrdS | NtrC1189 | >75 | |
| CrdS | T. maritima RR468 | >75 | |
| FrgB | CrdA | >75 | |
| FrgB | FrgC | 1.23 ± 0.37 | Yes |
| FrgB | NtrC1189 | >75 | |
| FrgB | T. maritima RR468 | >75 | |
| HK1190 | CrdA | >35 | |
| HK1190 | FrgC | >35 | |
| HK1190 | NtrC1189 | 1.23 ± 0.37 | Yes |
| HK1190 | T. maritima RR468 | >55 | |
| T. maritima HK853 | CrdA | >75 | |
| T. maritima HK853 | FrgC | >55 | |
| T. maritima HK853 | NtrC1189 | >55 | |
| T. maritima HK853 | T. maritima RR468 | 1.16 ± 0.18 | Yes |
| CrdS6 | CrdA | >75 | |
| CrdS6 | NtrC1189 | 2.09 ± 1.11 | Yes |
Binding affinities (dissociation constants) for 4 distinct TCS HK-RR protein interactions, with cognate and noncognate partners, were determined by ITC. Cognate TCS pairs that were found to interact and display phosphotransfer and phosphatase specificity are in bold. Noncognate proteins were found to have Kd values above the upper limit detectable in this assay.
Values are the averages of four separate measurements (± standard deviations).
Specificity residues dictate preferential binding for two-component systems.
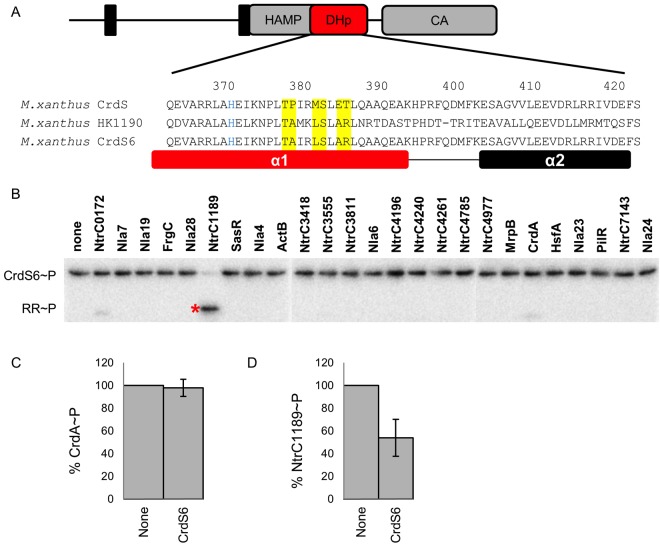
The results above provide strong support for the hypothesis that binding affinity dictates protein interactions for cognate signaling systems. Based on the previous work demonstrating that phosphotransfer specificity is controlled by a discrete set of residues within the HK and RR (36, 44), we wished to directly assess the role of those specificity residues for their effect on binding affinity for a subset of proteins tested above. Therefore, we replaced those residues within CrdS to mimic those of HK1190 (Fig. 5) and predicted that the chimeric protein CrdS/HK1190 (referred to here as CrdS6) would display specificity similar to that of HK1190 for its cognate partner, RR NtrC1189. We tested this possibility by determining the binding affinity for CrdS6 and NtrC1189 via ITC and also assayed kinetic preference using phosphotransfer and phosphatase assays.
FIG 5 .
Switching specificity determinants alters binding, phosphotransfer, and phosphatase affinity. (A) Domain topology of the prototypical histidine kinase CrdS with a DHp α1 helix sequence alignment for CrdS, HK1190, and the chimeric CrdS6. (B) Phosphotransfer profiling demonstrates that the chimeric CrdS6 protein mimics the phosphotransfer specificity of HK1190. Three separate gels were run and processed simultaneously to enable profiling for all 25 RRs. The resulting images were assembled into one row as shown. Phosphatase assays demonstrate that specificity for CrdS6 has been switched from CrdA~P (C) to NtrC1189~P (D).
To identify the correct specificity residues, we aligned the relevant sequences for CrdS and HK1190 spanning the DHp α1 and α2 helixes, as described previously (27). Only two of six specificity residues are identical between HK1190 and CrdS (Fig. 5A). Using site-directed mutagenesis, we changed the remaining four specificity residues within CrdS (resulting in the substitutions P379A, M382L, E385A, and T386R) to generate the CrdS6 mutant protein. We verified that CrdS6 was active as a kinase as measured by the ability to autophosphorylate using ATP (see Fig. S5 in the supplemental material). To confirm that CrdS6 mimicked the phosphotransfer specificity for RR NtrC1189 rather than CrdA, we utilized the phosphotransfer profiling assay. CrdS6~P was incubated with equimolar amounts of each NtrC homolog as described above. CrdS6 was found to mimic the specificity of HK1190, as was evident by a clear preference for phosphotransfer to NtrC1189 without significant phosphotransfer to CrdA (Fig. 5B). To quantify the observed kinetic preference, we measured the relative kcat/Km ratios as described previously (36). CrdS6 displays a 103-fold increase in kinetic preference for NtrC1189 relative to CrdA, similar to the kinetic preference displayed by wild-type CrdS for CrdA (6). In the reciprocal phosphatase assay, CrdS6 was unable to dephosphorylate CrdA~P (Fig. 5C) yet could dephosphorylate NtrC1189~P (Fig. 5D). Thus, the results clearly indicate that the chimeric CrdS6 protein switches its kinetic preference from CrdA to NtrC1189.
We then measured binding affinity using ITC to test pairwise interactions between CrdS6 and NtrC1189 or CrdA (Table 2). The CrdS6-NtrC1189 interaction displayed a dissociation constant of 2.1 ± 1 µM, while no measurable binding was observed between CrdS6 and CrdA. Moreover, the Kd for CrdS6-NtrC1189 is close to the observed Kd for each of the cognate pairs HK1190-NtrC1189 and CrdS-CrdA (Table 2). Furthermore, we tested binding between CrdS6 and CrdA at 10°C and observed no interaction (data not shown), confirming that CrdS6 lacks the capacity to bind CrdA. Lastly, using intrinsic tryptophan fluorescence, no binding was observed between CrdS6 and CrdA (Kd > 200 µM), further indicating these proteins do not interact (see Fig. S4 in the supplemental material). Together, the data lead us to conclude that the CrdS6 chimeric protein mimics HK1190 for phosphotransfer specificity, phosphatase specificity, and binding affinity with RR NtrC1189. Overall, these data indicate that TCS specificity residues dictate binding affinity, which imparts kinetic preference and signaling fidelity.
DISCUSSION
Specificity residues dictate binding affinity for two-component signaling systems.
For two-component systems, fidelity of phosphotransfer is predicated on specific protein-protein interactions between the HK and RR. Using ITC analysis, we tested four two-component systems and demonstrated that cognate signaling proteins had measurable binding affinities (Kd) of approximately 1 µM (Table 2), similar to values determined previously for EnvZ-OmpR, CheA-CheY, CheA-CheB, and CphA-RcpA (57–59). Importantly, we observed no measurable binding for all noncognate protein interactions assayed by ITC. The results from the binding assays correlated directly with those from both phosphotransfer and phosphatase assays, supporting the conclusion that signaling fidelity depends critically on binding affinity.
Previous work demonstrated that phosphotransfer activity is dependent on a set of six specificity residues (36). Based on those studies, we generated a chimeric protein which resulted in redirected phosphotransfer and phosphatase specificity as well as binding affinity between CrdS6 and NtrC1189. Furthermore, CrdS6 displayed neither any measurable binding affinity nor a kinetic preference for CrdA in the phosphotransfer and phosphatase assays. Therefore, the six specificity residues described previously are sufficient to impart both preferential binding between HK-RR cognate pairs and to regulate phosphorylation and dephosphorylation of the correct target. In support of this view, recent work demonstrated that engineering noncognate HK-RR pairs with eukaryotic scaffolding proteins promotes phosphotransfer and binding both in vitro and in vivo, even in the absence of the known specificity residues (60). Lastly, hybrid HKs which contain covalently linked REC domains typically lack specificity residues, presumably because those domains are spatially tethered (61).
It is known, however, that cross talk can occur when either the cognate kinase or response regulator is absent (37, 39, 62). Thus, without a preferential interacting binding partner, homologous TCS proteins are able to engage in cross talk, as demonstrated during phosphotransfer profiling (53). Similarly, we observed here that CrdS could phosphorylate three additional targets in vitro, NtrC0172, NtrC1189, and orphan NtrC3555 (see Fig. S2 in the supplemental material), but only at longer time points, approaching 1 h of incubation. Importantly, CrdS and NtrC1189 showed no detectable interaction via ITC until the specificity residues were altered to mimic those of HK1190 (Table 2). These results substantiate previous findings indicating that cross talk is enabled in vitro but is not likely relevant in vivo. Overall, we conclude that binding affinity coupled with relatively fast phosphotransfer kinetics is the critical parameter to effectively eliminate unwanted cross talk within the cell and that binding affinity is dictated by specificity residues for cognate two-component signaling proteins.
Signaling fidelity for TCS in M. xanthus.
As a general rule, a given HK displays an overwhelming kinetic preference for its cognate RR. For instance, EnvZ has a 103-fold kinetic preference for its cognate OmpR over the noncognate CpxR (44). The kinase VanS also exhibits a 104-fold kinetic preference for VanR over the noncognate PhoB (63). Previous work in our laboratory demonstrated that CrdS has a 105-fold kinetic preference for the cognate CrdA relative to the noncognate NtrC1189 (6). Thus, in vitro analysis by phosphotransfer profiling serves as an effective way to map relevant signaling pathways.
M. xanthus possesses 27 NtrC homologs, many of which have been demonstrated to be involved in regulation of the developmental program (Table 1). Because many mutations within this family of highly similar signaling proteins led to similar developmental phenotypes, we thought it possible that cross talk may occur or that bona fide cross-regulation may occur, as described for the NarXL-NarQP systems in E. coli (64). The results presented here, however, strongly suggest that HK-RR systems in M. xanthus are insulated at the molecular level, regardless of the additional complexity presented by their position within a hierarchy of gene regulation controlling development. Our results do not rule out cross regulation in vivo but strongly suggest that signaling fidelity is maintained based on the presence of a unique set of specificity residues for each HK associated with a given RR within the family of NtrC homologs.
The system-wide (profiling) approach used in this study allowed us to assess kinetic preferences during both kinase and phosphatase reactions for ten distinct M. xanthus TCS, verifying the fidelity of CrdSA, FrgBC, HsfBA, and SasSR in vitro. The results allow us to conclude that specificity residues dictate binding affinity and phosphorylation kinetics to maintain signaling fidelity in M. xanthus. It is therefore likely that complex signaling cascades regulating the developmental process in M. xanthus display limited cross talk and that binding affinity is the critical parameter for system-wide fidelity during signal transduction.
MATERIALS AND METHODS
Bacterial growth and DNA manipulations.
For routine cloning, E. coli strains were grown in Luria broth with antibiotic concentrations of 40 µg/ml kanamycin or 100 µg/ml ampicillin when selection was required. All proteins were expressed using the E. coli strain BL21(DE3) with the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector pET28a (Novagen) and were grown in Terrific Broth (TB). Strains are listed in Table S1 in the supplemental material. PCR was used to amplify genes encoding each HK and RR by using genomic DNA isolated from M. xanthus strain DZ2 as a template (65). Primers for these reactions are listed in Table S2 in the supplemental material. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent), and all constructs were verified by sequencing.
Protein purification.
All proteins used in kinase, phosphotransfer profiling, and phosphatase profiling assays were purified using a standard batch purification method, with the exception of HK0938, SasS, and HK3812, which were purified using a urea solubilization protocol. For batch purification, each strain was grown in 100 ml Terrific Broth in a 250-ml Erlenmeyer flask at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8. Protein expression was induced with 1 mM IPTG. After overnight growth at 20°C, cells were collected in a Beckman Coulter JLA-16 250 rotor at 5,000 × g for 10 min. Cell pellets were stored at −20°C until purification. For purification, cell pellets were thawed and suspended in 10 ml of cell lysis buffer (25 mM Tris [pH 7.6], 125 mM NaCl, 5 mM imidazole, 1% [vol/vol] Triton X-100, and 0.625 g of CelLytic Express from Sigma-Aldrich plus Complete Mini EDTA-free protease inhibitor from Roche) and incubated for 1 h before lysates were clarified by centrifugation at 5,000 × g. Two milliliters of His-Select cobalt affinity resin (Sigma-Aldrich) was equilibrated in wash buffer (25 mM Tris [pH 7.6], 125 mM NaCl, 5 mM imidazole, 1% [vol/vol] Triton X-100) and incubated with each lysate for 2 h at 4°C with mild agitation. His-resin was then pelleted by a 1-min spin at 200 × g, and the supernatant was discarded. The resin was washed 3 times with 10 ml of wash buffer and eluted with 5 ml of elution buffer (25 mM Tris [pH 7.6], 125 mM NaCl, 500 mM imidazole). Samples were dialyzed overnight at 4°C against dialysis buffer (25 mM Tris [pH 7.6], 125 mM NaCl, 1 mM dithiothreitol [DTT], 1% [vol/vol] Triton X-100, 50% [vol/vol] glycerol, 0.5 mM EDTA). Purified protein was assayed by standard denaturing gel electrophoresis, and the concentration was determined using the Bradford reagent. All purified proteins were stored in dialysis buffer at −20°C.
Proteins used for ITC analysis were prepared from cells grown to an OD600 of 0.8 in 2 liters of Terrific Broth, induced with 0.5 mM IPTG and shaken overnight at 20°C. Cells were pelleted by centrifugation and stored at −20°C until purification. For purification, cell pellets were thawed and suspended in 25 ml of ITC cell lysis buffer (25 mM Tris [pH 7.6], 125 mM NaCl, 0.1% [vol/vol] Triton X-100, 1 mg/ml lysozyme, Complete Mini EDTA-free protease inhibitor [Roche]) and lysed by passage through an LV02 Microfluidizer (Microfluidics). Sample lysates were then clarified by centrifugation at 40,000 × g and passage through a 0.45-µm filter disk. Lysates were loaded onto a 5-ml Hi-Trap HP (GE) column using an ÄKTA fast protein liquid chromatography (FPLC) system (GE). The column was washed with 7.5 column volumes (CV) of buffer A (25 mM Tris [pH 7.6], 125 mM NaCl, 10 mM imidazole, 0.1% [vol/vol] Triton X-100) and eluted with a 15-CV linear gradient to 100% buffer B (25 mM Tris [pH 7.6], 125 mM NaCl, 500 mM imidazole, 0.1% [vol/vol] Triton X-100). Fractions containing protein of interest were combined and dialyzed overnight against ITC buffer (25 mM Tris [pH 7.6], 150 mM NaCl, 10% glycerol, 0.1% [vol/vol] Triton X-100, 0.5 mM EDTA). Proteins were then assayed for purity using SDS-PAGE. If there were any apparent contaminating proteins, samples were further purified using a Superdex S75 (GE) gel filtration column equilibrated in ITC buffer.
Kinase assays.
Kinase assays were performed as described previously (16). Briefly, 5 µl of 50 µM HK stock was added to 5 µl 10× kinase buffer (250 mM Tris [pH 7.6], 500 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 10 mM CaCl2 and 10 mM dithiothreitol) and 35 µl distilled H2O. Reaction mixtures were incubated at room temperature and started by the addition of 5 µl of an ATP mix (250 µM ATP, 3 µM [γ-32P]ATP). Aliquots were removed at various time points and stopped by the addition of an equal volume of 2× SDS loading buffer. Samples were resolved by electrophoresis on 12% SDS-polyacrylamide gels. The dye front, containing unincorporated ATP, was removed, and then gels were exposed for 2 to 4 h on a phosphor screen and visualized using a Typhoon Imager (GE). ImageQuant v 5.1 was used to determine integrated pixel density and subtract background.
Phosphotransfer profiling.
Histidine kinases were allowed to autophosphorylate, as detailed above, until maximal phosphorylation levels were reached (10 min to 2 h, depending on the kinase). A 5-µl aliquot was mixed with 5 µl of purified RR (10 µM stock) in kinase buffer, and reactions were stopped at various times by addition of 2× SDS loading buffer. Incorporation of labeled phosphoryl groups was analyzed as detailed above. Each phosphotransfer profiling experiment was run on three separate gels. Gel images were handled identically and assembled into figures using Microsoft PowerPoint.
Phosphatase profiling.
For phosphatase profiling experiments, each RR was radiolabeled with acetyl phosphate ([32P]AcP) as described previously (6, 16). Each RR~P was then incubated in kinase buffer for 5 min with equimolar amounts of each HK (final concentration of 5 µM for each HK and RR). RR~P incubated with buffer alone was set to 100%. Samples were separated by SDS-PAGE and visualized as detailed above. Bar graphs show averages and standard deviations from three experiments.
Binding measurements using isothermal titration calorimetry.
Measurements were performed using a VP-ITC titration calorimeter from Microcal, Inc. Each histidine kinase and response regulator was dialyzed overnight against the same ITC buffer. The VP-ITC cell was filled with the histidine kinase protein at concentrations of ~10 to 15 µM in ITC buffer. The syringe was filled with ~150 to 200 µM response regulator in ITC buffer. The exact concentration of each protein was measured using a bicinchoninic acid protein assay kit (Thermo Scientific). The ITC cell and syringe solutions were extensively degassed. The chamber was kept under constant stirring at 350 rpm, and all experiments were performed at 25°C except where noted. Control injections were also performed to determine heats of dilution. The control injection data were subtracted from the raw data using the Origin ITC analysis package provided with the microcalorimeter. The heat of dilution not eliminated by controls was determined by the average of the last 3 to 5 injections and subsequently subtracted. The data were analyzed using the single-site binding model provided with the ITC analysis package. Results in Table 2 are the averages from at least four separate runs from two separate protein preparations.
Intrinsic tryptophan fluorescence measurements of binding affinities.
Intrinsic tryptophan fluorescence was used to monitor the binding of cognate and noncognate response regulators with histidine kinases. For each pair, the response regulator (~100 µM) was titrated into 1.3 ml of 5 µM histidine kinase contained in a stirred 1-cm quartz cuvette until saturation. Fluorescence intensity measurements were recorded at 25°C on a Fluorolog 3 (Jobin Yvon, Horiba) spectrofluorimeter. For each measurement, excitation was at 294 nm (5-nm slit width). The fluorescence emission spectrum was collected in the range 300 to 400 nm (5-nm slit width). Individual measurements were integrated for 30 s. An equal volume of buffer was titrated into the histidine kinase, and intensity measurements were collected under the conditions described above. These measurements were then subtracted from the corresponding response regulator titration to correct for the change in intensity due to dilution. The Kd for each titration was determined by fitting buffer-corrected data using nonlinear regression (Sigma Plot; SPPS, Inc.). The resulting dissociation constant was the average of three independent experiments.
SUPPLEMENTAL MATERIAL
Autophosphorylation of histidine kinases. (A) Incorporation of phosphoryl groups by ten purified HK proteins in the presence of [γ-32P]ATP is shown. Kinase activity was quantified as described in Materials and Methods. Reactions were allowed to proceed for 1, 5, 15, 30, or 60 min. (B) Quantification of autophosphorylation (pmol of phosphorylated HK over time) for each HK~P shown in panel A. Download
Extended phosphotransfer profiling for CrdS. Phosphotransfer profiling for CrdS with each of the 27 NtrC homologs from M. xanthus is shown. CrdS was incubated with all proteins for 10 s (top) or 60 min (bottom). Specific phosphotransfer from CrdS to CrdA is indicated by an asterisk. Nonspecific phosphotransfer to NtrC0172, NtrC1189, and NtrC3555 at 60 min is indicated by the red arrow. Download
T. maritima HK853 phosphotransfer profiling with M. xanthus NtrC homologs. Phosphotransfer profiling demonstrates that HK853 is able to phosphorylate only the cognate RR for HK853, T. maritima RR468 (*). Download
Intrinsic tryptophan fluorescence assay for binding affinity. Representative binding curves for the cognate CrdS-CrdA pair (closed circles; Kd, 2.28 ± 0.66 µM) and noncognate CrdS6-CrdA pair (open circles; Kd, >200 µM) were determined by tryptophan fluorescence. Download
Autokinase activity of the CrdS6 chimera. The autokinase assay demonstrates that the CrdS6 mutant protein is functional and likely retains overall proper structure, as measured by the ability to autophosphorylate. Download
Strains used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
Support for this work was provided by NSF MCB-1244021 to J.K. Additional support for J.W.W. was provided by NIH T32 GM077973. Partial support was provided by NSF MCB-0918807 and University of Iowa, Carver College of Medicine, Department of Biochemistry, to E.J.F.
DNA sequencing was performed by Nevada Genomics Center (University of Nevada, Reno, NV). We thank Cindy Darnell and the reviewers for critical reading of the manuscript.
The content is the responsibility of the authors and does not represent the official views of NIH or NSF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Willett JW, Tiwari N, Müller S, Hummels KR, Houtman JCD, Fuentes EJ, Kirby JR. 2013. Specificity residues determine binding affinity for two-component signal transduction systems. mBio 4(6):e00420-13. doi:10.1128/mBio.00420-13.
REFERENCES
- 1. Huntley S, Hamann N, Wegener-Feldbrügge S, Treuner-Lange A, Kube M, Reinhardt R, Klages S, Müller R, Ronning CM, Nierman WC, Søgaard-Andersen L. 2011. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol. Biol. Evol. 28:1083–1097 [DOI] [PubMed] [Google Scholar]
- 2. Giglio KM, Caberoy N, Suen G, Kaiser D, Garza AG. 2011. A cascade of coregulating enhancer binding proteins initiates and propagates a multicellular developmental program. Proc. Natl. Acad. Sci. U. S. A. 108:E431–E439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giglio KM, Eisenstatt J, Garza AG. 2010. Identification of enhancer binding proteins important for Myxococcus xanthus Development. J. Bacteriol. 192:360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862–872 [DOI] [PubMed] [Google Scholar]
- 5. Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:2008–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Willett JW, Kirby JR. 2011. CrdS and CrdA Comprise a two-component system that is cooperatively Regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2(4):e00110-11. 10.1128/mBio.00110-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo D, Wu Y, Kaplan HB. 2000. Identification and characterization of genes Required for Early Myxococcus xanthus Developmental Gene Expression. J. Bacteriol. 182:4564–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gronewold TM, Kaiser D. 2002. Act Operon Control of Developmental Gene Expression in Myxococcus xanthus. J. Bacteriol. 184:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi X. 2009. An analysis of two-component regulatory systems in Myxococcus xanthus. Philipps-Universität Marburg, Marburg, Germany [Google Scholar]
- 10. Ueki T, Inouye S. 2001. Transcriptional activation of a heat shock gene, lonD, of Myxococcus xanthus by a two-component His–Asp phosphorelay system. J. Biol. Chem. 277:6170–6177 [DOI] [PubMed] [Google Scholar]
- 11. Spormann AM, Kaiser D. 1999. Gliding mutants of Myxococcus xanthus with High Reversal Frequencies and Small Displacements. J. Bacteriol. 181:2593–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stephenson K, Hoch JA. 2002. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46:297–304 [DOI] [PubMed] [Google Scholar]
- 14. Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63:45–59 [DOI] [PubMed] [Google Scholar]
- 15. Rao CV, Kirby JR, Arkin AP. 2004. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2:E49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willett JW, Kirby JR. 2012. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 8:e1003084. 10.1371/journal.pgen.1003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dutta R, Yoshida T, Inouye M. 2000. The critical role of the Conserved Thr247 Residue in the functioning of the osmosensor EnvZ, a histidine kinase/phosphatase, in Escherichia coli. J. Biol. Chem. 275:38645–38653 [DOI] [PubMed] [Google Scholar]
- 18. Gutu AD, Wayne KJ, Sham LT, Winkler ME. 2010. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: Dependence of WalKSpn (VicK) phosphatase Activity on its PAS Domain. J. Bacteriol. 192:2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huynh TN, Noriega CE, Stewart V. 2010. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc. Natl. Acad. Sci. U. S. A. 107:21140–21145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huynh TN, Stewart V. 2011. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol. Microbiol. 82:275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Y, Qin L, Yoshida T, Inouye M. 2000. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl. Acad. Sci. U. S. A. 97:7808–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kramer G, Weiss V. 1999. Functional dissection of the transmitter module of the histidine kinase NtrB in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skarphol K, Waukau J, Forst SA. 1997. Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. J. Bacteriol. 179:1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsing W, Russo FD, Bernd KK, Silhavy TJ. 1998. Mutations that alter the kinase and phosphatase activities of the two-component Sensor EnvZ. J. Bacteriol. 180:4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atkinson MR, Ninfa AJ. 1993. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB). J. Bacteriol. 175:7016–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pioszak AA, Ninfa AJ. 2003. Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its Regulation by the PII Signal Transduction Protein. J. Bacteriol. 185:1299–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. 2010. Systematic dissection and Trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling Pathways. PLoS Genet. 6:e1001220. 10.1371/journal.pgen.1001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qin L, Cai S, Zhu Y, Inouye M. 2003. Cysteine-scanning analysis of the dimerization domain of EnvZ, an osmosensing histidine kinase. J. Bacteriol. 185:3429–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casino P, Rubio V, Marina A. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal Transduction. Cell 139:325–336 [DOI] [PubMed] [Google Scholar]
- 30. Gao R, Stock AM. 2013. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc. Natl. Acad. Sci. U. S. A. 110:672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keener J, Kustu S. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. U. S. A. 85:4976–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 34. Podgornaia AI, Laub MT. 2013. Determinants of specificity in two-component signal transduction. Curr. Opin. Microbiol. 16:156–162 [DOI] [PubMed] [Google Scholar]
- 35. Podgornaia AI, Casino P, Marina A, Laub MT. 2013. Structural basis of a rationally Rewired Protein-protein interface critical to bacterial Signaling. Structure 21:1636–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. 2008. Rewiring the specificity of two-component signal transduction Systems. Cell 133:1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol. Microbiol. 70:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siryaporn A, Perchuk BS, Laub MT, Goulian M. 2010. Evolving a robust signal transduction pathway from weak cross-talk. Mol. Syst. Biol. 6:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. 2009. Kinetic buffering of cross talk between bacterial two-component sensors. J. Mol. Biol. 390:380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szurmant H, Hoch JA. 2010. Interaction fidelity in two-component signaling. Curr. Opin. Microbiol. 13:190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schug A, Weigt M, Hoch JA, Onuchic JN, Hwa T, Szurmant H. 2010. Computational modeling of phosphotransfer complexes in two-component signaling. Methods Enzymol. 471:43–58 [DOI] [PubMed] [Google Scholar]
- 42. Lunt B, Szurmant H, Procaccini A, Hoch JA, Hwa T, Weigt M. 2010. Inference of direct residue contacts in two-component signaling. Methods Enzymol. 471:17–41 [DOI] [PubMed] [Google Scholar]
- 43. Capra EJ, Perchuk BS, Skerker JM, Michael LT. 2012. Adaptive mutations that prevent Crosstalk enable the expansion of paralogous signaling protein Families. Cell 150:222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways Regulating growth and cell cycle progression in a Bacterium: A System-level Analysis. PLoS Biol. 3:e334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laub MT, Shapiro L, McAdams HH. 2007. Systems biology of Caulobacter. Annu. Rev. Genet. 41:429–441 [DOI] [PubMed] [Google Scholar]
- 46. Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38:D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stock JB, Ninfa AJ, Stock AM. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho K, Treuner-Lange A, O’Connor KA, Zusman DR. 2000. Developmental aggregation of Myxococcus xanthus requires frgA, an frz-related gene. J. Bacteriol. 182:6614–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun H, Shi W. 2001. Genetic studies of mrp, a Locus Essential for cellular Aggregation and Sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sarwar Z, Garza AG. 2012. The Nla6S protein of Myxococcus xanthus is the prototype for a new family of bacterial histidine kinases. FEMS Microbiol. Lett. 335:86–94 [DOI] [PubMed] [Google Scholar]
- 52. Sarwar Z, Garza AG. 2012. The Nla28S/Nla28 two-component signal transduction system regulates sporulation in Myxococcus xanthus. J. Bacteriol. 194:4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Laub MT, Biondi EG, Skerker JM. 2007. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 423:531-548 [DOI] [PubMed] [Google Scholar]
- 54. Park H, Saha SK, Inouye M. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 95:6728–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component Signal Transduction Systems from Escherichia coli. J. Biol. Chem. 280:1448–1456 [DOI] [PubMed] [Google Scholar]
- 56. Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277:24155–24161 [DOI] [PubMed] [Google Scholar]
- 58. Li J, Swanson RV, Simon MI, Weis RM. 1995. The response Regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34:14626–14636 [DOI] [PubMed] [Google Scholar]
- 59. Sharda S, Koay MS, Kim YJ, Engelhard M, Gärtner W. 2009. A non-hydrolyzable ATP Derivative generates a stable complex in a Light-inducible two-component System. J. Biol. Chem. 284:33999–34004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whitaker WR, Davis SA, Arkin AP, Dueber JE. 2012. Engineering robust control of two-component system phosphotransfer using modular scaffolds. Proc. Natl. Acad. Sci. U. S. A. 109:18090–18095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Capra EJ, Perchuk BS, Ashenberg O, Seid CA, Snow HR, Skerker JM, Laub MT. 2012. Spatial tethering of kinases to their substrates relaxes evolutionary constraints on specificity. Mol. Microbiol. 86:1393–1403 [DOI] [PubMed] [Google Scholar]
- 62. Wanner BL, Latterell P. 1980. Mutants affected in alkaline phosphatase, expression: evidence for Multiple positive Regulators of the phosphate Regulation in Escherchia coli. Genetics 96:353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fisher SL, Kim SK, Wanner BL, Walsh CT. 1996. Kinetic comparison of the specificity of the vancomycin Resistance kinase VanS for two Response Regulators, VanR and PhoB. Biochemistry 35:4732–4740 [DOI] [PubMed] [Google Scholar]
- 64. Noriega CE, Lin HY, Chen LL, Williams SB, Stewart V. 2010. Asymmetric cross-regulation between the nitrate-responsive NarX–NarL and NarQ–NarP two-component regulatory systems from Escherichia coli K-12. Mol. Microbiol. 75:394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Müller S, Willett JW, Bahr SM, Darnell CL, Hummels KR, Dong CK, Vlamakis HC, Kirby JR. 2013. Draft genome sequence of Myxococcus xanthus Wild-Type Strain DZ2, a Model Organism for Predation and Development. Genome Announc. 1:e00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Autophosphorylation of histidine kinases. (A) Incorporation of phosphoryl groups by ten purified HK proteins in the presence of [γ-32P]ATP is shown. Kinase activity was quantified as described in Materials and Methods. Reactions were allowed to proceed for 1, 5, 15, 30, or 60 min. (B) Quantification of autophosphorylation (pmol of phosphorylated HK over time) for each HK~P shown in panel A. Download
Extended phosphotransfer profiling for CrdS. Phosphotransfer profiling for CrdS with each of the 27 NtrC homologs from M. xanthus is shown. CrdS was incubated with all proteins for 10 s (top) or 60 min (bottom). Specific phosphotransfer from CrdS to CrdA is indicated by an asterisk. Nonspecific phosphotransfer to NtrC0172, NtrC1189, and NtrC3555 at 60 min is indicated by the red arrow. Download
T. maritima HK853 phosphotransfer profiling with M. xanthus NtrC homologs. Phosphotransfer profiling demonstrates that HK853 is able to phosphorylate only the cognate RR for HK853, T. maritima RR468 (*). Download
Intrinsic tryptophan fluorescence assay for binding affinity. Representative binding curves for the cognate CrdS-CrdA pair (closed circles; Kd, 2.28 ± 0.66 µM) and noncognate CrdS6-CrdA pair (open circles; Kd, >200 µM) were determined by tryptophan fluorescence. Download
Autokinase activity of the CrdS6 chimera. The autokinase assay demonstrates that the CrdS6 mutant protein is functional and likely retains overall proper structure, as measured by the ability to autophosphorylate. Download
Strains used in this study.
Primers used in this study.