ABSTRACT
We report that establishment and maintenance of the Drosophila melanogaster microbiome depend on ingestion of bacteria. Frequent transfer of flies to sterile food prevented establishment of the microbiome in newly emerged flies and reduced the predominant members, Acetobacter and Lactobacillus spp., by 10- to 1,000-fold in older flies. Flies with a normal microbiome were less susceptible than germfree flies to infection by Serratia marcescens and Pseudomonas aeruginosa. Augmentation of the normal microbiome with higher populations of Lactobacillus plantarum, a Drosophila commensal and probiotic used in humans, further protected the fly from infection. Replenishment represents an unexplored strategy by which animals can sustain a gut microbial community. Moreover, the population behavior and health benefits of L. plantarum resemble features of certain probiotic bacteria administered to humans. As such, L. plantarum in the fly gut may serve as a simple model for dissecting the population dynamics and mode of action of probiotics in animal hosts.
IMPORTANCE
Previous studies have defined the composition of the Drosophila melanogaster microbiome in laboratory and wild-caught flies. Our study advances current knowledge in this field by demonstrating that Drosophila must consume bacteria to establish and maintain its microbiome. This finding suggests that the dominant Drosophila symbionts remain associated with their host because of repeated reintroduction rather than internal growth. Furthermore, our study shows that one member of the microbiome, Lactobacillus plantarum, protects the fly from intestinal pathogens. These results suggest that, although not always present, the microbiota can promote salubrious effects for the host. In sum, our work provides a previously unexplored mechanism of microbiome maintenance and an in vivo model system for investigating the mechanisms of action of probiotic bacteria.
INTRODUCTION
Recent innovations in metagenomics and microbial ecology have sparked an explosion of research on the human microbiome (1, 2). Resident microbes aid in digestion of complex substrates (3, 4) and protect the gut epithelium from damage by pathogenic bacteria (5, 6). Dysbiosis of the human gut microbiome is associated with many chronic diseases, such as diabetes (7, 8), obesity (9–11), colon cancer (12), and depression (13). Although details of the relationship between the microbiome and disease are still emerging, a surge of interest in manipulating the composition of gut communities has generated a need for model systems in which to learn the principles that govern the dynamics and function of microbial communities.
Due to its short rearing time and cultivable microbiota, Drosophila melanogaster presents an ideal model to study dynamics of the microbiota throughout the life span of its host. In spite of these advantages, little is known about the dynamics of the Drosophila microbiota. Previous studies using culture-independent techniques have shown that representations of members of the microbiome differ between younger and older adults (14). Another study found that total bacterial populations increased over the lifetime of the fly (15). However, Drosophila strategies for microbiome maintenance have been overlooked. This process has been well characterized in invertebrates, such as aphids, which maintain an obligate bacterial endosymbiont (16), and in the bobtail squid, which is colonized daily by its symbiont, Vibrio fischeri (17). In humans, it is thought that microbial growth within the intestine is sufficient to maintain many commensal species (18). Another strategy may involve repeated reintroduction of bacteria into the host from an environmental source. Since Drosophila feeds on decomposing foods and ingests polymicrobial mixtures of bacteria, we hypothesized that external sources may serve as a means to repopulate its microbiome.
RESULTS
Microbiome composition and establishment in Drosophila.
Tag pyrosequencing of 16S rRNA genes revealed that Lactobacillus and Acetobacter spp. comprise 94% of the microbiome in our lab colony of Drosophila, which is consistent with a previous description of the microbiome (Fig. 1A) (14). To distinguish Acetobacter and Lactobacillus spp. by colony morphology, Drosophila homogenates were cultured on semiselective and differential media; 4 tan colonies on Acetobacter (Ace) medium and 10 white colonies on de Man, Rogosa, and Sharpe (MRS) medium were identified as Acetobacter and Lactobacillus spp., respectively, by Sanger sequencing of the 16S rRNA gene from each colony. Subsequent analysis of 40 tan and white colonies has consistently discriminated the two genera based on media type and colony morphology (data not shown).
FIG 1 .
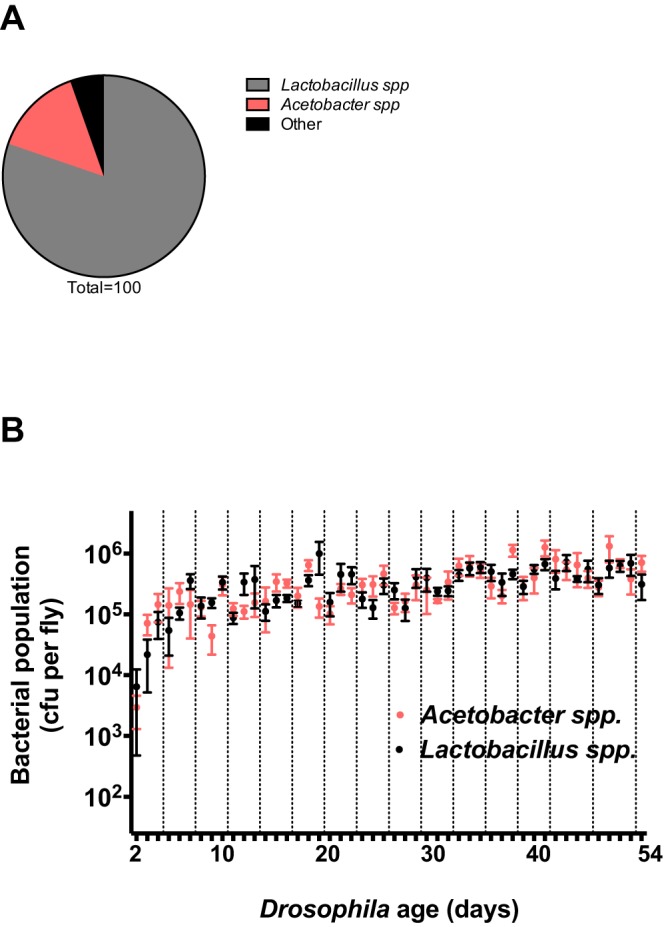
Populations within the adult Drosophila microbiome. (A) One- to 2-day-old surface-sterilized adults were analyzed for bacterial community membership by 454 tag pyrosequencing. The proportions of the community represented by the two most abundant taxa, Lactobacillus and Acetobacter, which represent 94% of the community, are shown. (B) Two- to 54-day-old adult Drosophila were sampled daily (points at 43, 45, 50, and 51 days were omitted). Five adult females were sampled at most time points (with three samples on days 14, 21, 25, and 44 and four samples on days 16, 17, 19, 20, 26, 28, and 46). Values represent mean bacterial population per fly ± the standard error of the mean as estimated by culturing. The limit of detection is 25 CFU per fly. Dashed lines represent transfer of flies to sterile vials.
We sought to measure the dynamics of the dominant bacterial members of the Drosophila microbiome. Acetobacter and Lactobacillus spp. were assessed in newly emerged flies and the environment in which they emerged. Although they emerged from pupae into an environment laden with bacteria, 9 out of 10 adult flies did not contain detectable, culturable bacteria 1 h after emergence from the pupal case as adults (see Fig. S1A and B in the supplemental material). This observation is consistent with a previous report (14). Within 24 h of eclosion, 6 out of 10 flies contained detectable bacteria (see Fig. S1B). Collectively, our results show that newly emerged flies harbor low microbial populations, suggesting that the members of the microbiome of adult Drosophila come from their environment.
The abundance of bacteria in the Drosophila microbiome increased over the lifetime of the fly, reflecting the amount of time that the flies spent on the same food source (Fig. 1B). Throughout the 54-day time course, flies were maintained on the same food source for 3 days and then transferred to fresh food. Populations of the bacterial members declined when flies were transferred to new food and peaked when Drosophila were maintained on the same food source for 2 to 3 days. We propose that upon transfer, flies seed the sterile food with members of the microbiome that grow to high abundance and repopulate the microbiome when consumed by the fly.
Fly food as a bacterial reservoir for the Drosophila microbiome.
To test whether fly food itself supports the growth of microbiome members, we measured the population of bacteria on fly food over time. As flies remained on food, the bacterial population increased, peaking at 72 h (see Fig. S2 in the supplemental material). These data demonstrate that Drosophila inoculate their food with bacteria that then multiply on the food.
Establishment of the Drosophila microbiome.
To assess the time scale of microbiota acquisition in Drosophila, we transferred newly emerged flies to fresh food daily, every 3rd day, or not at all for 7 days, evaluating bacterial populations daily. Flies that were not transferred harbored larger bacterial populations than those that were transferred (Fig. 2). The dynamics of these populations were similar in males and females, and bacterial population size corresponded to the time spent on the same food source (see Fig. S3 and S4 in the supplemental material). These results collectively support the idea that establishment of the Drosophila microbiome requires access to and consumption of exogenous bacteria.
FIG 2 .
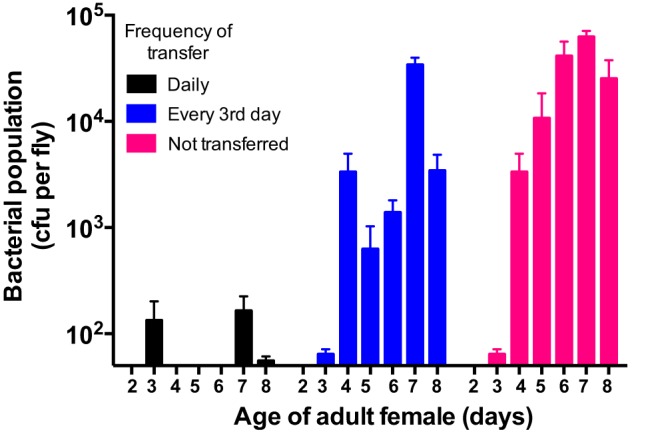
Establishment of the Drosophila microbiome requires frequent consumption of bacteria. Cultured populations of Acetobacter and Lactobacillus spp. in newly emerged Drosophila subjected to various transfer regimens were measured. (B) Following eclosion, flies were transferred to fresh food daily (black), every 3rd day (blue), or not at all (pink) for 7 days. A total of nine females obtained from three vials (three/vial) were sampled daily throughout the 7-day period. The height of each bar represents the mean bacterial population (Lactobacillus and Acetobacter spp.) in flies sampled that day ± the standard error of the mean.
Maintenance of the Drosophila microbiome.
We measured microbial populations in 16-day-old conventionally reared flies that were transferred twice daily to either sterilized food or food inoculated with Lactobacillus plantarum and Acetobacter pasteurianus. Flies feeding on the amended medium contained more bacteria than those feeding on sterilized food, although populations in the flies on sterilized food were not completely eliminated and varied in size (Fig. 3). These results indicate that bacterial populations in the Drosophila microbiome are influenced by fly access to exogenous bacteria. Without repeated consumption of bacteria, established microbiota populations decline and are much smaller than those achieved when Drosophila has access to an environmental reservoir of bacteria. Reduction of bacterial population size is detectable within 6 h of transfer to fresh food or under starvation conditions (see Fig. S5 in the supplemental material), although bacterial populations in starved flies were larger than those in flies transferred to sterile food. This is consistent with the observation that starvation reduces the rate of defecation by Drosophila, thereby slowing the loss of bacteria from the gut (19).
FIG 3 .
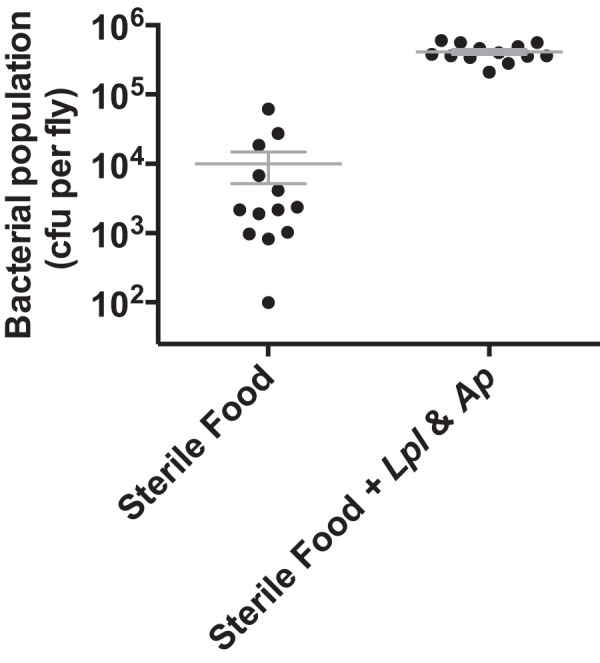
Maintenance of the Drosophila microbiome requires frequent consumption of bacteria. Cultured populations of Acetobacter and Lactobacillus spp. in conventionally reared flies introduced into environments with or without exogenous bacteria. Sixteen-day-old flies were transferred twice daily to sterile food or sterile food amended with A. pasteurianus and L. plantarum. Three to five female Drosophila were sampled every 3rd day for 9 days after transfer to either environment. Data points represent the mean total bacterial population (Lactobacillus and Acetobacter spp.) ± the standard error of the mean in flies sampled on days 3, 6, and 9. An unpaired two-tailed t test revealed significant differences (P < 0.0001) between bacterial populations under the two rearing regimens.
Impact of the community on fly fitness.
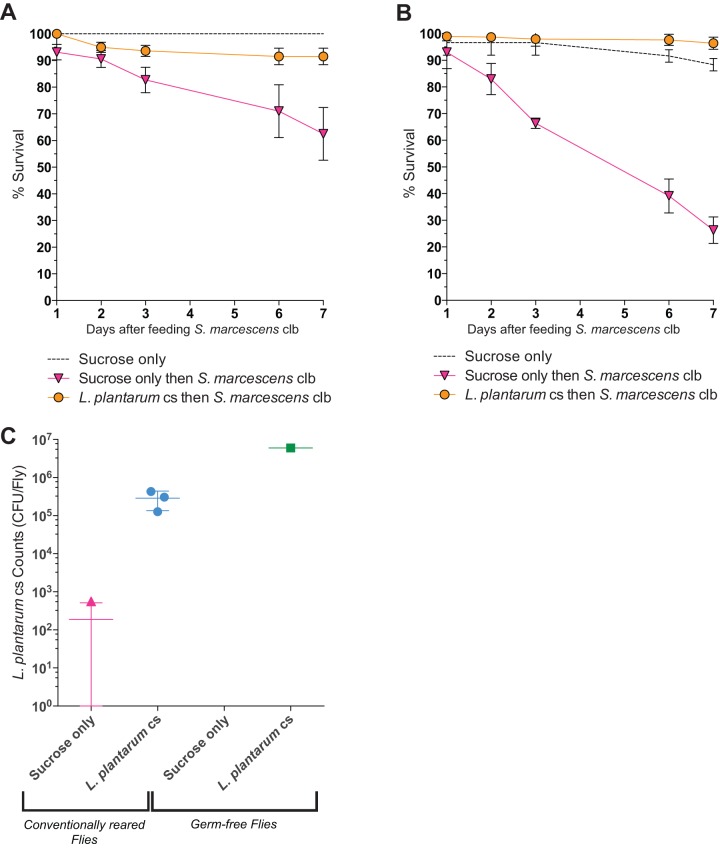
To test whether the abundance of organisms in the Drosophila microbiome has functional consequences for host fitness, we fed L. plantarum to conventionally reared and germfree flies and then challenged them with Serratia marcescens, a Drosophila and nosocomial human pathogen, and assessed fly mortality. L. plantarum, a member of the native gut microbiome, protected flies from mortality induced by S. marcescens (Fig. 4A). Feeding L. plantarum reduced mortality more in germfree than in conventionally reared flies (Fig. 4B). Overall, the level of protection corresponded to the size of the L. plantarum population detected in the fly (Fig. 4C).
FIG 4 .
Lactobacillus plantarum protects flies from Serratia marcescens infection. (A and B) Conventionally reared (A) or germfree (B) flies were fed L. plantarum for 3 days prior to S. marcescens challenge. Flies were fed a sucrose suspension of S. marcescens for 2 days and then transferred to clean bottles with sterile sucrose solution. Fly mortality was recorded over time. n was 20 flies per treatment. Error bars represent the standard deviation of the mean for three replicate groups per treatment. (C) Cultured populations of L. plantarum in conventionally reared and germfree flies were measured 3 days after feeding. L. plantarum cs is an isolate from Canton-S Drosophila in our laboratory; S. marcescens clb is an isolate from cottonwood leaf beetle.
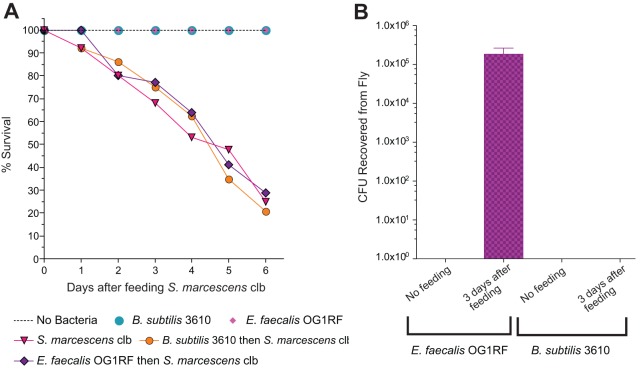
To assess the specificity of protection, conventionally reared flies were fed Enterococcus faecalis or Bacillus subtilis 3 days prior to challenge with S. marcescens.
Although E. faecalis was recovered from flies 3 days after feeding, B. subtilis was not. Neither E. faecalis nor B. subtilis reduced S. marcescens-induced mortality, demonstrating that protection by L. plantarum is specific and is not achieved with populations of all bacterial species (Fig. 5A). These data show that a prominent member of the Drosophila microbiome protects its host from intestinal infection.
FIG 5 .
Enterococcus faecalis and Bacillus subtilis do not protect flies from Serratia marcescens infection. (A) Conventionally reared flies were fed E. faecalis or B. subtilis for 3 days prior to S. marcescens challenge. Flies were fed a sucrose solution containing S. marcescens for 2 days and then transferred to clean bottles with sterile sucrose solution. Fly mortality was recorded over time. n was 20 flies per treatment. (B) Conventionally reared flies were fed E. faecalis, B. subtilis, or sterile sucrose solution (“no feeding”) for 3 days. Cultured populations of E. faecalis or B. subtilis were measured 3 days after feeding. n was 20 per treatment; data from one representative of three experiments are shown. Error bars represent the standard deviation of the mean of five flies.
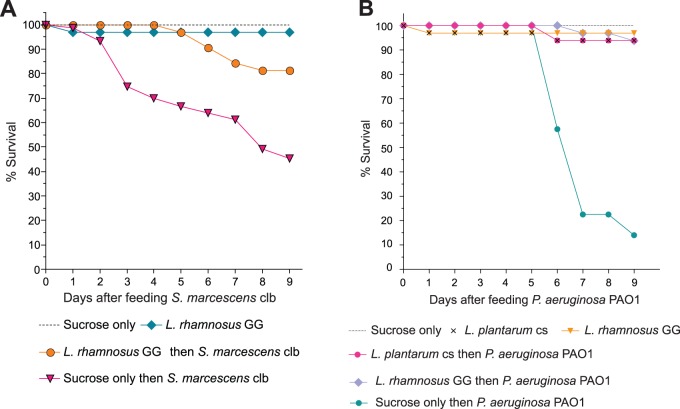
Many members of the Lactobacillus genus besides L. plantarum are used as human probiotics—formulations of live bacteria ingested for their health benefits. Therefore, we investigated whether other probiotic strains could be studied in Drosophila. The commonly used human probiotic Lactobacillus rhamnosus GG protected conventionally reared flies from infection by S. marcescens and Pseudomonas aeruginosa PAO1, another nosocomial pathogen of humans (Fig. 6A and B). These results suggest that Drosophila may serve as a model system for studying human probiotics.
FIG 6 .
The probiotic fly model extends to other probiotics and pathogens. Lactobacillus plantarum and L. rhamnosus GG reduce fly death from Serratia marcescens and Pseudomonas aeruginosa when fed prior to infection. (A) Conventionally reared flies were fed L. rhamnosus GG for 3 days prior to S. marcescens challenge. Flies were fed a sucrose suspension of S. marcescens for 2 days and then transferred to clean bottles with sterile sucrose solution. Fly mortality was recorded over time. n was 20 per treatment; one representative experiment of three is shown. (B) Conventionally reared flies were fed L. plantarum or L. rhamnosus GG for 3 days prior to P. aeruginosa challenge. Flies were fed a sucrose suspension of P. aeruginosa for the duration of the assay. Fly mortality was recorded over time. n was 20 per treatment; data from one representative of three experiments are shown.
DISCUSSION
Our results show that Drosophila establishes and maintains its microbiome by frequently consuming bacteria, highlighting the contribution of external inputs, rather than internal maintenance, to sustaining the microbiome and revealing a novel facet of host-microbe interactions in this model system. We demonstrated that the lifetime abundance of the Drosophila microbiota was associated with the abundance of environmental microbes and that the microbiome was not established in newly emerged adults or maintained in flies with existing microbial populations when flies were deprived of exogenous bacteria. The influence of rearing regimen could extend to microbiome composition, as members may or may not be transferred to fresh food sources, perhaps revealing the cause of the variation in composition of the Drosophila microbiome among research labs (20). In light of these findings, we predict that both abundance and composition of Drosophila gut communities vary with differences in Drosophila husbandry techniques in various laboratory settings.
This work will advance several lines of inquiry about the Drosophila microbiome. First, the results raise questions about the mechanisms by which Drosophila and its symbionts come to be associated with one another. We speculate that innate or learned behaviors may enable the fly to replenish its microbiome. Recent work has highlighted an olfactory circuit used by Drosophila to avoid harmful microorganisms (21). Future work may identify an analogous circuit dedicated to detecting beneficial microorganisms. In this way, Drosophila would be attracted to and preferentially consume specific bacterial species, enabling it to maintain a beneficial microbiome. Though the population sizes of many host-associated microbial communities are dictated by host immunity and bacterial growth rates, the Drosophila system may represent an alternative mutualism strategy that we term “quotidian replenishment,” which is intended to indicate the need for daily replenishment to obtain a consistent community in the animal. In this model, the symbiotic community in Drosophila is maintained through frequent ingestion from an external reservoir of bacteria, as suggested by Storelli and colleagues (22).
Perhaps Drosophila rids its gut of most microbes to minimize the risk of acquiring and maintaining potential pathogens. Drosophila undoubtedly encounters both beneficial and pathogenic microbes in its natural environment, and if the fly provided an environment for a more permanent, actively growing microbial community, then it might also be more vulnerable to colonization by pathogens as well. Another possibility is that the microbiome may compete with its Drosophila host for nutrients in the gut. If so, we would predict that maintaining a bacterial population internally would present a cost to the host by reducing nutrients available to it. Further study of the relationship between the fly and its microbiome may reveal novel features of Drosophila mutualism and general principles of host-microbe relationships.
Our evidence indicates that Drosophila seeds its food with commensal organisms by depositing fecal matter on its food source on which the bacteria grow. In this way, Drosophila could be cultivating an inoculum with which to replenish the microbiome. A similar system is the symbiosis strategy employed by the Acromyrmex leaf-cutting ant, which cultivates fungal mats that provide the insect with a rich food source (23). Like the leaf-cutter ant, Drosophila may digest its resident bacteria as a food source. Regardless of other similarities and differences between these biological systems, they both may represent examples of microbial farming by which animals cultivate beneficial microorganisms (24).
The discovery that both Drosophila-associated and human-administered probiotic strains protect Drosophila from infection provides a foundation for the use of the Drosophila system to study probiotic strains in a host that can be genetically altered and manipulated experimentally. Variation in human disease susceptibility and responses to treatment may be in part a consequence of variation between individuals’ microbiomes. As such, managing the microbiome is an essential component of treatments intended to alter the host microbiome, including probiotics. Consumption of probiotics can alleviate symptoms of antibiotic-associated diarrhea (25), lactose intolerance (26), and childhood irritable bowel syndrome (27). However, the complexity of the human gut microbiome, the cost of clinical trials, and the limits of appropriate experimental procedures in human subjects have precluded elucidation of the in vivo mechanisms leading to these health benefits. Such understanding is needed to address the inconsistent performance of probiotics (28–30) and to direct their use in a targeted and precise manner.
The basis of probiotic action and failure will be advanced by studying a host model harboring a relatively simple microbial community, such as Drosophila melanogaster. The fly is a particularly attractive model in which to study probiotics because L. plantarum, a species formulated as a probiotic for humans, is a prominent symbiont of wild and laboratory-reared flies (14, 31–36). Drosophila and humans may share a mode of interaction with Lactobacillus: both hosts benefit from certain Lactobacillus strains, but the bacteria do not persist in either host, thereby necessitating quotidian replenishment in Drosophila (37). The lack of persistent colonization by probiotic lactobacilli has produced skepticism about their health-promoting effects on humans, but it may represent a common feature of animal-host symbioses. The Drosophila model provides a system that overcomes many of the experimental challenges of studying such interactions in the human gut, providing a path to understanding probiotics as well as diverse host-microbe interactions.
MATERIALS AND METHODS
Fly stocks and culture.
Drosophila melanogaster Canton-S flies were reared at 25°C on medium containing 10% dextrose, 5% heat-killed yeast, 7% cornmeal, 0.6% propionic acid, and 0.6% agar. No microorganisms could be isolated from this sterile food. Flies raised in this manner are described as conventionally reared. A germfree Canton-S line was created by washing <12-h-old embryos successively in ethanol, 50% bleach, and ethanol followed by transfer of embryos to sterile medium in a sterile biosafety cabinet as described previously (34). Germfree status was checked by plating adult flies on MRS, Ace, and nutrient agar plates and sequencing the 16S rRNA gene (see Fig. S6A and B in the supplemental material). To maintain their sterility, germfree flies were transferred to autoclaved food within a sterilized biosafety cabinet.
Culture-independent identification of bacteria in Drosophila.
DNA isolated from surface-sterilized flies (1 to 2 days old) was used to survey the Drosophila microbiome. Amplifications were performed in 25-µl reaction mixtures with Qiagen HotStar Taq master mix (Qiagen Inc., Valencia, CA), 1 µl of each 5 µM primer, and 1 µl of template. Reactions were performed on ABI Veriti thermocyclers (Applied Biosystems, Carlsbad, CA) using the following conditions: 95°C for 5 min and then 35 cycles of 94°C for 30 s, 54°C for 40 s, and 72°C for 1 min, followed by one cycle of 72°C for 10 min and a 4°C hold. Amplification products were visualized with eGels (Life Technologies, Grand Island, NY). Products were then pooled at equimolar ratios, and each pool was cleaned with Diffinity RapidTip (Diffinity Genomics, West Henrietta, NY) and selected by size using Agencourt AMPure XP (Beckman, Coulter, Indianapolis, IN) according to Roche 454 protocols (454 Life Sciences, Branford, CT). Size-selected pools were then quantified, and 150 ng of DNA was hybridized to Dynabeads M-270 (Life Technologies, Carlsbad, CA) to create single-stranded DNA according to Roche 454 protocols (454 Life Sciences). Single-stranded DNA was diluted and used in emulsion PCRs (emPCRs) which were performed, and the reaction mixtures were subsequently enriched. Sequencing followed the manufacturer’s protocols (454 Life Sciences).
Culture-dependent identification of bacteria in Drosophila.
Flies were washed with 10% bleach, 70% ethanol, and phosphate-buffered saline (PBS) in succession. Homogenates from surface-sterilized flies were cultured on de Man, Rogosa, and Sharpe (MRS) agar (Fisher Scientific, Hampton, NH), nutrient agar (Becton Dickinson, Franklin Lakes, NJ), and Ace agar (0.8% yeast extract, 1.5% peptone, 1% dextrose, 0.3% acetic acid, 0.5% ethanol, and 0.01% cycloheximide) at 28°C.
To identify Lactobacillus and Acetobacter spp. in the fly microbiome, colonies with distinct morphologies were selected from each medium type and placed in a 25-µl PCR mix containing HotStar HiFidelity DNA polymerase (Qiagen), water, MgSO4, glycerol, and deoxynucleoside triphosphates (dNTPs) and amplified for 35 cycles in a thermocycler. 16S rRNA amplicons were cleaned using a QIAquick PCR purification kit (Qiagen), visualized by gel electrophoresis, gel purified using a QIAex II gel extraction kit (Qiagen), and sequenced using 8F and 1492R primers. Sequences were aligned using the Ribosomal Database Project. The resulting community profile was consistent with previous studies (14, 33).
Estimation of bacterial population size in Drosophila.
Adult flies were collected within 24 h of emergence and placed in vials at a density of 30 flies per vial; these were designated 1-day-old flies. Flies were transferred to fresh food every 3rd day for the duration of the experiment. Homogenates of surface-sterilized flies were cultured on MRS, nutrient, and Ace agar using a Spiral Plating System Autoplater (Advanced Instruments Inc., Norwood, MA). Nutrient agar was used to monitor the growth of the culturable bacterial community. Plates were incubated at 28°C for 2 to 4 days, and bacterial CFU were estimated using a QCount automated colony counter (Advanced Instruments Inc., Norwood, MA) or manually.
Establishment and maintenance of the Drosophila microbiome.
To assess establishment of the Drosophila microbiome, flies were collected within 24 h of eclosion, divided into three groups, and transferred to fresh fly food. One group remained on the fly food for the duration of the experiment, the second group was transferred every 3rd day, and the third group was transferred daily. Bacterial populations in three males and three females from each of three vials in each experimental group were sampled.
To assess maintenance of the Drosophila microbiome, 16-day-old flies were transferred twice daily either to fresh food or to fresh food amended with Lactobacillus plantarum and Acetobacter pasteurianus. L. plantarum was cultured for 24 h at 28°C, and A. pasteurianus was cultured in broth for 48 h at 28°C with shaking at 200 rpm. Five to nine females from each experimental treatment were sampled every 3 days for 9 days.
Probiotic feeding and pathogen infection assays.
L. plantarum cs, an isolate from Canton-S Drosophila in our laboratory, and Lactobacillus rhamnosus GG were cultured overnight in MRS medium at 37°C. Serratia marcescens clb, an isolate from cottonwood leaf beetle; Pseudomonas aeruginosa PAO1; and Bacillus subtilis 3610 were cultured overnight in LB at 37°C with shaking at 225 rpm. Enterococcus faecalis OG1RF was cultured in brain heart infusion medium (BHI) (VWR International) at 37°C with shaking at 225 rpm.
One-day-old flies were added to a glass bottle and fed a 5% sucrose suspension of either S. marcescens or P. aeruginosa on a sterile paper disc. In S. marcescens killing assays, flies were removed from the bacterium-sucrose suspension after 3 days and placed in new bottles with sterile sucrose solution. In P. aeruginosa killing assays, flies were reared on the bacterium-sucrose mixture for the duration of the experiment. Mortality was assessed daily.
To determine whether bacteria protected Drosophila from infection, 1-day-old flies were fed a sucrose solution inoculated with B. subtilis, E. faecalis, or Lactobacillus spp. for 3 days and then fed either S. marcescens or P. aeruginosa. When fed S. marcescens, the flies remained on the bacterial suspension for 2 days and then were transferred to bottles with sterile sucrose solution; when fed P. aeruginosa, the flies were transferred every 3 days.
Statistical analysis.
All data analysis was performed using GraphPad Prism 6.0b software.
SUPPLEMENTAL MATERIAL
Newly emerged adults harbor low bacterial populations despite emerging into an environment laden with bacteria. (A) Food into which Drosophila adults emerged from the pupal case was sampled. Each column represents the mean of 15 food samples taken from five separate vials ± the standard error of the mean. (B) Ten flies were sampled at 1, 4, and 24 h after emergence from the pupal case. Data points are represented as the mean of individual flies ± the standard error of the mean. MRS and Ace media captured Lactobacillus and Acetobacter populations, respectively, and nutrient agar medium was used as a general bacterial growth medium. The vials used for food and fly sampling were the same. Download
Populations of Acetobacter spp. and Lactobacillus spp. on food of Drosophila adults increased with time that flies spent on the food. Food was sampled on which Drosophila remained for 15, 24, 48, 72, and 144 h. Horizontal lines represent the mean bacterial population of three to nine replicates from separate vials, and the vertical bars represent the standard errors of the means. The limit of detection is 500 CFU per g of fly food. Download
Establishment of the Drosophila microbiome in males requires frequent consumption of bacteria. Following eclosion, flies were transferred to fresh food daily, every 3rd day, or not at all for 7 days. Nine males from three separate vials were sampled daily throughout the 7-day period. The height of each bar represents the mean bacterial population in flies sampled that day ± the standard error of the mean. Bacterial populations from flies transferred to sterile food daily are shown in black, those from flies transferred to sterile food every 3rd day are shown in blue, and those from flies that were not transferred for the duration of the experiment are shown in pink. Download
Populations of Acetobacter and Lactobacillus spp. on food of Drosophila increase with time that flies spend on the food. Following eclosion, flies were transferred to fresh food daily (black), every 3rd day (blue), or not at all for 7 days (pink). Food from three separate vials was sampled daily throughout the 7-day period. The height of each bar represents the mean bacterial population of food sampled that day ± the standard error of the mean. Download
Transfer to fresh food or starvation reduces populations in the Drosophila microbiome. Cultured populations of Drosophila microbiome members were sampled from flies before and following transfer to fresh food or to a vial containing sterile water (starvation condition). Data points represent the mean bacterial population of seven female flies ± the standard error of the mean. Download
Assessment of germfree Drosophila. (A) The left side of the photograph shows MRS, Ace, and nutrient agar plates (left to right), which have no visible colonies after plating of a PBS suspension of 3 germfree flies; the right side of the photograph shows positive control (3 conventionally reared flies) cultured on the same media with robust bacterial populations. (B) Gel photograph showing PCR products using the 16S primers 8F-1492R and 357F-907R. The top half of the gel shows products using the 8F-1492R primer pair. Lanes 1, 5, 9, and 13 are marker lanes. Lanes 2 to 4 are biological replicates of germfree fly DNA as input. Lanes 6 to 8 are DNA extraction negative controls. Lanes 10 to 12 have conventionally reared fly DNA as input and show a strong band at ~1.5 kb. Lanes 14 to 16 are negative controls (PCR reagents). The bottom half of the gel has the same lane assignments as the top half but uses the 357–907-R primer pair. Samples from the germfree flies have a band at ~750 bp, whereas the conventionally reared flies have the same band but also a 540-bp band as predicted from successful amplification of the bacterial DNA using the primer pair. Download
ACKNOWLEDGMENTS
We thank Kenneth Raffa (University of Wisconsin—Madison), Gary Dunny (University of Minnesota), and Barbara Iglewski (University of Rochester) for Serratia marcescens clb, Enterococcus faecalis OG1RF, and Pseudomonas aeruginosa, respectively. Lactobacillus rhamnosus LGG ATCC 53103 was provided by Katie Mason (University of Michigan), and the Canton-S Drosophila strain was provided by John Carlson (Yale University). We thank Jesse Morin (Yale University) for comments on the manuscript.
This work was supported by National Institutes of Health grants NIDDK RC1DK086831-0, RO1GM099563, 2T32GM007499-36, and 5T32HG003198-10.
Footnotes
Citation Blum JE, Fischer CN, Miles J, Handelsman J. 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4(6):e00860-13. doi:10.1128/mBio.00860-13
REFERENCES
- 1. Morgan XC, Segata N, Huttenhower C. 2013. Biodiversity and functional genomics in the human microbiome. Trends Genet. 29:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Microbiome Human, Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venema K. 2010. Role of gut microbiome in the control of energy and carbohydrate metabolism. Curr. Opin. Clin. Nutr. Metab. Care 13:432–438 [DOI] [PubMed] [Google Scholar]
- 4. Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283–307 [DOI] [PubMed] [Google Scholar]
- 5. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 7. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. 2011. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 6:e25792. 10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. 2010. Gut microbiome in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085. 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 10. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fei N, Zhao L. 2013. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7:880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rowland IR. 2009. The role of the gastrointestinal microbiota in colorectal cancer. Curr. Pharm. Des. 15:1524–1527 [DOI] [PubMed] [Google Scholar]
- 13. Maes M, Kubera M, Leunis JC, Berk M. 2012. Increased IgA and IgM responses against gut commensals in chronic depression: further evidence for increased bacterial translocation or leaky gut. J. Affect. Disord. 141:55–62 [DOI] [PubMed] [Google Scholar]
- 14. Wong CN, Ng P, Douglas AE. 2011. Low diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13:1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren C, Webster P, Finkel SE, Tower J. 2007. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6:144–152 [DOI] [PubMed] [Google Scholar]
- 16. Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465 [DOI] [PubMed] [Google Scholar]
- 17. McFall-Ngai MJ, Ruby EG. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1494 [DOI] [PubMed] [Google Scholar]
- 18. Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. 2009. The gut microbiota in inflammatory bowel disease. Curr. Pharm. Des. 15:1528–1536 [DOI] [PubMed] [Google Scholar]
- 19. Cognigni P, Bailey AP, Miguel-Aliaga I. 2011. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13:92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7:1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stensmyr MC, Dweck HK, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, Wicher D, Sachse S, Knaden M, Becher PG, Seki Y, Hansson BS. 2012. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151:1345–1357 [DOI] [PubMed] [Google Scholar]
- 22. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14:403–414 [DOI] [PubMed] [Google Scholar]
- 23. Zhang MM, Poulsen M, Currie CR. 2007. Symbiont recognition of mutualistic bacteria by Acromyrmex leaf-cutting ants. ISME J. 1:313–320 [DOI] [PubMed] [Google Scholar]
- 24. Stallforth P, Brock DA, Cantley AM, Tian X, Queller DC, Strassmann JE, Clardy J. 2013. A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. Proc. Natl. Acad. Sci. U. S. A. 110:14528–14533 DOI:10.1073/pnas.1308199110. PubMed. [DOI] [PMC free article] [PubMed]
- 25. Szajewska H, Ruszczynski M, Radzikowski A. 2006. Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J. Pediatr. 149:367–372 [DOI] [PubMed] [Google Scholar]
- 26. Pelletier X, Laure-Boussuge S, Donazzolo Y. 2001. Hydrogen excretion upon ingestion of dairy products in lactose-intolerant male subjects: importance of the live flora. Eur. J. Clin. Nutr. 55:509–512 [DOI] [PubMed] [Google Scholar]
- 27. Gawronska A, Dziechciarz P, Horvath A, Szajewska H. 2007. A randomized double-blind placebo-controlled trial of Lactobacillus GG for abdominal pain disorders in children. Aliment. Pharmacol. Ther. 25:177–184 [DOI] [PubMed] [Google Scholar]
- 28. Rijkers GT, Bengmark S, Enck P, Haller D, Herz U, Kalliomaki M, Kudo S, Lenoir-Wijnkoop I, Mercenier A, Myllyluoma E, Rabot S, Rafter J, Szajewska H, Watzl B, Wells J, Wolvers D, Antoine JM. 2010. Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J. Nutr. 140:671S–676S. 10.3945/jn.109.113779 [DOI] [PubMed] [Google Scholar]
- 29. Boyle RJ, Bath-Hextall FJ, Leonardi-Bee J, Murrell DF, Tang ML. 2009. Probiotics for the treatment of eczema: a systematic review. Clin. Exp. Allergy 39:1117–1127 [DOI] [PubMed] [Google Scholar]
- 30. Doherty GA, Bennett GC, Cheifetz AS, Moss AC. 2010. Meta-analysis: targeting the intestinal microbiome in prophylaxis for post-operative Crohn’s disease. Aliment. Pharmacol. Ther. 31:802–809 [DOI] [PubMed] [Google Scholar]
- 31. Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DE. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73:3470–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7(9):e1002272. 10.1371/journal.pgen.1002272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75:1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. 2008. Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science 319:777–782 [DOI] [PubMed] [Google Scholar]
- 35. Bakula M. 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Invertebr. Pathol. 14:365–374 [DOI] [PubMed] [Google Scholar]
- 36. Staubach F, Baines JF, Kuenzel S, Bik EM, Petrov DA. 2013. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural Environment. PLoS One 8:e70749. 10.1371/journal.pone.0070749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3:106ra106. 10.1126/scitranslmed.3002701 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Newly emerged adults harbor low bacterial populations despite emerging into an environment laden with bacteria. (A) Food into which Drosophila adults emerged from the pupal case was sampled. Each column represents the mean of 15 food samples taken from five separate vials ± the standard error of the mean. (B) Ten flies were sampled at 1, 4, and 24 h after emergence from the pupal case. Data points are represented as the mean of individual flies ± the standard error of the mean. MRS and Ace media captured Lactobacillus and Acetobacter populations, respectively, and nutrient agar medium was used as a general bacterial growth medium. The vials used for food and fly sampling were the same. Download
Populations of Acetobacter spp. and Lactobacillus spp. on food of Drosophila adults increased with time that flies spent on the food. Food was sampled on which Drosophila remained for 15, 24, 48, 72, and 144 h. Horizontal lines represent the mean bacterial population of three to nine replicates from separate vials, and the vertical bars represent the standard errors of the means. The limit of detection is 500 CFU per g of fly food. Download
Establishment of the Drosophila microbiome in males requires frequent consumption of bacteria. Following eclosion, flies were transferred to fresh food daily, every 3rd day, or not at all for 7 days. Nine males from three separate vials were sampled daily throughout the 7-day period. The height of each bar represents the mean bacterial population in flies sampled that day ± the standard error of the mean. Bacterial populations from flies transferred to sterile food daily are shown in black, those from flies transferred to sterile food every 3rd day are shown in blue, and those from flies that were not transferred for the duration of the experiment are shown in pink. Download
Populations of Acetobacter and Lactobacillus spp. on food of Drosophila increase with time that flies spend on the food. Following eclosion, flies were transferred to fresh food daily (black), every 3rd day (blue), or not at all for 7 days (pink). Food from three separate vials was sampled daily throughout the 7-day period. The height of each bar represents the mean bacterial population of food sampled that day ± the standard error of the mean. Download
Transfer to fresh food or starvation reduces populations in the Drosophila microbiome. Cultured populations of Drosophila microbiome members were sampled from flies before and following transfer to fresh food or to a vial containing sterile water (starvation condition). Data points represent the mean bacterial population of seven female flies ± the standard error of the mean. Download
Assessment of germfree Drosophila. (A) The left side of the photograph shows MRS, Ace, and nutrient agar plates (left to right), which have no visible colonies after plating of a PBS suspension of 3 germfree flies; the right side of the photograph shows positive control (3 conventionally reared flies) cultured on the same media with robust bacterial populations. (B) Gel photograph showing PCR products using the 16S primers 8F-1492R and 357F-907R. The top half of the gel shows products using the 8F-1492R primer pair. Lanes 1, 5, 9, and 13 are marker lanes. Lanes 2 to 4 are biological replicates of germfree fly DNA as input. Lanes 6 to 8 are DNA extraction negative controls. Lanes 10 to 12 have conventionally reared fly DNA as input and show a strong band at ~1.5 kb. Lanes 14 to 16 are negative controls (PCR reagents). The bottom half of the gel has the same lane assignments as the top half but uses the 357–907-R primer pair. Samples from the germfree flies have a band at ~750 bp, whereas the conventionally reared flies have the same band but also a 540-bp band as predicted from successful amplification of the bacterial DNA using the primer pair. Download