SUMMARY
A balance between Six2-dependent self-renewal and canonical Wnt signaling-directed commitment regulates mammalian nephrogenesis. Intersectional studies using chromatin immunoprecipitation and transcriptional profiling identified direct target genes shared by each pathway within nephron progenitors. Wnt4 and Fgf8 are essential for progenitor commitment; cis-regulatory modules flanking each gene are co-bound by Six2 and β-catenin, and dependent on conserved Lef/Tcf binding sites for activity. In vitro and in vivo analyses suggest that Six2 and Lef/Tcf factors form a regulatory complex that promotes progenitor maintenance while entry of β-catenin into this complex promotes nephrogenesis. Alternative transcriptional responses associated with Six2 and β-catenin co-binding events occur through non-Lef/Tcf DNA binding mechanisms highlighting the regulatory complexity downstream of Wnt signaling in the developing mammalian kidney.
INTRODUCTION
Assembly of the mammalian kidney is driven by epithelial-mesenchymal interactions between stem/progenitor cells within the ureteric epithelium and the adjacent cap mesenchyme (CM) (Costantini and Kopan, 2010; Dressler, 2009). GDNF and FGF signals, predominantly secreted by the CM, induce repetitive branching of the ureteric epithelium elaborating the renal collecting duct network (Sánchez et al., 1996; Moore et al., 1996; Vega et al., 1996; Michos et al., 2010). In contrast, Wnt9b secreted by the ureteric epithelium induces CM progenitors to transition to epithelial renal vesicles (RVs), each RV giving rise to a single nephron (Carroll et al., 2005).
Progenitors located adjacent to the medullary face of branch tips cluster to form pretubular aggregates before transitioning to epithelial RVs whereas those in the outermost kidney cortex remain undifferentiated. Importantly, the maintenance of CM progenitors ensures continued ureteric branching through the production of branching factors, and the cellular template for new waves of nephrogenesis until the progenitor population is exhausted in the prenatal or early postnatal period.
Nephron progenitors express the transcriptional regulator Six2 (Kobayashi et al., 2008). The Six2+ population comprises self-renewing, multi-potent nephron progenitors, and Six2 is essential for maintaining the progenitor state; CM prematurely undergoes an ectopic mesenchymal to epithelial transition (MET) in Six2 mutants, rapidly depleting the nephron progenitors (Self et al., 2006; Kobayashi et al., 2008). Normal nephrogenesis and ectopic nephrogenesis in Six2 mutants require a Wnt9b signal from the ureteric epithelium (Carroll et al., 2005; Kobayashi et al., 2008). A second Wnt-family member, Wnt4, and an FGF family member, Fgf8, act downstream of Wnt9b in the transition of induced CM to RVs (Stark et al., 1994; Grieshammer et al., 2005; Perantoni et al., 2005). Canonical Wnt signaling directed by β-catenin is necessary and sufficient to mediate the essential early inductive actions of Wnt9b and Wnt4, though Wnt4 likely utilizes an alternative mechanism in the final phase of epitheliarization (Park et al., 2007; Tanigawa et al., 2011; Burn et al., 2011). Although Wnt/β-catenin and Six2 pathways have opposing actions, commitment and self-renewal of progenitors, respectively, recent studies also suggest Six2 and Wnt9b collaborate at some level in maintaining expression of a subset of CM-specific genes (Karner et al., 2011).
Here, we identified a prominent set of cis-regulatory modules (CRMs) co-bound by both Six2 and β-catenin containing transcriptional complexes. A functional analysis of CRMs and regulatory transcriptional complexes provide evidence for both opposing and collaborative interactions between Six2 and Wnt-driven regulatory programs through distinct DNA-binding mechanisms at shared enhancers within the nephron progenitor population.
RESULTS
Transient activation of canonical Wnt signaling induces MET within isolated Six2+ nephron progenitor cells
We previously showed that expression of a dominant active form of β-catenin in nephron progenitors induces ectopic expression of early markers of nephrogenesis including Fgf8 and Wnt4. However, induced cells retained a mesenchymal phenotype, failing to transition to an E-cadherin producing renal epithelium (Park et al., 2007). In contrast, transient elevation of β-catenin levels and canonical Wnt pathway activity through pharmacological suppression of glycogen synthase kinase-3 (GSK3) with BIO (6-bromoindirubin-3’-oxime; Meijer et al., 2003) within isolated kidney mesenchyme – a heterogeneous population comprising nephron, interstitial, and vascular progenitors - stimulates the MET of induced nephron progenitors (Kuure at al., 2007).
To determine the specific action of BIO-directed canonical Wnt pathway activation on isolated nephron progenitors, we used fluorescence-activated cell sorting (FACS) to isolate Six2 progenitors from mice expressing a GFP transgene under Six2 regulatory control (Kobayashi et al., 2008). When Six2+ progenitors were aggregated and cultured for 48 hours, the first 24 hrs in BIO supplemented medium, the second 24 hrs in vehicle carrier (DMSO), progenitors activated E-cadherin (Figure 1A) and a post-epithelial specific tubule marker, Jag1 (data not shown). When Six2+ progenitors were cultured continuously in BIO (Figure 1A), or in dispersed culture with transient exposure to BIO (Figure S1), Six2+ cells failed to activate E-cadherin. Thus, a transient elevation of β-catenin levels within isolated, aggregates of Six2+ progenitor cells, in the absence of other cellular inputs, is sufficient to initiate the induction of nephrogenesis mirroring the normal action of Wnt9b in the mammalian kidney. Quantitative analysis of expression markers suggests that there is no substantial change in cell number in DMSO or BIO treated populations over 24 hrs (data not shown).
Figure 1. Transient activation of canonical Wnt signaling induces nephron progenitors to undergo a mesenchymal-to-epithelial transition.
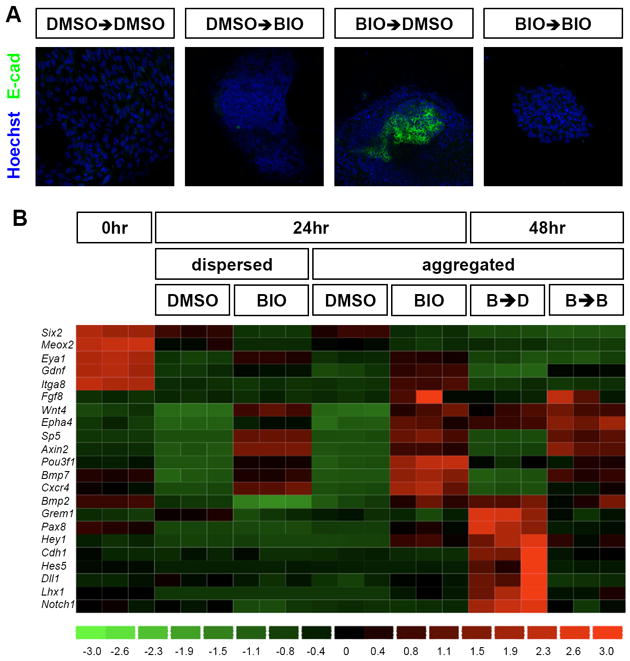
(A) Aggregated Six2-GFP+ nephron progenitors were incubated for 24 hrs with either BIO or DMSO (vehicle) and further incubated for an additional 24 hrs with either BIO or DMSO. (B) Transcriptional profiles of nephron progenitors after activation of canonical Wnt signaling under dispersed or aggregated culture conditions. Aggregated nephron progenitors were exposed to either transient activation (B➔D, 24 hrs with BIO followed by another 24 hrs without BIO) or constitutive activation of canonical Wnt signaling (B➔B, 24 hrs with BIO and another 24 hrs with BIO).
To examine the global transcriptional changes occurring on Wnt pathway activation, Six2+ progenitor aggregates were incubated with BIO and examined by transcriptional profiling. Figure 1B highlights a selected subset of key genes whose expression and activity is linked to the regulation of kidney development; a detailed analysis of the transcriptional responses is presented in Table S1. As expected, markers of the CM, including Six2, Meox2, and Eya1, were highly expressed in freshly isolated nephron progenitors. Interestingly, Six2 and Meox2 showed significantly lower expression levels in BIO versus DMSO treated control aggregates, consistent with canonical Wnt signaling antagonizing their expression. In contrast, Eya1, a gene essential for appropriate specification of the metanephric mesenchyme in the kidney anlagen (Xu et al., 1999; Sajithlal et al., 2005), showed elevated levels in BIO treated samples at 24 hrs suggesting a positive role for canonical Wnt signaling in maintaining normal levels of this regulatory factor within the CM.
Axin2 and Sp5, broad targets of canonical Wnt signaling in multiple tissue types, and Wnt4, a specific transcriptional readout of a Wnt9b-dependent nephrogenic response, showed a strong activation in BIO treated cells independent of cell density at 24 hrs (Figure 1B). However, expression of most early nephrogenic inductive markers including Fgf8 (Grieshammer et al., 2005; Perantoni et al., 2005), Pax8 (Carroll et al., 2005), Bmp2 (Georgas et al., 2009), Bmp7 (Dudley et al., 1995), and Cxcr4 (Ueland et al., 2009) was only observed in aggregate cultures highlighting the importance of cell density dependent interactions in the inductive response. Expression of these genes was maintained on extension of BIO treatment for an additional 24 hrs whereas the withdrawal of BIO resulted in a downregulation of general Wnt targets, and the activation of E-cadherin (Cdh1) and a number of Notch pathway components (Notch1, Hes5, and Dll1), consistent with an epithelial transition and the initiation of proximal nephron patterning pathways, respectively, on transient Wnt signaling in Six2 progenitors (Chen and Al-Awqati, 2005; Cheng et al., 2007).
In summary, the in vitro BIO-mediated model of Six2-GFP+ cells replicates many features of the in vivo Wnt-mediated induction of Six2+ cells. Further, the in vitro data provides evidence for additional roles for β-catenin action, and potentially canonical Wnt signaling, in both abrogating (Six2) and supporting (Eya1) expression of key regulatory factors within the CM. Finally, 80% of the genes reported downregulated in CM of Wnt9b mutants at E11.5 (Karner et al., 2011) were up regulated on BIO treatment of FACS isolated Six2 cells at E16.5, in good agreement with a Wnt9b-driven canonical Wnt pathway (Table S1). Differences between the data sets may reflect temporal differences in the cellular responses or the modifying role of other cell-types present in the E11.5 kidney analysis.
Genomic mapping of β-catenin and Six2 binding sites in nephron progenitor cells
Canonical Wnt signaling is mediated by four Lef/Tcf family members: each member is reported to generate multiple protein isoforms (Arce et al., 2006). Lef/Tcf factors associate with DNA targets in repressive complexes independent of Wnt signaling. High quality antibodies with broad specificity for all isoforms of each Lef/Tcf factor have not been described. In order to identify direct transcriptional targets specific to a Six2-mediated pathway of nephron progenitor maintenance and a canonical Wnt pathway driven program of nephron induction, we isolated Six2-GFP+ nephron progenitors by FACS and examined Six2 and β-catenin association with DNA targets by chromatin immunoprecipitation (ChIP) and high-throughput DNA sequencing (ChIP-seq).
At a False Discovery Rate (FDR) of 0.01, we detected 569 β-catenin ChIP peaks and 1359 Six2 ChIP peaks using a two-sample iterative peak caller (Ma and Wong, 2011) (Figure 2A and Table S2). Analysis of the distribution of peaks showed that Six2 bound peaks generally lie closer to the transcriptional start site (TSS) of the nearest gene than those bound by β-catenin (Figure 2B). When compared to 10,000 random control regions, Six2 peaks are significantly enriched in 5’ UTR regions (p-value, 7.6E-19), intron regions (p-value, 2.8E-19), and 1 kb upstream regions (p-value, 1.8E-60) while β-catenin peaks are enriched in intronic regions (p-value, 3.8E-3) (Table S2).
Figure 2. Peak Statistics, Motif Analyses and GREAT annotations of β-catenin and Six2 ChIP-seq datasets.
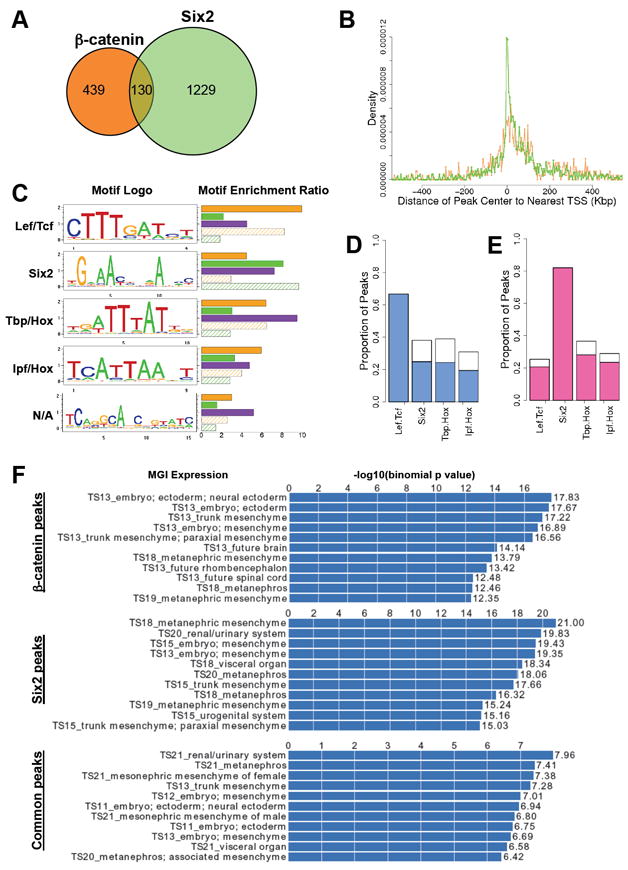
(A) Venn-diagram of the overlap between β-catenin (orange circle) and Six2 (green circle) bound ChIP products with a 0.01 FDR cutoff.
(B) Density graph of the peak locations in relation to the nearest transcriptional start sites (TSS). X-axis represents distance from the peak center to the nearest TSS, where 0 is the position of the TSS. Y-axis represents proportion of detected bindings that are located within each distance interval (distance increasing at 10 kb intervals). Orange line, β-catenin peaks; Green line, Six2 peaks.
(C) Top five enriched de novo motifs recovered from ChIP-seq peak regions. Separate runs of de novo motif discovery were performed on genomic regions bound by β-catenin, Six2, or both. The motif logos display nucleotide frequencies (scaled relative to the information content) at each position. The horizontal bars on the right side represent the motif enrichment in peak regions (r3, defined in Supplemental Experimental Procedures). For each motif, we calculate its enrichment in five different sets of peak regions: β-catenin peak regions (orange bar), Six2 peak regions (green bar), common peak regions (purple bar), β-catenin distinctive peak regions (orange shadowed bar) and Six2 distinctive peak regions (green shadowed bar). N/A, not applicable.
(D) De novo motif occupancies in β-catenin peak regions. The blue bars are for the peaks containing de novo Lef/Tcf motif sites.
(E) De novo motif occupancies in Six2 peak regions. The pink bars are for the peaks containing de novo Six2 motif sites.
(F) MGI expression annotations of β-catenin, Six2 and common peaks analyzed by GREAT. A sample of 11 representative annotations are shown for each peak category. Histogram represents the –log10 (binomial p-value) for each annotation.
Interestingly and unexpectedly, 130 (22.84%) of the β-catenin peaks overlapped with Six2 peaks (9.57%; Figure 2A and Table S2), a significant result (p-value <1E-360). A number of other β-catenin associated sites showed statistically significant binding in only one biological replicate of the Six2 ChIP, but a clear trend of Six2 association in the weaker sample. Estimating that only 1.0% of the genome of mouse kidney cells is accessible for transcription factor binding (Song et al., 2011), the intersection of β-catenin and Six2 peaks was highly significant (p-value, <1E-156).
To further annotate and assign potential function to β-catenin, Six2, and co-bound DNA regions, a GREAT (Genomic Region Enrichment of Annotation Tool) analysis (McLean et al., 2010) was performed on each data set (Figure 2F, Table S2). The recovery of anatomical annotation terms in each grouping shows a strong association with the target tissue (Figure 2F). Analysis of Gene Ontology (GO) Biological Processes shows a correlation for all groupings with processes involving development and morphogenesis, with enrichments in co-bound regions for kidney and renal system development (Table S2). All regions also show strong correlations with genes related to kidney specific phenotypes, such as abnormal kidney development and renal hypoplasia (Table S2). These data indicate that Six2 and β-catenin bind near and regulate expression of genes that are necessary for proper kidney development.
De novo motif analysis uncovers Tcf, Six2, and Hox motifs in β-catenin and Six2 bound regions
To understand the interplay between β-catenin and Six2, and to identify other regulatory factors that may collaborate in protein-DNA interactions, de novo motif recovery was performed on β-catenin binding regions, Six2 binding regions, and co-bound regions, separately, using the CisGenome package (Ji et al., 2008). Five de novo motifs were significantly enriched in our peak sets (Figure 2C). These motifs were compared with TRANSFAC (TRANScription regulatory FACtors) (Matys et al., 2003) and UniProbe (Universal PBM Resource for Oligonucleotide-Binding Evaluation) (Badis et al., 2009; Berger et al., 2008) databases, and data in the literature.
The most enriched motif in the β-catenin peak dataset is highly related to the Lef/Tcf prediction in the TRANSFAC and UniProbe databases. In contrast, the predicted de novo Six2 motif does not match any known motif in the TRANSFAC database, and differs from the motif identified in microarray binding studies (Berger et al., 2008) but matches a verified Six2 motif within a regulatory region proximal to the Six2 promoter (Brodbeck et al., 2004, Figure S2). We examined Six2 binding to these motifs by electrophoretic mobility shift assays (EMSA). Strong Six2 binding was observed to the recovered predicted Six2 motif, and binding was not effectively competed by the Six2 motif predicted though protein-DNA microarray studies (Figure S2). Thus, the de novo Six2 motif recovered here likely represents a genuine target site for Six2 interaction in vivo.
The de novo Tbp/Hox motif is similar to the TRANSFAC TBP motif and almost identical to the published Hoxc9 motif recovered from ChIP-seq analysis of ES-cell derived motor neurons (Jung et al., 2010). The de novo Ipf/Hox motif is similar to the TRANSFAC Ipf and UniProbe Hoxc9 motif predictions. The recovery of Hox motifs in our ChIP-seq data suggests that Hox factors are likely an additional component within the gene regulatory networks of a set of Six2/β-catenin targets, consistent with genetic analysis illustrating a critical role for Hox11 paralogs in kidney development (Wellik et al., 2002).
Of the β-catenin bound peak regions, 380 out of 569 (66.78%) were predicted to encode at least one Lef/Tcf site, a highly significant enrichment over matched control regions (p-value <1E-149), consistent with Lef/Tcf family members acting as the primary binding partners for β-catenin (Figure 2D). In addition, 38.14%, 39.02%, and 30.03% of the β-catenin peaks contain predicted Six2, Tbp/Hox, and Ipf/Hox motifs, respectively. 60-65% of β-catenin peaks with a Six2, Tbp/Hox or Ipf/Hox motif are also predicted to contain a Lef/Tcf motif (Figure 2D). The de novo Six2 motif is the most significantly enriched motif in the Six2 ChIP-seq peak regions (p-value <1E-595): 1116 (82.12%) of the 1359 Six2 peak regions contain the recovered Six2 motif sites, a significantly higher frequency than that observed for Lef/Tcf (25.39%; p-value for enriched occupancy in data set, <1E-3), Tbp/Hox motif (36.72%; p-value for enriched occupancy in data set, <1E-35), or Ipf/Hox motif (29.07%; p-value for enriched occupancy in data set, <1E-27) predictions (Figure 2E). A similar percentage of Lef/Tcf (81.74%), Tbp/Hox (76.95%) and lpf/Hox (81.27%) recovered motif containing peaks also contain the recovered Six2 motif (Figure 2E). Taken together, these observations suggest potential interactions among β-catenin, Six2, Lef/Tcf, and Hox family members to regulate self-renewal and differentiation of nephron progenitors.
Identification of direct targets of Six2 and β-catenin during early nephrogenesis
To examine the relationship between Six2 and β-catenin binding and gene expression changes in response to BIO treatment, we identified the nearest neighboring gene to each ChIP region for each data set, then intersected these genes with those displaying BIO-dependent expression changes on in vitro culture of the Six2+ nephrogenic compartment (Table S3). To access the significance of the association between Six2 and β-catenin binding and the expression patterns of the target genes, we randomly selected 10,000 regions from the genome as the control, and calculated the p-value of enrichment using one-proportion z-test. For Six2, 693 peaks associated with genes whose expression decreased when aggregated progenitor cultures treated with BIO for 24 hrs were compared to freshly sorted cells (p-value for enrichment <1E-71); while 193 of the β-catenin bound regions mapped to neighboring genes that showed elevated expression when BIO treated aggregates were compared to control DMSO cultures (p-value for enrichment <1E-48). In addition, we also observed a significant correlation of β-catenin binding about genes whose expression decreased on culture in BIO when compared with the starting population (237 binding regions; p-value for enrichment <1E-10). Thus, β-catenin has a strong association with distinct transcriptional outcomes within the Six2+target population.
To functionally interrogate putative CRMs predicted by Six2 and β-catenin ChIP-seq, we focused on the subset of co-bound regions that lie adjacent to genes encoding three key signaling factors whose expression was modified by BIO treatment (Figure 3 and Table S3). Fgf8 and Wnt4 are essential for the transition of pretubular aggregates to RVs and are amongst the earliest responses to ureteric Wnt9b signaling (Grieshammer et al., 2005; Perantoni et al., 2005; Stark et al., 1994; Carroll et al., 2005). Bmp7 is expressed in the CM, differentiating nephron progenitors, and the ureteric epithelium (Dudley et al., 1997; Godin et al., 1998). Loss of Bmp7 results in a complex phenotype part of which reflects the failure to maintain nephron progenitors (Dudley et al., 1995, 1999; Blank et al., 2009).
Figure 3. Identification of common interaction sites for Six2 and β-catenin adjacent to genes encoding key nephrogenic regulatory factors.
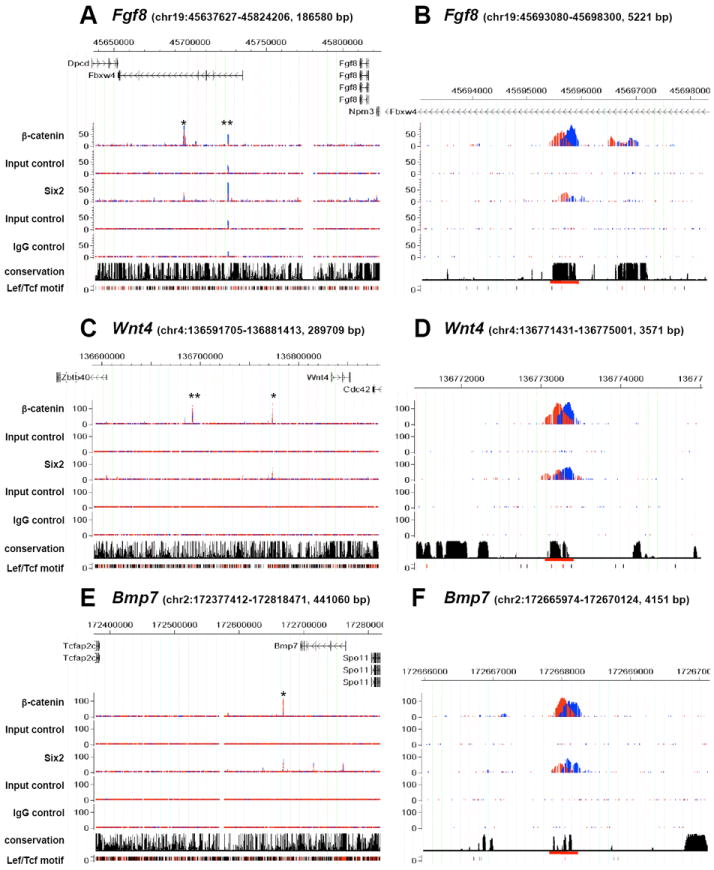
(A-F) Binding of Six2 and β-catenin viewed in the CisGenome Browser (Jiang et al., 2010) within the indicated genomic intervals (mm9 coordinates). ChIP product sequences on Watson and Crick strands are shown in red and blue; binding sites are predicted to be at the intersection of these peaks. Input control represents sequencing data of chromatin that was not subjected to immunoprecipitation. IgG control represent sequencing data from mock immunoprecipitates with rabbit IgG. Conservation denotes placental mammal basewise conservation by Phastcons score. Lef/Tcf motif predictions are displayed; overlap with conserved DNA regions is indicated in red and non-conserved regions in black. Putative CRMs tested in transgenic analyses (Figure 4) are marked by asterisks (*) in A, C, and E (global view) and by a red underline in B, D, and F (viewed in higher magnification). The peak marked by ** in (A) is an artifact detected in both Input and IgG controls. The β-catenin peak marked by ** in (C) was tested in transgenic analysis but no expression of the reporter was observed in kidneys of the transgenic embryos. The Six2 bound region downstream of Fgf8 showed statistically significant association of Six2 in only one biological replicate.
Table S4 shows the position of co-bound regions relative to the transcriptional start site (TSS) of the nearest target genes showing a BIO-dependent increase in expression within Six2+ aggregate cultures. Each binding region overlies a conserved block of DNA, a considerable distance from the TSS of the putative target gene. In the case of Fgf8, binding was localized within the intron of Fbxw4, a gene displaying weak, non-specific expression unaltered by BIO treatment (data not shown). Interestingly, deletions and duplications within intronic regions of Fbxw4 are associated with split-hand/foot malformation 3 (SHFM3), a human disease whose phenotypes correlate closely with Fgf8-dependent developmental processes (Sidow et al., 1999; Friedli et al., 2008). Together these data are consistent with a cis-regulatory action of these intronic regions on Fgf8 transcription.
To examine the regulatory activity of conserved blocks of DNA incorporating the identified Six2/β-catenin binding regions, DNA fragments were tested for cis-regulatory activity in the E15.5 mouse kidney (Table S4). Test regions comprising the conserved block of DNA that incorporates the binding regions were cloned upstream of a minimal promoter driven-lacZ reporter cassette and reporter expression was examined in G0 transgenic embryos (Figure 4).
Figure 4. Transgenic validation of cis-regulatory modules associated with nephrogenic determinants that are co-bound by Six2 and β-catenin.
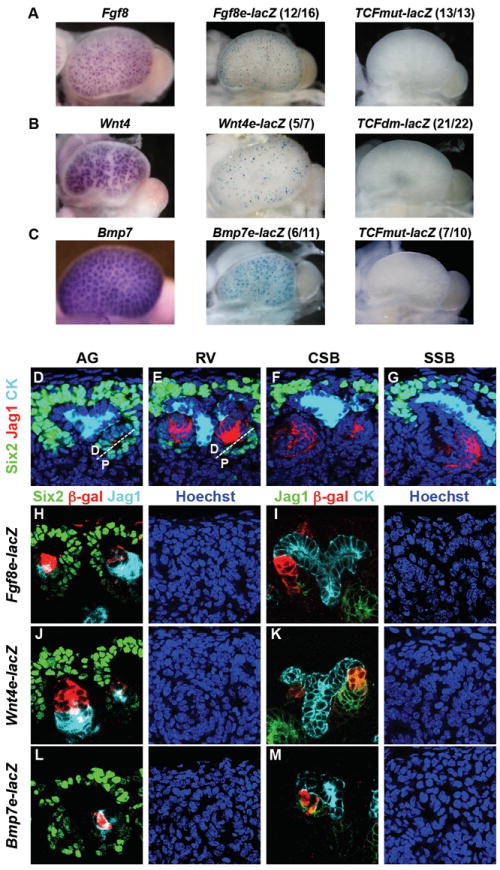
(A-C) Whole mount in situ hybridization detects expression of endogenous genes (left panels). β-galactosidase activity of transgenic reporter driven by the putative CRMs identified through Six2 and β-catenin co-binding (middle panels). β-galactosidase activity driven by the CRMs with mutated Lef/Tcf binding sites (right panels). The ratio indicates the number of embryos showing the illustrated expression pattern over the total number of transgenic progeny, each from a unique founder. (D-G) Expression of Six2 and Jag1 in the pretubular aggregate (AG), renal vesicle (RV), comma-shaped body (CSB), and S-shaped body (SSB) stages of nephrogenesis in the mammalian kidney. The AG and RV are shown divided into the distal and proximal parts by a white dashed line. Jag1 is expressed in the lumen of AG, at the distal part of RV, and at the medial segment of SSB. Six2 is expressed in undifferentiated nephron progenitors and downregulated in AG and RV. Cytokeratin (CK) marks the ureteric epithelium. (H-M) Expression of reporters driven by CRMs co-bound by Six2 and β-catenin in E15.5 transgenic kidneys.
Each of the three putative CRMs supported robust, reproducible, transgene expression in over 50% of G0 transgenic embryos within the RV and RV derivatives (Figure 4A-C). To verify transgene expression in early Wnt-induced derivatives of the Six2 population, we compared expression of each transgene with Six2, and Jag1, a marker of differentiating RV derivatives. Six2 was highly expressed in the CM and downregulated when nephron progenitors aggregated on Wnt9b induction, persisting transiently in the proximal region of the early RV (Figure 4D and 4E). As previously reported (Georgas et al., 2009), Jag1 was detected in the distal region of the newly formed RV (Figure 4E), expression resolving to the mid-segment of the S-shaped body during later stages of tubule development (Figure 4F and 4G). The three putative CRMs about Fgf8, Wnt4 and Bmp7 showed similar reporter gene expression in distal regions of the RV-derivative overlapping Jag1 (Figure 4H-M). Expression at later S-shaped body stages remained distally restricted; in contrast, Jag1 expression was confined to the mid-section at these stages. In summary, transgene expression in the RV is polarized at the outset of RV formation, consistent with reported polarized expression of Fgf8 (Grieshammer et al., 2005) and Wnt4 (Georgas et al., 2009; Mugford et al., 2009). Similar overall findings were obtained for conserved co-bound regions adjacent to Cxcr4, a gene linked to kidney development (Takabatake et al., 2009;Ueland et al., 2009) (Figure S3).
As discussed earlier, β-catenin does not bind directly to DNA but complexes with Lef/Tcf family members to activate transcription of target genes in a canonical Wnt signaling response. A strong Lef/Tcf binding motif was recovered through de novo motif discovery of β-catenin bound DNA regions (Figure 2C). To address the role of Lef/Tcf interactions, we mutated conserved Lef/Tcf motifs in each of the CRMs associated with Wnt4, Fgf8, and Bmp7: a single motif in Fgf8 and Bmp7 regulatory elements, and paired motifs for Wnt4 (Table S4). In each case, three-base changes in the core Lef/Tcf binding region (CTTTG > ATGGG) abolished binding of TCF4B to its target site in EMSA assays (Figure S2), and led to the complete loss of enhancer activity of each of the CRMs in the kidneys of transgenic mice (Figure 4A-C). In addition, we mutated a conserved Six2 motif in the CRM associated with Wnt4 to address the role of Six2. Seven out of 12 transgenic kidneys showed transgene expression (data not shown). Of these, six showed expression within the RV derivatives while two of the six also showed weaker broader expression throughout the kidney. The seventh was ectopically activated within the interstitial mesenchyme progenitors. These results contrast with five of six transgenics expressing the reporter under the control of the wild-type Wnt4 enhancer element all of which displayed a RV derivative restricted pattern. The variable results on mutation of this sequence may reflect more complex interactions than simply the loss of a Six2 regulatory input.
In summary, each target shows BIO-dependent activation in vitro and Wnt9b-dependent activation in vivo within differentiating Six2 derivatives (Carroll et al., 2005). The putative enhancers drive reporter gene expression specifically within the RV. Whereas the putative targets are expressed in other kidney compartments, only the RV derivative is labeled as expected. Enhancer activity was dependent on evolutionary conserved Lef/Tcf binding sites in each enhancer, consistent with β-catenin acting in conjunction with Lef/Tcf factors in a conventional, canonical Wnt signaling pathway. Thus, each tested CRM likely mediates a component of the endogenous Wnt regulatory response underlying the expression of these essential regulators of kidney development. Although each tested region was selected for co-binding to Six2 in nephron progenitors, the reporter gene expression was turned on after Six2 is downregulated and Six2 is not required for the inductive response but for suppression of inductive activity (Self et al., 2006); thus, Six2 binding correlates with inactivity of these CRMs. Six2 is known to associate with members of groucho family of transcriptional repressors (Lopez-Rios et al., 2003). While none of the individual elements precisely replicated expression of the target gene, additional Six2 and β-catenin binding sites flanking each target gene are predicted to contribute to the overall transcriptional output.
Six2 and β-catenin directly regulate the CM-specific expression of Six2 and Eya1
Six2 is auto-regulated through a proximal enhancer; the Six2 binding region in this enhancer was recovered in the Six2 ChIP-seq data set but no co-association of β-catenin was observed at this site (marked by ** in Figure 5A, Brodbeck et al., 2004; Kuure et al., 2007). Interestingly, when the down-regulation of Six2 was compared in FACS isolated Six2+ cells cultured for 24 hrs in DMSO or BIO, loss of Six2 expression was enhanced by Wnt pathway activation (Figure 1B and data not shown). We identified a region 60 kb upstream of the Six2 TSS that co-bound Six2 and β-catenin at multiple sites over a highly conserved, 1.1 kb block of DNA and we examined the potential regulatory role of this region in transgenic mice (Figure 5A and 5C). Reporter expression was very precisely directed to the endogenous Six2 expression domain (Figure 5E and 5G). Despite the presence of multiple β-catenin binding peaks, only one Lef/Tcf binding site was predicted in the enhancer (Figure 5C) and the mutation of this site did not alter transgene activity. Thus, the interaction of β-catenin at this Six2 enhancer correlates with the downregulation of Six2 expression but not through a direct Lef/Tcf factor-mediated DNA binding mechanism (Figure 5E).
Figure 5. Six2 and Eya1 genes are common targets of Six2 and β-catenin.
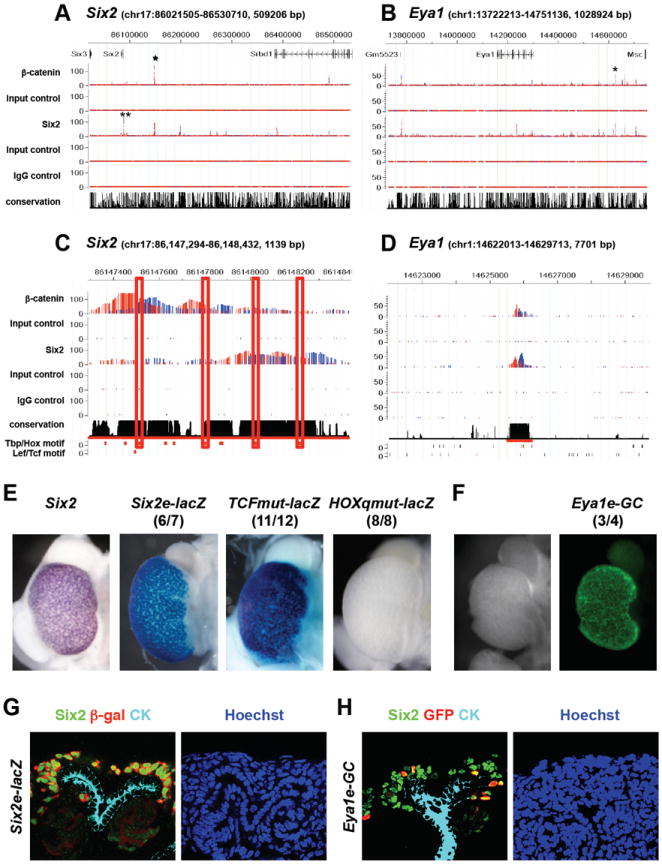
(A-B) Binding of Six2 and β-catenin in genomic intervals containing Six2 (A) and Eya1 (B). Putative CRMs tested in transgenic analyses (E-H) are marked with asterisks (*). (C) De novo motifs found in Six2 enhancer. Tbp/Hox motifs, rather than Lef/Tcf motif, were found to center on peaks of Six2 and β-catenin binding. (D) Putative CRM upstream of the Eya1 gene tested in transgenic analysis (shown in (F)) is underlined in red. (E) G0 transgenic analysis of Six2 enhancer. Endogenous expression of Six2 is shown on the left. Images of β-galactosidase expression driven by the Six2 enhancer (Six2e-lacZ) and by the Six2 enhancer carrying a mutated Tcf motif (TCFmut-lacZ) are shown in the middle. Expression of β-galactosidase driven by the Six2 enhancer carrying quadruple mutations of Tbp/Hox motifs (HOXqmut-lacZ) is shown on the right. (F) G0 transgenic analysis of Eya1 enhancer. (G) Expression of the transgenic reporter driven by Six2 enhancer. Note Six2 enhancer recapitulates expression of endogenous Six2. Endogenous Six2 (green) is localized in nuclei and β-galactosidase driven by Six2 enhancer cassette is present throughout the cell. (H) Expression of the reporter driven by putative Eya1 enhancer. Eya1 enhancer shows mosaic expression in a restricted domain that overlaps with endogenous Six2.
Alignment of other recovered motifs indicated a close correlation of the recovered Tbp/Hox motif and the predicted peak centers for β-catenin and Six2 binding (Figure 5C). This observation suggests that Hox proteins, rather than Lef/Tcf factors or Six2 itself, may mediate regulatory inputs within the identified enhancer region. Interestingly, Hoxa11, Hoxc11, and Hoxd11 play a combinatorial role in kidney development and Six2 expression is lost when all three of these paralogs are mutated (Wellik et al., 2002). In addition, Hox11 paralogs have been shown to regulate Six2 expression through the proximal enhancer where we detected Six2 binding (Gong et al., 2007; Yallowitz et al., 2009). Thus, Hox proteins may serve as binding partners for Six2, and possibly for β-catenin, to regulate expression of Six2. Consistent with this view, mutation of all predicted Tbp/Hox motifs, in the context of the 1.1 kb Six2 CRM, abolished expression of the reporter (Figure 5E). These results suggest that binding of β-catenin to a Hox/Six2 complex at the distal Six2 enhancer abrogates enhancer activity attenuating Six2 expression during nephron induction.
Eya and Six factors interact to regulate target gene expression (Ohto et al., 1999). Eya factors do not bind DNA, but modify the activity of their Six partners (Rebay et al., 2005; Jemc and Rebay, 2007). Eya1 displays similar expression to Six2 in the CM during kidney development suggesting that Eya1 may modulate Six2 activity (Mugford et al., 2009). In contrast to Six2, Eya1 is essential prior to ingrowth of the ureteric epithelium, a phenotype resembling Six1 mutants (Xu et al., 1999; Xu et al., 2003). A later collaborative role for Eya1 and Six2 in progenitor maintenance remains an open question. Whereas Six2 was downregulated by canonical Wnt signaling, Eya1 was upregulated on BIO stimulation relative to DMSO control samples at 24 hrs (Figure 1B and data not shown). These data suggest that Wnt action may promote nephron progenitor maintenance through Eya1.
The large number of Six2 and β-catenin associated binding regions around the Eya1 locus suggest a complex regulation by these two pathways. Our ChIP-seq data identified multiple β-catenin and Six2 binding sites associate with Eya1 upstream of the TSS and in intronic regions (Figure 5B), and two conserved regions co-associated with β-catenin and Six2; one in intron 9, the other 325 kb upstream of the Eya1 TSS. The potential regulatory action of the - 325kb region was examined in transgenic kidneys: although transgenics displayed mosaic expression, reporter expression was restricted to the Six2/Eya1 co-expressing CM (Figure 5F and 5H). The mutation of predicted Lef/Tcf motifs in this enhancer did not alter transgene expression (data not shown). Thus, β-catenin action at this distant enhancer element is not consistent with a canonical Wnt transcriptional response in which β-catenin association would depend on DNA binding of Lef/Tcf transcriptional partners.
Eya1 is one member of a group of genes predominantly expressed in the CM that are associated with β-catenin and Six2 binding regions, whose expression is maintained in cultured Six2+ cells through a BIO-dependent mechanism (Tables 1, S1, and S3). Transcriptional profiling of Six2 and Wnt9 mutant kidneys has also identified a number of genes positively regulated by both Six2 and Wnt9b input at the outset of kidney development (Karner et al., 2011): among these, Itga8 and Fam19a5 were identified in our ChIP-seq studies examining the E16.5 kidney. Collectively, our data indicate that Wnt signaling plays an active role in supporting nephron progenitor specific programs of gene regulation through a non-canonical mechanism with regard to β-catenin association at target sites (e.g. Eya1), and through a classic Lef/Tcf dependent canonical DNA association in the commitment of nephron progenitors (e.g. Wnt4 and Fgf8).
Table 1. Examples of three regulatory classes of genes co-bound by Six2 and β-catenin in nephron progenitors.
Class I and III genes are both up-regulated by canonical Wnt signaling when BIO treated cultures are compared with control DMSO treated cultures at 24 hrs; however, class III genes are expressed at high levels in the CM initially, while class I genes are more weakly expressed at this time and strongly induced on BIO treatment, correlating with induction of RV progenitors. Class II genes are expressed in the CM and repressed by canonical Wnt signaling upon differentiation in response to BIO relative to DMSO treated controls. The enhancers tested in transgenic analyses are indicated in bold.
| Fold change
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | RefSeq | Probe ID | BIO/DMSO | BIO/0hr | Peak location | Peak length | Highest score | Relative position | Distance to TSS |
|
Class I (DMSO@24hr < BIO@24hr AND 0hr < BIO@24hr)
| |||||||||
| Cxcr4 | NM_009911 | 1448710_at | 32.35 | 2.82 | chr1:130718705-130718922 | 218 | 51.3 | TSS-upstream | -229938 |
| Wnt4 | NM_009523 | 1441687_at | 10.26 | 4.79 | chr4:136773187-136773353 | 167 | 120.9 | TSS-upstream | -60279 |
| Fgf8 | NM_010205 | 1451882_a_at | 7.75 | 8.19 | chr19:45695446-45695814 | 369 | 55.0 | TES-downstream | 121560 |
| Bmp7 | NM_007557 | 1418910_at | 6.82 | 1.73 | chr2:172667933-172668149 | 217 | 90.4 | TES-downstream | 97752 |
| Aldh1a2 | NM_009022 | 1422789_at | 4.28 | 1.45 | chr9:71065207-71065805 | 599 | 62.3 | Intron | 1911 |
| Lhfpl2 | NM_172589 | 1434129_s_at | 4.52 | 1.30 | chr13:94853201-94853454 | 254 | 116.4 | Intron | 25577 |
|
| |||||||||
|
Class II (DMSO@24 > BIO@24hr AND 0hr > BIO@24hr)
| |||||||||
| Sox4 | NM_009238 | 1419156_at | -5.36 | -5.99 | chr13:28810434-28811092 | 659 | 50.6 | TES-downstream | 234787 |
| Dlc1 | NM_015802 | 1460602_at | -3.66 | -1.79 | chr8:37744738-37744926 | 189 | 51.1 | TSS-upstream | -67905 |
| Gas1 | NM_008086 | 1416855_at | -3.48 | -4.43 | chr13:60201125-60201558 | 434 | 45.0 | TES-downstream | 77554 |
| Mllt3 | NM_027326 | 1429205_at | -3.33 | -2.23 | chr4:87025468-87025844 | 377 | 53.3 | TES-downstream | 426561 |
| Zbtb20 | NM_019778 | 1437598_at | -2.86 | -5.94 | chr16:43361297-43361493 | 197 | 101.1 | Intron | 113999 |
| Six2 | NM_011380 | 1427436_at | -2.84 | -5.07 | chr17:86147429-86147586 | 158 | 114.5 | TSS-upstream | -59914 |
| Meis2 | NM_010825 | 1457632_s_at | -2.10 | -1.77 | chr2:115694660-115695137 | 478 | 40.9 | Intron | 195458 |
| Hmga2 | NM_010441 | 1450780_s_at | -2.02 | -3.06 | chr10:119857810-119858640 | 831 | 56.1 | Intron | 55765 |
|
| |||||||||
|
Class III (DMSO@24hr < BIO@24hr AND 0hr > BIO@24hr)
| |||||||||
| Gdnf | NM_010275 | 1419080_at | 4.12 | -1.74 | chr15:7647936-7648151 | 216 | 40.3 | TSS-upstream | -112967 |
| Itga8 | NM_001001309 | 1454966_at | 3.13 | -1.63 | chr2:11941243-11941615 | 373 | 76.6 | TES-downstream | 282117 |
| Itga8 | NM_001001309 | 1454966_at | 3.13 | -1.63 | chr2:11944818-11945605 | 788 | 59.5 | TES-downstream | 278335 |
| Sall1 | NM_021390 | 1437983_at | 3.17 | -1.15 | chr8:91936032-91936284 | 253 | 111.7 | TSS-upstream | -368098 |
| Sall1 | NM_021390 | 1437983_at | 3.17 | -1.15 | chr8:92130016-92130238 | 223 | 74.6 | TSS-upstream | -562067 |
| Eya1 | NM_010164 | 1457424_at | 1.89 | -1.93 | chr1:14235068-14235289 | 222 | 44.9 | Intron | 65101 |
| Eya1 | NM_010164 | 1457424_at | 1.89 | -1.93 | chr1:14625742-14625927 | 186 | 62.2 | TSS-upstream | -325555 |
| Hoxb4 | NM_010459 | 1460379_at | 1.73 | -1.23 | chr11:96180625-96180790 | 166 | 92.3 | Intron | 655 |
The Eya1 regulatory grouping - expression potentially enhanced by Six2 and β-catenin - is unlikely to reflect a homogeneous regulatory response. Whereas, Hoxb4 (Preger-Ben Noon et al., 2009; Dressler, 2009) and Gdnf (Sánchez et al., 1996; Pichel et al., 1996; Moore et al., 1996) show similar trends to Eya1 in our microarray and DNA binding studies, transgenic reporter assays analyzing conserved regions co-associated with Six2 and β-catenin showed that transgene expression was restricted to distal regions of the RV-derivative (Figure S4). Interestingly, Gdnf is the major signal regulating branching growth of the ureteric epithelium, the tissue source of the Wnt9b inductive signal: normal levels of GDNF expression depend on a reciprocal interaction with Wnt11 secreted at the ureteric tips (Majumdar et al, 2003). Classical experiments have long postulated a feedback in which nephron induction stimulates branching (Saxén, 1987). Our data suggest a Wnt pathway-specific regulatory input into Gdnf within newly induced cells consistent with a feedback response. The domain of transgene expression more closely resembles other aspects of a Wnt9b response, though a potential regulatory input by Wnt11 cannot be ruled out.
Six2 and HoxA11 can complex with β-catenin and each other
To further investigate the molecular mechanism of Six2 action, we examined the interaction of Six2 with β-catenin in nephron progenitor cells. As we show in Figures 4D and 4E, a low level of Six2 is present in early, differentiating nephron progenitors. Consequently, loss of Six2 is not essential for commitment of nephron progenitors (see discussion) and the Six2 population is heterogeneous containing a minor fraction of early-induced cells. In contrast, recent data indicates that Cited1 marks a subpopulation of Six2+ cells that consists of undifferentiated cells (Mugford et al., 2009; Figure S5; see also www.gudmap.org for a more extensive characterization). Interestingly, immunoprecipitation with anti-Six2 antibodies pulled down β-catenin in Six2+ cells isolated by FACS but not in the Cited1+ subset of the Six2+ population (Figure 6A). Thus, Six2 and β-catenin form a complex in vivo consistent with the overlap in their DNA association in ChIP studies. However, this complex appears to be restricted to the Wnt-induced population of committed nephron progenitors (Six2+Cited1-cells).
Figure 6. Six2 interacts with β-catenin, Tcf4, and HoxA11.
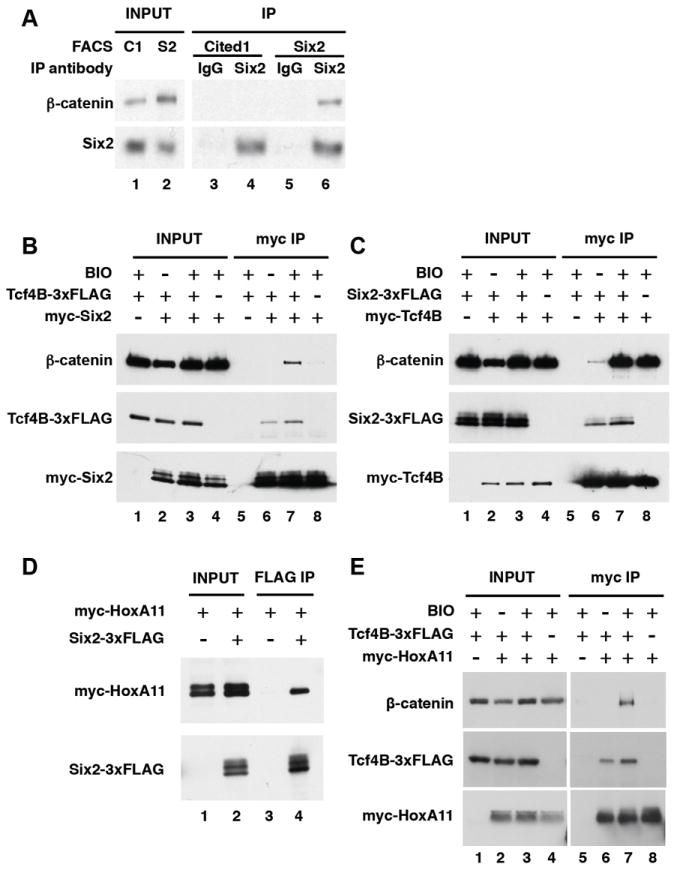
(A) Six2 interacts with β-catenin. Immunoprecipitation of Six2 was performed with nephron progenitors isolated from either Cited1-RFP+ or Six2-GFP+ embryonic kidneys and analyzed by Western blot to detect β-catenin. (B) Six2 forms a ternary complex with Tcf4B and β-catenin. HEK293T cells were transfected with plasmids as indicated. BIO was added to induce accumulation of β-catenin. Lysates were immunoprecipitated with anti-myc antibody and analyzed by Western blot to detect proteins as indicated. (C) Six2 and β-catenin do not compete for binding to Tcf4B. (D) Six2 interacts with HoxA11. (E) HoxA11 forms a complex with β-catenin in the presence of Tcf4B.
The interaction between Six2 and β-catenin was explored in more detail in Wnt responsive HEK293 kidney cells (Figure 6B-C). Immunoprecipitation of myc-tagged Six2 pulled down FLAG-tagged Tcf4B independent of BIO-stimulation: thus, Six2 and Tcf4B form a complex in the absence of canonical Wnt signaling (Figure 6B, lanes 6 and 7). In contrast, Six2 pull-down of β-catenin was Tcf4B dependent (Figure 6B, lanes 7 and 8) while Tcf4B association with Six2 or β-catenin was independent of the other factor (Figure 6C, lanes 6-8). Further, whereas BIO stimulation resulted in a large increase in β-catenin association with Tcf4B as expected, Six2 binding to Tcf4B was largely unaltered (Figure 6C, lanes 6-8). Given the prediction of Hox-binding sites, and known role of Hox11 paralogs in kidney development, we also examined potential interactions amongst HoxA11, Tcf4b, Six2 and β-catenin. In HEK 293 cells, Six2 can interact with HoxA11 (Figure 6D) and with β-catenin in the presence of Tcf4B (Figure 6E). Collectively, these data suggest that co-binding of Six2 and β-catenin at CRMs in vivo is mediated, at least in part, through a common interaction with Lef/Tcf family members. Further, Six2 does not compete with β-catenin for binding to a common Tcf binding partner. Finally, Hox11 members are likely interacting partners in aspects of the regulatory programs. Unfortunately, the absence of suitable antibodies precluded analysis of potential interactions between Hox11 paralogs and β-catenin containing complexes in the metanephric mesenchyme.
DISCUSSION
All stem/progenitor cell based systems must balance the maintenance of stem/progenitor cell types and their commitment to differentiated components of the mature organ. Utilizing an ex vivo inductive system, and intersecting transcriptional profiling data of Wnt response with genomic analysis of Six2 and β-catenin bound DNA targets, our study provides a comprehensive insight into the gene regulatory networks underpinning maintenance and commitment of nephron progenitors.
Gene regulation mediated by Six2 and β-catenin
A moderate number of target regions, 500-1,500 associate with, and likely mediate, the transcriptional actions of Six2 and β-catenin. Most regions are associated with one factor, but given evidence for opposing regulatory actions of the Six2 and Wnt pathway, our analysis focused on a smaller set of shared binding regions, and their putative target genes. We provide evidence for three distinct classes of transcriptional program amongst shared targets of Six2 and β-catenin transcriptional networks (Table 1).
Class I genes are induced by canonical Wnt signaling in differentiating RVs (Figure 7). Amongst this group are genes such as Fgf8 and Wnt4 encoding critical downstream regulatory signals that are essential for nephrogenesis. Class I genes require β-catenin activity in a canonical Lef/Tcf regulatory partnership at target sites (Park et al., 2007). Class I targets are silent in uninduced CM cells where Six2 is bound at target sites. Six2 binding likely reflects endogenous Six2/Lef/Tcf complexes from our analysis of protein-protein interactions in vitro. The simplest model is one in which Six2 binding to Lef/Tcf factors in the CM ensures that β-catenin is unable to broadly activate targets of the nephrogenic inductive response throughout the CM (Figure 7). Interestingly, cell culture and in vivo studies indicate that this is unlikely to reflect a competitive role for Six2 and β-catenin binding to a Tcf factor. Instead, the association of both Six2 and Lef/Tcf factors with groucho repressors (Arce et al., 2009; Lopez-Rios et al., 2003) suggests Six2 and Lef/Tcf factors may cooperate to silence early drivers of nephrogenesis. A more precise definition of this regulatory process will require improved Lef/Tcf antibodies with broad specificity recognizing all isoforms.
Figure 7. Regulation of class I genes by Six2 and β-catenin in nephron progenitors.
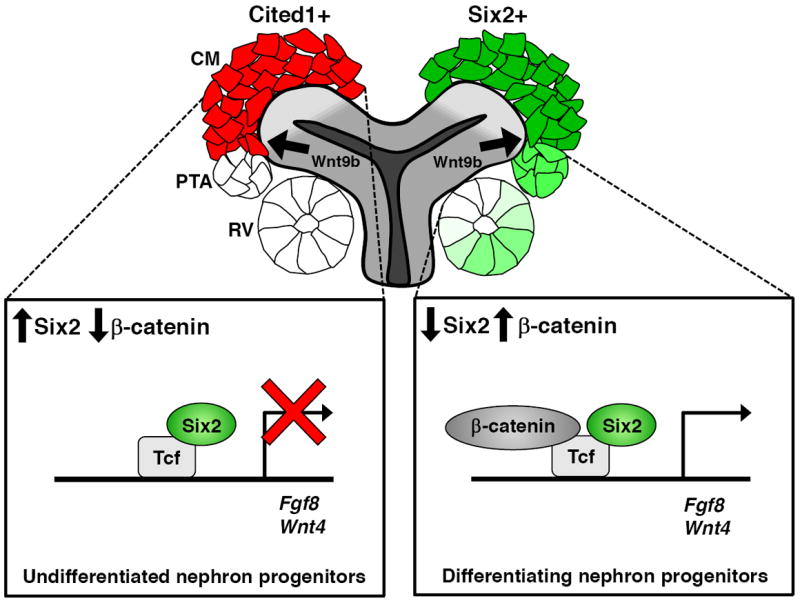
In undifferentiated nephron progenitors (Cited1+ cells), Six2/Lef/Tcf complexes prevents β-catenin from activating class I genes, such as Fgf8 and Wnt4. In differentiating nephron progenitor cells, a high dosage of Wnt9b activates class I genes by elevating β-catenin level and lowering Six2 level.
Six2+ cells are targeted by the Wnt9b-directed nephron inducing response. Further, Six2 is transiently present within cells that have activated Wnt4. Thus, the presence of Six2 is clearly not sufficient to prevent some cells from responding to Wnt9b. Rather, Six2 action likely ensures that at each round of induction, only a fraction of the potential progenitor pool undergoes a commitment to nephrogenesis. Consequently, the loss of Six2 function leads to all progenitor cells rapidly and quantitatively committing to a RV fate, through a Wnt9b-dependent mechanism (Self et al., 2006; Kobayashi et al., 2008). Together these findings imply a normal process that uncouples the inhibitory input of Six2 to enable a Wnt9b inductive process. One possible mechanism could be the level of Wnt9b signaling. Earlier work demonstrated close cellular contact was required between CM and Wnt9b producing cells to invoke a RV inductive response (Carroll et al., 2005), consistent with a high-threshold dependence for this inductive interaction. Once engaged, Wnt4 directed canonical Wnt signaling would be expected to act as a rapid feed-forward mechanism at an early stage in the RV induction process (Park et al., 2007).
Class II genes, represented by Six2 itself, are predominantly active in CM and downregulated by Wnt input in the inductive process. Our data identifies a distal enhancer for Six2 that mirrors the activity of a proximal regulatory element in driving reporter gene activity to the Six2 population much like the shadow enhancers recently described around many key developmental regulatory genes in Drosophila (Perry et al., 2010). Interestingly, the association of β-catenin at the distal but not the proximal enhancer correlates with the enhanced suppression of Six2 expression on Wnt9b mediated initiation of nephrogenesis suggesting that the presence of β-catenin within this regulatory complex attenuates Six2 transcription. Given the strong association of β-catenin with the activation of transcription, it is unlikely β-catenin is acting directly to recruit repressor factors but rather its presence within this complex may interfere with a Six2-directed activating response.
Class III genes show similar expression to Six2 in the undifferentiated CM but are positively regulated by Wnt signaling. Many of the genes identified in this category at the transcriptional level correspond with those displaying a dual requirement for Six2 and β-catenin at the outset of kidney development (Karner et al., 2011; Table S1). However, we see relatively little evidence of co-binding of Six2 and β-catenin around most of these genes. Thus, their regulation may reflect indirect regulatory responses that are themselves dependent on Six2 and β-catenin. In contrast, Eya1 displays a Class III regulatory pattern in the E16.5 kidney progenitors but not at E11.5 (Karner et al., 2011) and the intersection of Six2 and β-catenin binding predicts an enhancer with CM activity. Thus, co-regulatory positive inputs through Six2 and β-catenin most likely maintain essential gene regulatory programs in nephron progenitors
The finding that Six2 and β-catenin have opposing actions in the regulation of class I and II genes, but act cooperatively in maintaining class III gene activity raises an interesting question as to how these distinct regulatory actions are carried out. Analysis of target sites suggests distinct DNA-regulatory complex interactions: enhancers for Class I targets are predicted to be directly engaged through DNA binding by Lef/Tcf factors while those at Eya1 are postulated to utilize a distinct DNA-binding component. Thus, different protein complexes may guide distinct regulatory outputs to a β-catenin input. A continued focus on the regulatory mechanisms acting on the enhancers identified in the current work is expected to provide additional resolution to the regulatory principles governing mammalian nephrogenesis.
EXPERIMENTAL PROCEDURES
Mouse strains
The Six2TGC BAC transgenic was described previously (Kobayashi et al., 2008). In this strain, a GFP∷CRE fusion gene is expressed under the control of Six2 regulatory regions. Characterization of the Cited1-nuc-TagRFP-T BAC transgenic is available at http://gudmap.org.
Fluorescence activated cell sorting (FACS)
E16.5 kidneys from Six2TGC+ or Cited1-nuc-TagRFP-T+ embryos were trypsinized, dissociated by repetitive pipetting, and resuspended in PBS containing 2% FBS and 10mM EDTA. The resuspended cells were filtered through 40 μm Nylon Cell Strainer (BD Falcon) and kept on ice until FACS. The GFP+ or RFP+ cells were isolated using Dako cytomation MoFlo.
In vitro culture and transcriptional profiling of nephron progenitor cells
The FACS isolated Six2+ cells (about 40,000 cells per sample) were aggregated by centrifugation at 800 × g for 5 min and cultured in 10% FBS/DMEM in the presence of 4 μM BIO or DMSO. After 24 hrs, the media was changed as indicated. RNA from three biological replicates under each experimental condition was isolated by using Absolutely RNA Nanoprep kit (Stratagene) and amplified by using TargetAmp 2-Round Biotin-aRNA Amplification kit 3.0 (Epicentre). The biotinylated probes were hybridized to GeneChip Mouse Genome 430 2.0 (Affymetrix). The raw microarray data were processed and normalized using dChip (Li and Wong, 2001).
ChIP-seq analysis
ChIP was performed as previously described with some modification (Vokes, et al. 2007). A detailed procedure is provided in Supplemental Data.
Transgenic analyses
Each of the putative enhancer regions was cloned into p2xINS-Hsp68-lacZ (Vokes et al., 2007) or p2xINS-HSP68-GFPcre. In these constructs, a putative enhancer was inserted downstream of two copies of the chicken β-globin insulator and upstream of Hsp68 minimal promoter-driven β-galactosidase or GFPcre. The entire expression cassette was purified by electroelution. Pronuclear injections were performed at Harvard Genome Modification Facility. The embryos were harvested at E15.5. In the case of p2xINS-Hsp68-lacZ, the kidneys were stained with X-gal, fixed in 4% PFA/PBS, cleared in 80% glycerol/PBS, and photographed using a Nikon SMZ 1500 fluorescent microscope. In the case of p2xINS-HSP68-GFPcre, the kidneys were photographed using a Nikon Eclipse 90i epi-fluorescent microscope.
In situ hybridization
E15.5 kidneys were fixed in 4%PFA/PBS at 4°C overnight and dehydrated in methanol. Hybridized samples were developed in BM purple (Roche), cleared in 80% glycerol/PBS, and photographed using a Nikon DXM1200 digital camera.
Immunofluorescence
Embryonic kidneys were obtained from transgenic lines carrying p2xINS-Hsp68-lacZ or from G0 transgenic embryos carrying p2xINS-HSP68-GFPcre. E15.5 kidneys or in vitro cultured nephron progenitor cells were fixed in 4% PFA/PBS, incubated in 20% sucrose/PBS at 4°C overnight, and imbedded in OCT. 12 μm sections were incubated overnight with 5% heat inactivated sheep serum/PBST containing primary antibodies against E-cadherin (rat, 1:1000, Sigma) β-galactosidase (rabbit, 1:15000, MP bio or mouse IgG2a, 1:1000, Promega), GFP (chick, 1:500, AvesLabs), cytokeratin (mouse IgG1, 1:200, Sigma), Jag1 (rat, 1:20, DSHB), Six2 (rabbit, 1:500, Proteintech). The secondary antibodies were conjugated with Cy2 (chick), Alexa 488 (rat or mouse IgG2a), Alexa 555 (rabbit), Alexa 568 (rat), or Alexa 633 (rat or mouse IgG1). Nuclei were stained with Hoechst 33342 (Invitrogen) before mounting. Confocal images were acquired on a Zeiss 710 inverted confocal microscope.
Pull down assay
The nuclear extracts of FACS isolated cells were incubated with Dynabeads (Invitrogen) coupled with either normal rabbit antibody or Six2 antibody (Proteintech) at 4°C for overnight. The beads were washed four times with TBS containing 0.25% TX-100. HEK293 cells were transfected with plasmids as indicated. After 24 hrs of transfection, cells were treated with either 4 μM of BIO or DMSO as indicated. Additional 24 hrs later, cells were lysed in lysis buffer (50mM HEPES, pH7.5, 140mM NaCl, 1mM EDTA, 0.5% NP-40, 0.25% TX-100, 1x complete mini protease inhibitor (Roche)). Lysates were incubated with anti-myc antibody (9E10, DSHB) coupled dynabeads at 4°C for overnight. The beads were washed three times with lysis buffer. The western blot was performed with anti-Six2 antibody (Proteintech), anti-FLAG antibody (M2, Sigma), anti-myc antibody (Cell Signaling Technology), or anti β-catenin (Cell Signaling Technology) antibodies.
Supplementary Material
Highlights.
Identification of gene regulatory networks in nephron progenitors
Maintenance and commitment pathways integrated at cis-regulatory modules
Multiple roles for β-catenin complexes in progenitor programs
Acknowledgments
We thank Brian Tilton, Patricia Rogers, Christopher Daly, and Jennifer Couget for their technical support and members of the HSCI Genome Modification Facility for transgenic studies. Work in APM’s laboratory was supported by a grant from the NIH (R37 DK054364). JSP was supported by research fellowships from the National Kidney Foundation and Charles A. King Trust, Bank of America, Co-Trustee (Boston, MA). LLO was supported by a Ruth L. Kirschstein postdoctoral research fellowship from the NIH/NIDDK (F32 DK085959). Work by WM and WHW was supported by NIH grants (R01 HG005717 and R01 HG003903).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Arce L, Pate KT, Waterman ML. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer. 2009;9:159. doi: 10.1186/1471-2407-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Brown A, Adams DC, Karolak MJ, Oxburgh L. BMP7 promotes proliferation of nephron progenitor cells via a JNK-dependent mechanism. Development. 2009;136:3557–3566. doi: 10.1242/dev.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck S, Besenbeck B, Englert C. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech Dev. 2004;121:1211–1222. doi: 10.1016/j.mod.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Burn SF, Webb A, Berry RL, Davies JA, Ferrer-Vaquer A, Hadjantonakis AK, Hastie ND, Hohenstein P. Calcium/NFAT signalling promotes early nephrogenesis. Dev Biol. 2011;352:288–298. doi: 10.1016/j.ydbio.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chen L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol. 2005;288:F939–952. doi: 10.1152/ajprenal.00369.2004. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Robertson EJ. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev Dyn. 1997;208:349–362. doi: 10.1002/(SICI)1097-0177(199703)208:3<349::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Friedli M, Nikolaev S, Lyle R, Arcangeli M, Duboule D, Spitz F, Antonarakis SE. Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3. Mamm Genome. 2008;19:272–278. doi: 10.1007/s00335-008-9106-0. [DOI] [PubMed] [Google Scholar]
- Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, et al. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- Godin RE, Takaesu NT, Robertson EJ, Dudley AT. Regulation of BMP7 expression during kidney development. Development. 1998;125:3473–3482. doi: 10.1242/dev.125.17.3473. [DOI] [PubMed] [Google Scholar]
- Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu Rev Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang F, Dyer NP, Wong WH. CisGenome Browser: a flexible tool for genomic data visualization. Bioinformatics. 2010;26:1781–1782. doi: 10.1093/bioinformatics/btq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF, Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S, Popsueva A, Jakobson M, Sainio K, Sariola H. Glycogen synthase kinase-3 inactivation and stabilization of beta-catenin induce nephron differentiation in isolated mouse and rat kidney mesenchymes. J Am Soc Nephrol. 2007;18:1130–1139. doi: 10.1681/ASN.2006111206. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P. Six3 and Six6 activity is modulated by members of the groucho family. Development. 2003;130:185–195. doi: 10.1242/dev.00185. [DOI] [PubMed] [Google Scholar]
- Ma W, Wong WH. The analysis of ChIP-Seq data. Methods Enzymol. 2011;497:51–73. doi: 10.1016/B978-0-12-385075-1.00003-2. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D’Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carver-Moore K, Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Yu J, Kobayashi A, McMahon AP. High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population. Dev Biol. 2009;333:312–323. doi: 10.1016/j.ydbio.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichel JG, Shen L, Sheng HZ, Granholm AC, Drago J, Grinberg A, Lee EJ, Huang SP, Saarma M, Hoffer BJ, et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- Preger-Ben Noon E, Barak H, Guttmann-Raviv N, Reshef R. Interplay between activin and Hox genes determines the formation of the kidney morphogenetic field. Development. 2009;136:1995–2004. doi: 10.1242/dev.035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284:323–336. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- Saxén L. Organogenesis of the kidney. Cambridge Cambridgeshire; New York: Cambridge University Press; 1987. [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. Embo J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Birren BW, Altshuler D, Jaenisch R, Johnson KR, Lander ES. A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet. 1999;23:104–107. doi: 10.1038/12709. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Takabatake Y, Sugiyama T, Kohara H, Matsusaka T, Kurihara H, Koni PA, Nagasawa Y, Hamano T, Matsui I, Kawada N, et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J Am Soc Nephrol. 2009;20:1714–1723. doi: 10.1681/ASN.2008060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa S, Wang H, Yang Y, Sharma N, Tarasova N, Ajima R, Yamaguchi TP, Rodriguez LG, Perantoni AO. Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev Biol. 2011;352:58–69. doi: 10.1016/j.ydbio.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland J, Yuan A, Marlier A, Gallagher AR, Karihaloo A. A novel role for the chemokine receptor Cxcr4 in kidney morphogenesis: an in vitro study. Dev Dyn. 2009;238:1083–1091. doi: 10.1002/dvdy.21943. [DOI] [PubMed] [Google Scholar]
- Vega QC, Worby CA, Lechner MS, Dixon JE, Dressler GR. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci U S A. 1996;93:10657–10661. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yallowitz AR, Gong KQ, Swinehart IT, Nelson LT, Wellik DM. Non-homeodomain regions of Hox proteins mediate activation versus repression of Six2 via a single enhancer site in vivo. Dev Biol. 2009;335:156–165. doi: 10.1016/j.ydbio.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


