Abstract
Brain abnormalities such as white matter lesions (WMLs) are not only linked to cerebrovascular disease, but also with normal aging, diabetes and other conditions increasing the risk for cerebrovascular pathologies. Obtaining quantitative measures which assesses the degree or probability of WML in patients is important for evaluating disease burden, and for evaluating its progression and response to interventions. In this paper, we introduce a novel approach for detecting the presence of WMLs in periventricular areas of the brain using manifold-constrained embeddings. The proposed method uses locally linear embedding (LLE) to create “normality” distributions in 12 locations of the brain where deviations from the manifolds are estimated by calculating geodesic distances along locally linear planes in the embedding. A smooth mapping function approximating the relationship between ambient and manifold spaces as a joint distribution maps unseen test images in the intrinsic space. We create a set of low-dimensional embeddings from 876 patches of healthy tissue in 73 subjects and test it on 396 patches imaging both WML and healthy areas in 33 subjects with diabetes. Experiments highlight the need of nonlinear techniques to learn the studied data with detection rates over 85% in true-positives, and the relevance of the computed distance for comparing individuals to a specific pathological pattern.
Index Terms: Manifold embedding, locally linear embedding, white matter lesions, brain MRI, diabetes
1. INTRODUCTION
The prevalence of cerebral white matter lesions (WMLs) has been extensively studied for the past decade in the elderly population, patients with cardiovascular risk factors or from patients with central nervous system disorders, including cerebrovascular disease and dementia. With such an increased interest towards the development of improved diagnosis and prognosis possibilities for patients with cardiovascular symptoms, specific biologic markers linking the extent of WML to the severity of the the pathology have prevailed enormous promise. In this respect, the use of an automated detection and segmentation method for WMLs exhibiting high sensitivity and specificity, which are demonstrated in a quantitative and objective fashion, could be advantageous to confidently distinguish between healthy and diseased tissue.
Towards this end, precise measurement of such pathology from magnetic resonance imaging (MRI), and more importantly measuring the evolution of a pathology over time, becomes a crucial aspect for detecting and monitoring the disease, as well as for evaluating the risk of progression. While a number of protocols are being researched to best visualize these lesions, WML are shown as hyper-intense regions in fluid-attenuated inversion recovery (FLAIR) T2 images. Consequently, clarity of the lesions located in periventricular areas is increased in FLAIR images compared with other images.
In the literature, a wealth of methods have been used to segment multiple sclerosis lesions from FLAIR images using support vector machine classifier, fuzzy inference or Bayesian classification [1]. However, few have aimed to demonstrate the relevance of describing each pathological pattern as a deviation from healthy brain images along a manifold structure, allowing the computation of an appropriate distance between individuals and each pathological pattern. The definition of an optimal space for the comparison of different populations was already addressed by dimensionality reduction techniques, such as principal component analysis (PCA), Kernel-PCA or linear discriminant analysis [2]. The main limitation of the above mentioned methods lies on the assumption the high dimensionality and complex data falls onto a linear distribution. This is not necessarily suited for the non-linear underlying structure of images of the brain cortex and where the local similar topologies variations are not particulary taken into account. Manifold learning techniques intrinsically take under consideration the inherent geometry representation, and allow relevant comparison of individuals to the studied population through a low-dimensional Euclidean space map. Manifold representation has been applied for segmentation, physiological testing [3] or respiratory compensation [4]. In [5], the authors assessed morphological brain variations by calculating distances in manifold space based on metrics related to reconstruction errors, but did not treat the problem of divergence from normality.
In this work, we propose a novel framework which extends manifold learning techniques to help to discriminate brain lesions from various normal tissue image profiles established as patterns of normal appearances in healthy patients (Fig. 1). These multiple nonlinear embeddings obtained from a set of training images are used to create “normality” distributions where the distances of new mapped images are considered as “deviations” from healthy areas. Towards this end, we present a smooth mapping function estimating the relationship between ambient and manifold spaces as a joint distribution to map unseen test images. A collection of pairwise locally linear patches linking the new point finally estimates the geodesic distance from the normal manifold distribution. The originality of our method resides in the training of healthy tissue images, which allows to detect the presence of WML in test images and the definition of physiological difference from normal images within manifold space. We present WML detection results on FLAIR MRI scans of elderly patients with diabetes obtained in a clinical study.
Fig. 1.

Principle of the manifold-based white matter lesion detection method, embedding new sample points in the low-dimensional space to estimate their deviation from a learned healthy tissue distribution.
2. METHODS
The computation of a distance between unseen image patches and a given population considered as a healthy distribution of periventricular areas, seen as a pathologic deviation from normality, consists of two steps: (1) the embedding of multiple high-dimensional datasets of ventricle images using a recently proposed weighted graph similarity measure to create locally linear manifold spaces, followed by (2) the projection of new unseen image patches onto the appropriate manifold space and determine the geodesic of the low-dimensional point to the tangential plane of the manifold.
2.1. Manifold Embedding of Periventricular Images
Let us consider a set of N images in the ambient space ℜD such that
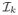 = {I0, …, IN] ∈ Vk, extracted from a given periventricular area Vk where each image patch Ii is of dimensionality D. The aim is to create a low-dimensional manifold consisting of N points Yi, Yi ∈ ℜd, i ∈ [1, N] where d ≪ D based on locally linear embedding [6]. In such a framework, if an adequate number of data points is available, then the underlying manifold
= {I0, …, IN] ∈ Vk, extracted from a given periventricular area Vk where each image patch Ii is of dimensionality D. The aim is to create a low-dimensional manifold consisting of N points Yi, Yi ∈ ℜd, i ∈ [1, N] where d ≪ D based on locally linear embedding [6]. In such a framework, if an adequate number of data points is available, then the underlying manifold
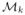 is considered to be “well-sampled”. Therefore, it can represent the underlying healthy population structure. In the sub-cluster corresponding to a pathological population, each individual point of the training set and its neighbours would lie within a locally linear patch on the manifold.
is considered to be “well-sampled”. Therefore, it can represent the underlying healthy population structure. In the sub-cluster corresponding to a pathological population, each individual point of the training set and its neighbours would lie within a locally linear patch on the manifold.
To select the K closest neighbor for each image patch, we adopt an approach similar to a fuzzy block matching approach which avoids the constraint of a strict one-to-one pairing [7]. One can assume over the image domain ℜD, a weighted graph wij that links together pixels x of the input image Ii and pixels y of the image Ij with a weight wij(x, y), which is computed as follows:
| (1) |
where PIi(x)is a 2D ROI of the image Ii centered at pixel x and the parameter Zij,x is a constant of normalization such as: ∀x ∈ ℜD, Σy∈ℜD
wij|(x, y) = 1; m is the number of pixels of the image ROI; σ is the standard deviation of the noise and β is a smoothing parameter. This weighted graph wij is a representation of non-local interactions between the input image Ii and the image Ij of the training dataset. In the context of ventricular image regions in the brain, the location of the brain structures is not highly variable and it is then not desirable to go through the entire image domain to find a good match. In this work, we constrain the search of non-local structures by using a limited number of neighbors around x: w = {w(x, y), ∀x ∈ ℜD, y ∈
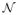 (x)}, where
(x)}, where
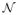 (x) is the neighborhood of the pixel x. The size of the considered neighborhood
(x) is the neighborhood of the pixel x. The size of the considered neighborhood
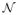 is linked to variability in the periventricular region. Hence, an ambient similarity measure is defined as:
, thereby reflecting the global interactions between Ii and Ij by summing the most significant weights for each neighborhood
is linked to variability in the periventricular region. Hence, an ambient similarity measure is defined as:
, thereby reflecting the global interactions between Ii and Ij by summing the most significant weights for each neighborhood
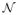 in Ij.
in Ij.
The manifold reconstruction weights are estimated by assuming the local geometry of the neighborhood patches in Ii can be described by linear coefficients that permit the reconstruction of every model point from its neighbours. The value of the weights are therefore determined by minimizing the following reconstruction errors:
| (2) |
| (3) |
Here, ε(W) sums the squared distances between all data points and their corresponding reconstructed points. The weights Wij represent the importance of the jth data point to the reconstruction of the ith element. It is easy to show that each weight can be calculated individually [6]. Each sample Ii and weight Wij contributes to ε:
| (4) |
with Q as a K × K matrix, where each element is defined as
with r as an individual image region taken from the K samples of
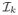 . Wij are solved by a least squares problem given the constraint in (3). The algorithm maps each high-dimensional Ii to a low-dimensional Yi. These internal coordinates are found with a cost function minimizing the reconstruction error:
. Wij are solved by a least squares problem given the constraint in (3). The algorithm maps each high-dimensional Ii to a low-dimensional Yi. These internal coordinates are found with a cost function minimizing the reconstruction error:
| (5) |
with M as a sparse and symmetric N × N matrix enclosing the reconstruction weights Wij such that M = (I − W)T(I − W), and Y spanning the Yi’s. By constraining Y to identity, the problem becomes a straightforward one with min tr(YMYT) given the constraint . The optimal embedding, up to a global rotation, is obtained from the bottom d + 1 eigenvectors of M and helps to minimize the cost function Φ(Y) as a simple eigenvalue problem.
2.2. Normality Distances of Manifold Projected Images
To obtain the low-dimensional manifold position of a new image from the ambient space (image domain), one has to infer intrinsic coordinates based on it’s neighborhood representation in high-dimensional space. We first assume an explicit mapping f: ℜD →
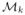 from the ambient space ℜD to the manifold space
from the ambient space ℜD to the manifold space
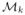 . The forward mapping of Ii is then performed by estimating the relationship between ℜD and
. The forward mapping of Ii is then performed by estimating the relationship between ℜD and
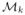 as a joint distribution, such there exists a smooth functional which belongs to a local neighborhood. Theoretically the manifold should follow the conditional expectation:
as a joint distribution, such there exists a smooth functional which belongs to a local neighborhood. Theoretically the manifold should follow the conditional expectation:
| (6) |
which captures the overall trend of the data in d-space. Here, both
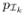 (Ii) (marginal density of
(Ii) (marginal density of
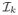 ) and p(Ii, Yi) (joint density) are un-known. Using Nadaraya-Watson kernel regression, densities are replaced by kernel functions under a conditional expectation setting [8]. We replace the variable with
and
. The Gaussian regression kernels G require the neighbors Yj of j ∈
) and p(Ii, Yi) (joint density) are un-known. Using Nadaraya-Watson kernel regression, densities are replaced by kernel functions under a conditional expectation setting [8]. We replace the variable with
and
. The Gaussian regression kernels G require the neighbors Yj of j ∈
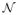 (i) to determine the bandwidths h, g so it includes all K data points (
(i) to determine the bandwidths h, g so it includes all K data points (
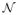 (i) representing the neighborhood of i). By substitution in (6), this gives:
(i) representing the neighborhood of i). By substitution in (6), this gives:
| (7) |
By assuming G is symmetric about the origin and generalize the expectation such that the observations
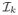 are defined in terms of a metric
are defined in terms of a metric
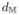 in manifold space d, we obtain:
in manifold space d, we obtain:
| (8) |
which integrates the distance
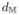 (Yi, Yj) defined below in Eq.(9) and updates fNW(Ii) using the closest neighbors of point Ii in ℜD. This constrains the regression to be valid for similar data points in its vicinity since locality around Ii preserves locality in Yi.
(Yi, Yj) defined below in Eq.(9) and updates fNW(Ii) using the closest neighbors of point Ii in ℜD. This constrains the regression to be valid for similar data points in its vicinity since locality around Ii preserves locality in Yi.
Using the smooth function fNW: ℜD →
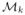 , a new manifold coordinate Ynew is created such that fNW(Inew) → Ynew, based on it’s high-dimensional representation. From the intrinsic coordinates of Ynew, we approximate it’s deviation from the healthy distribution by calculating the geodesic distance based on the shortest path to the tangent plane of the manifold medoid point Ymed (center of the manifold). We achieve this by summing a series of pairwise distances between n locally linear patches Ωi, linking the new data point in
, a new manifold coordinate Ynew is created such that fNW(Inew) → Ynew, based on it’s high-dimensional representation. From the intrinsic coordinates of Ynew, we approximate it’s deviation from the healthy distribution by calculating the geodesic distance based on the shortest path to the tangent plane of the manifold medoid point Ymed (center of the manifold). We achieve this by summing a series of pairwise distances between n locally linear patches Ωi, linking the new data point in
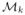 to Ymed, with Ynew ∈ Ω0 and Ymed ∈ Ωn−1. For a 2-chain patch (i and it’s neighboring patch j), the point Yij belonging to the edge shared by Ωi and Ωj is determined via convex programming [9]. Generalizing along the entire path, the estimated distance gives:
to Ymed, with Ynew ∈ Ω0 and Ymed ∈ Ωn−1. For a 2-chain patch (i and it’s neighboring patch j), the point Yij belonging to the edge shared by Ωi and Ωj is determined via convex programming [9]. Generalizing along the entire path, the estimated distance gives:
| (9) |
where the estimated geodesic distance
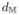 is calculated from the low-dimensional mapping and {Yi ∈ Ωi, Yj ∈ Ωj, Yij ∈ (Ωi ∩ Ωj)} are vertices along the manifold path from Ynew to Ymed.
is calculated from the low-dimensional mapping and {Yi ∈ Ωi, Yj ∈ Ωj, Yij ∈ (Ωi ∩ Ωj)} are vertices along the manifold path from Ynew to Ymed.
3. EXPERIMENTS AND RESULTS
Data images used in this study were obtained from a diabetic population of 106 elderly patients. The patient mean age was 60 ± 4.7 (range 54 – 77, median 59). Forty-eight were female and 55 were male. MRIs were performed during the baseline period on enrollment into the study (TR = 8000 TR, TE = 100). All participant exams consisted of transaxial T1-w, T2-w, PD, and FLAIR scans. All scans except T1-w were performed with a 3-mm slice thickness, no slice gap, a 240 × 240mm field of view and a 256 × 256 matrix. From these brain images, 12 periventricular locations were identified and for each of these locations, 73 healthy training and 33 test image patches (39×39) were extracted. Lesions were manually segmented by a trained rater on the test images and classified as healthy or with WMLs according to the total lesion load. From the 396 patches, 263 were classified as healthy, and 133 as with WMLs. All images were co-registered to a common template using HAMMER [10].
The training patches were embedded in the landmark specific-manifold (12 separate manifolds for each of the 12 landmarks) using the approach in 2.1. Then for each test image patch, we would determine the corresponding manifold
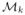 based on the weighted similarity measure
based on the weighted similarity measure
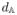 (It, Imed) in 2.1, where It is the test image and Imed is the training image corresponding to manifold medoid Ymed in
(It, Imed) in 2.1, where It is the test image and Imed is the training image corresponding to manifold medoid Ymed in
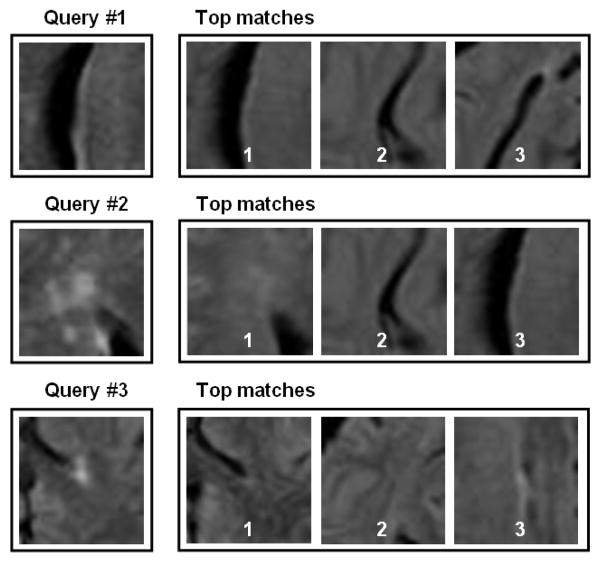
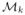 . The test image is then mapped onto the selected manifold to compute the patchwise geodesic distance to manifold center Ymed. We tested the manifold selection process used to map each test image patch into the appropriate manifold. Fig. 2 illustrates some results to identify the correct training dataset for a series of test images.
. The test image is then mapped onto the selected manifold to compute the patchwise geodesic distance to manifold center Ymed. We tested the manifold selection process used to map each test image patch into the appropriate manifold. Fig. 2 illustrates some results to identify the correct training dataset for a series of test images.
Fig. 2.
Results from the manifold assignment scheme based on the weighted graph block matching method. It compares the test image to each of the 12 manifold cluster medoids.
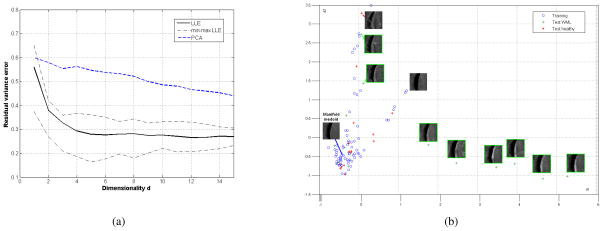
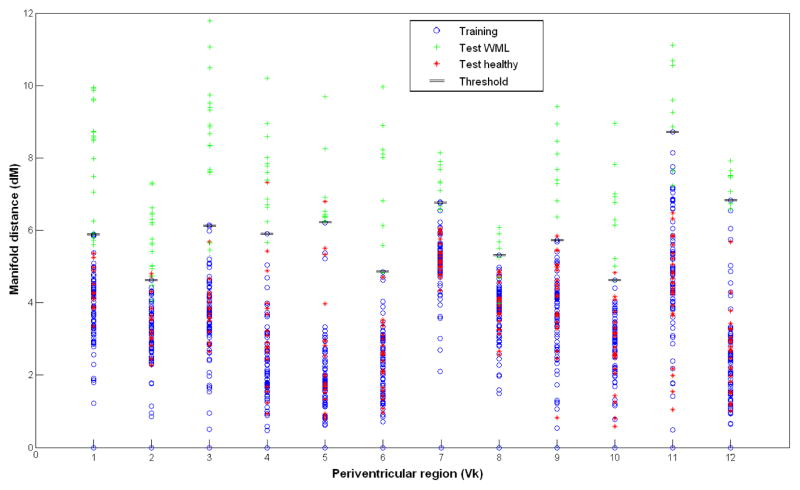
The optimal neighborhood size was found at K = 10 for all manifolds in the training set, which guaranteed that all the images from the manifold dataset were connected among themselves. The optimal manifold dimensionality was estimated by computing the residual reconstruction error between the ambient and intrinsic spaces for each periventricular population, as shown in Fig. 3a. The manifold dimensionality range was d = [4, 8] depending on the selected training set. One can observe the non-linear trend of the curve when comparing the evolution to a linear approximation of the manifold based on PCA. A 2D embedding of the computed manifold (output of LLE) is represented in Fig. 3b, showing the group of training healthy patients in the left portion of the 2D map and the test images which either fall onto the normal tissue manifold or deviate from the manifold in an orthogonal fashion. The bandwidths for the kernels Gh and Gg were set as average distances over the local neighborhoods in ambient and manifold space, respectively. Fig. 4 represents the projected distances between all the images (training and testing) involved in this study and the medoids for each of the 12 manifolds. Images of healthy tissue obtain lower geodesic distances, while most of the images with WMLs demonstrate larger values spanning outside the training cluster. By defining a manifold-specific threshold based on the furthest healthy point to identify the presence of WMLs in the image, the total WML detection rate was of 85.7% from all 396 test images, with a false positive rate of 2.2%.
Fig. 3.
(a) Evolution of the residual variance error against dimensions d, comparing PCA and LLE. (b) 2D LLE manifold embedding of images from the parietal lobe and corpus callosum areas, according to its two first dimensions (d1, d2). Blue points correspond to the training images of healthy tissue, while green crosses and red stars represent testing images for WMLs and healthy tissue respectively.
Fig. 4.
Distribution of geodesic distances for all images in the study, assessing the presence of WMLs by a deviation to the manifold.
4. CONCLUSION
We have proposed a method for detecting the presence of periventricular white matter lesions in FLAIR images of the brain using a manifold-constrained embedding approach. The set of training manifolds from 12 ventricle locations represents the distribution of healthy images in high dimensional space. A weighted graph is implemented as a non-local similarity measure to select local neighborhoods in the datasets, while a Gaussian-based regression kernel is subsequently used to map unseen image patches onto the training manifold created by LLE. Finally, a chain of locally linear patches in manifold space is applied to assess the approximate geodesic distance to the midpoint of a given pathological pattern. Experiments demonstrate the need of nonlinear embedding of the learning data, and the relevance of the proposed method for stratifying different stages of white matter lesion progression. In the context of multiple sclerosis, the method can improve for the early detection of the disease with promising classification rates based on ground-truth knowledge. Future work will improve the deviation threshold selection by cross-validation and investigate into fully automated WML segmentation for monitoring and therapy applications.
References
- 1.Mortazavi D, Kouzani AZ, Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: a review. Neuroradiology. 2011 doi: 10.1007/s00234-011-0886-7. [DOI] [PubMed] [Google Scholar]
- 2.Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. Neuro Image. 2004;23:S189–S195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 3.Duchateau M, De Craene M, Piella G, Frangi AF. Characterizing pathological deviations from normality using constrained manifold-learning. MICCAI. 2011:256–63. doi: 10.1007/978-3-642-23626-6_32. [DOI] [PubMed] [Google Scholar]
- 4.Wachinger C, Yigitsoy M, Navab N. Manifold learning for image-based breathing gating with application to 4D ultrasound. MICCAI. 2010;II:26–3. doi: 10.1007/978-3-642-15745-5_4. [DOI] [PubMed] [Google Scholar]
- 5.Gerber S, Tasdizen T, Fletcher P, Joshi S, Whitaker R. Manifold modeling for brain population analysis. Med Image Anal. 2010;14:643–653. doi: 10.1016/j.media.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roweis S, Saul L. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–6. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 7.Rousseau F, Habas P, Studholme C. Human brain labeling using image similarities. CVPR. 2011:1081–8. [Google Scholar]
- 8.Davis B, Fletcher P, Bullitt E, Joshi S. Population shape regression from random design data. Int J Comput Vis. 2010;90:255–266. [Google Scholar]
- 9.Meng D, Leung Y, Xu Z, Fung T, Zhang Q. Improving geodesic distance estimation based on locally linear assumption. Patt Reco Letters. 2008;29:862–871. [Google Scholar]
- 10.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE TMI. 2002;21:1421–39. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]