Abstract
Light chain amyloidosis is a devastating disease where immunoglobulin light chains form amyloid fibrils, resulting in organ dysfunction and death. Previous studies have shown a direct correlation between the protein thermodynamic stability and the propensity for amyloid formation for some proteins involved in light chain amyloidosis. Here we investigate the effect of somatic mutations on protein stability and in vitro fibril formation of single and double restorative mutants of the protein AL-103 compared to the wild type germline control protein. A scan rate dependence and hysteresis in the thermal unfolding and refolding was observed for all proteins. This indicates that the unfolding/refolding reaction is kinetically determined with different kinetic constants for unfolding and refolding even though the process remains experimentally reversible. Our structural analysis of AL-103 and AL-103 delP95aIns suggests a kinetic coupling of the unfolding/refolding process with cis-trans prolyl isomerization. Our data reveal that the deletion of Proline 95a (AL-103 delP95aIns), which removes the trans cis diproline motif present in the patient protein AL-103, results in a dramatic increment in the thermodynamic stability and a significant delay in fibril formation kinetics with respect to AL-103. Fibril formation is pH dependent; all proteins form fibrils at pH 2; reactions become slower and more stochastic as the pH increases up to pH 7. Based on these results, we propose that in addition to thermodynamic stability, kinetic stability (possibly influenced by the presence of cis Proline 95a), plays a major role in the AL-103 amyloid fibril formation process.
Introduction
Light chain (AL) amyloidosis is a fatal, progressive disease characterized by extracellular deposition of light chains into amyloid fibrils, resulting in multiple organ dysfunction and death. AL amyloidosis is caused by an abnormal proliferation of monoclonal plasma cells that secrete a high amount of free immunoglobulin light chains into the bloodstream. These light chains misfold and deposit as insoluble amyloid fibrils in various organs, ultimately causing organ failure and death. Current treatments target the plasma cell population and are not curative. AL amyloidosis patients have a median survival of 12-40 months after diagnosis.1 Immunoglobulin light chains are composed of a variable and a constant domain. The structure of the immunoglobulin light chain variable domain consists of nine β-strands packed tightly against each other in two antiparallel β-sheets forming a β-sandwich. The β-strands are labeled A, B, C, C′, C″, D, E, F, and G from the N- to C- termini. These strands form the framework regions (FRs). Strands B and C; C′ and C″; F, and G are connected by unstructured loops called complementarity determining regions or CDRs that determine the specificity of the antigen-antibody interactions (Fig. 1a).2,3 From these three loops, CDR3 is the region with the highest sequence and length variability within the variable domain.
Fig. 1. Structure, sequence, and spectroscopic properties of AL-103.
(a) Structural alignment of x-ray crystal structures of κI O18/O8 germline (gold), AL-103 (red) and AL-103 del95aIns (blue) (PDB codes: 2Q20, 3DVI and 4K07 respectively). Highly conserved single Tryptophan residue at position 35 (black sticks) and disulfide bridge (gray sticks) are shown. (b) Structure-based multiple sequence alignment of κI O18/O8 and AL-103. AL-103 somatic mutations are highlighted. Three of the four AL-103 mutations are clustered in the CDR3 and β-strand G, while the fourth (N34I) is in the dimer interface on β-strand C. (c) Fluorescence (native, filled circles; unfolded, open circles) and (d) Far-UV CD spectra of AL-103.
Numerous experimental reports have implicated thermodynamic stability as one of the major modulators of light chain amyloidogenicity. Studies using light chain proteins from AL amyloidosis patients4-12 have shown that mutations (somatic and otherwise destabilizing) that reduce thermodynamic stability make the proteins more prone to form amyloid fibrils. Based on these reports, an empirical rule has been proposed that amyloidogenic light chains are less thermodynamically stable than their non-amyloidogenic counterparts.10,13,14 It has been suggested that the formation of amyloid fibrils generally does not begin from the protein’s native state, but more likely from partially folded or intermediate states.15
In the case of immunoglobulin light chains, mutations as well as changes in solution conditions that lead to destabilization of the native state (e.g., low pH values, chemical denaturants such as urea or guanidinium hydrochloride, salts, glycosaminoglycans, lipids/detergents, and high temperatures), promote partial unfolding and amyloid fibril formation in vitro.13,16-27
We previously reported a biophysical and biochemical comparison of two patient-derived light chain proteins, AL-09 and AL-103. Both proteins are derived from the germline gene product, κI O18/O8 (also known as IGKV 1-33) and they share more than 90% sequence identity.27,28 AL-103 has four somatic mutations: N34I (β-strand B), D92H (β-strand F), the insertion of a Proline after residue 95 called P95aIns (CDR3), and Q100P (β-strand G). All of AL-103 somatic mutations are non-conservative. AL-09 has seven somatic mutations, three of which are non-conservative changes (N34I (β-strand B), K42Q (loop C-C′), and Y87H (β-strand F)). Both AL-09 and AL-103 have similar thermodynamic stability; both are less stable than the germline protein κI O18/O8. Depending on the species that are enriched at the beginning of the fibril formation reaction (monomers or dimers), AL-09 and AL-103 can present similar fibril formation kinetics (in the case of reactions initiated with enriched dimers) or AL-09 can form fibrils under a broad variety of conditions and with faster kinetics compared to AL-103 (in the case of reactions initiated with enriched monomers).25,27
Restorative mutations are defined as the amino acid change from the somatic mutation found in the amyloidogenic protein to the amino acid found in the germline (wild type) protein. Using a systematic restorative mutational analysis of the non-conservative mutations of AL-09 protein - by restoring the residues found in the corresponding germline sequence κI O18/O8- we found a link between the protein’s thermodynamic stability and their kinetics of amyloid fibril formation. In addition, we found that the total number of mutations in AL proteins was less important than their location.4,29,30
In this work, four AL-103 single mutants and one double restorative mutant were characterized to determine the role of non-conservative mutations in the protein stability and amyloid fibril formation kinetics.
Results
κI O18/O8, AL-103, and the five AL-103 restorative mutants share more than 96% sequence identity (Fig. 1b). κI O18/O8 was expressed and purified from the bacterial periplasmic space, while AL-103 and its mutants form inclusion bodies (see Material and Methods) as has been reported before.4,28 After solubilization of inclusion bodies, every protein was dialyzed and allowed to refold before purification. After purification, Tryptophan fluorescence and far-UV circular dichroism (CD) spectra were obtained under native conditions to ensure correct refolding and recovery of native conformation.
Native tryptophan fluorescence emission spectrum of the highly conserved single Tryptophan at position 35 showed low fluorescence counts for all proteins, with a maximum emission spectrum of 340 nm. This previously reported characteristic of immunoglobulin light chain proteins is due to the fact that the Tryptophan fluorescence is quenched in the native state by the conserved disulfide bridge in positions 23--88, (Fig. 1c shown for AL-103). Urea-induced denaturation curves produced an increase in fluorescence intensity along with a 10 nm red shift in the emission fluorescence maxima (Fig. 1c) due to the unfolding of the protein, as has been reported before.17,24 Likewise, all proteins showed the characteristic Far UV-CD spectrum observed for AL amyloidosis variable domains with two minima, one at 215-217 nm (typical of β-sheet rich structure) and a second at ~230 nm (due to aromatic residues that are optically active in this region)17,20,22,24,27 (Fig. 1d).
In previous studies, we found that κI O18/O8 and AL-103 equilibrium unfolding can be described in terms of the classical two-state unfolding behavior 4,27,28. As mentioned before, AL-103 is less thermodynamically stable than κI O18/O8. In addition, there are no significant structural differences observed between AL-103 and κI O18/O8.28
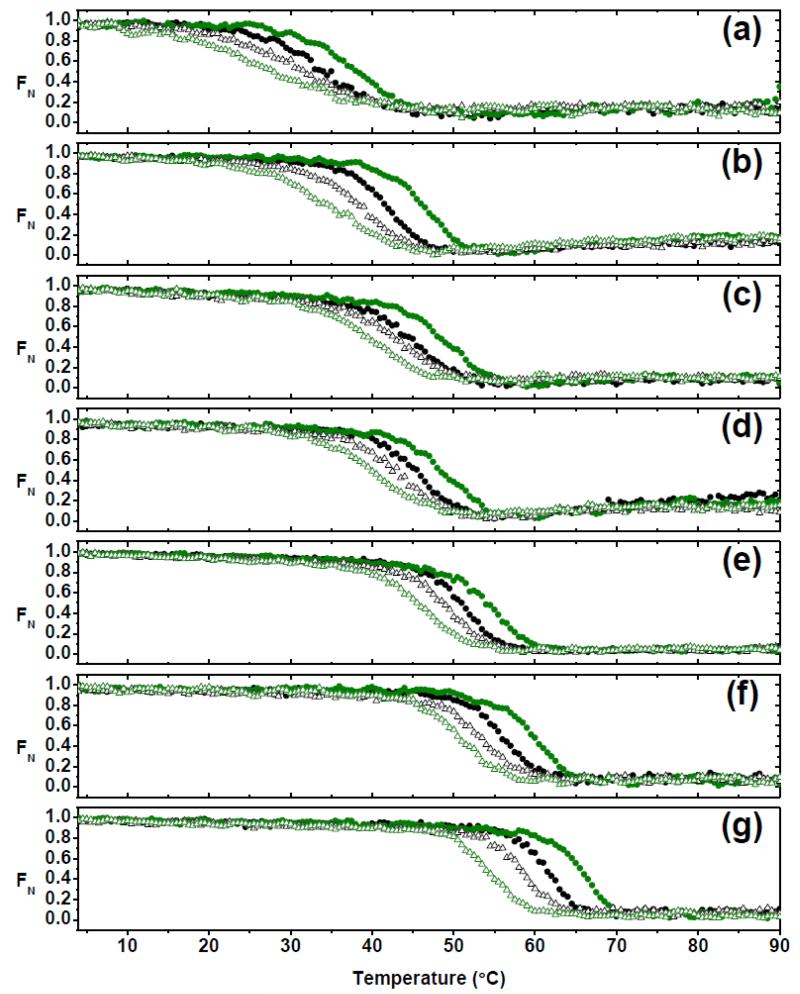
In order to evaluate the role of each AL-103 non-conservative somatic mutation in the protein’s stability, thermal denaturation experiments following the change in molar ellipticity at 217 nm 29 were carried out at different scan rates (0.5 −2.5°C/min). The seven proteins display similar behavior in the unfolding/refolding transitions: clearly monophasic sigmoidal and fully reversible transitions (~99% the original CD signal after refolding). Interestingly, in all cases, unfolding and refolding transitions show hysteresis and a strong scan rate (heating/cooling) dependence, which is evidence that a folding reaction is kinetically controlled.31 As can be seen in Fig. 2, as well as in supplementary Fig. S1, different proteins show differential effects in the scan rate dependence and hysteresis. From visual inspection of thermal transitions and the calculation of the change in the melting temperature values between unfolding and refolding transitions at the fastest scan rate (apparent ΔTm), it is easy to note that AL-103 delP95aIns displays the highest hysteresis and scan rate dependence (Fig. 3g), followed by AL-103 (Fig. 3b) and AL-103 H92D (Fig. 3a).
Fig. 2. Hysteresis in thermal unfolding/refolding transitions as a function of scan rate.
Thermal unfolding (filled circles) and refolding (open triangles) transitions were obtained at different scan rates (0.5°C/min, black; 2.5°C/min; olive green) of (a) AL-103 H92D, (b) AL-103, (c) AL-103 P100Q, (d) AL-103 H92D-I34N, (e) AL-103 I34N, (f) κI O18/O8 and (g) AL-103 del95aIns following the change in molar ellipticity at 217nm.
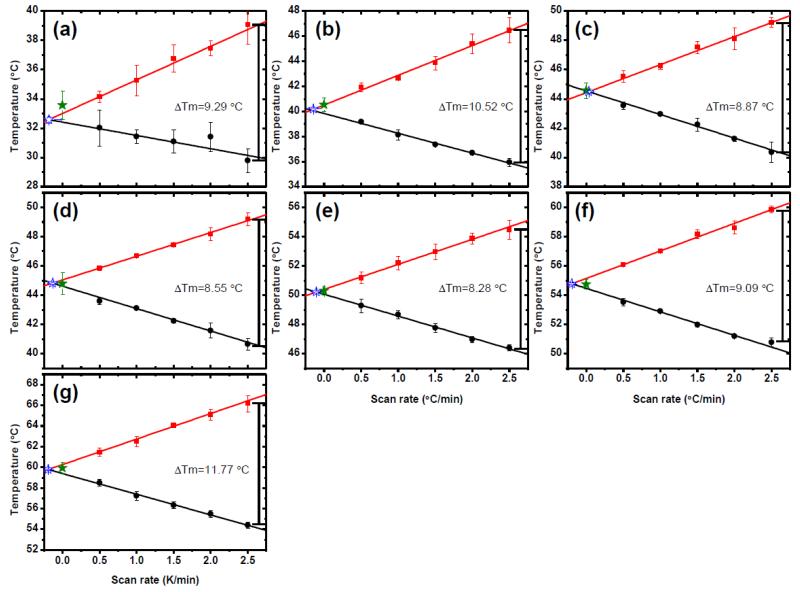
Fig. 3. Scan rate dependence of thermal unfolding/refolding.
(a) AL-103 H92D, (b) AL-103, (c) AL-103 P100Q, (d) AL-103 H92D-I34N, (e) AL-103 I34N, (f) κI O18/O8 and (g) AL-103 del95aIns. Apparent Tm values calculated from unfolding experiments are shown in red, those corresponding to refolding experiments are shown in black. ΔTm was calculated as the difference between the apparent Tm from the unfolding transition- apparent Tm from the refolding transition from the fastest scan rate values for each protein. Tm values obtained from kinetic model analysis (filled star) are in agreement with values obtained from model-free analysis (open star) within the error. Continuous lines represent the best fit to a straight line (n=30).
To evaluate the effect of each restorative mutation, we fit the thermal unfolding curves at different scan rates using a simple kinetic model for scan rate dependence within a reversible two-state process (fitting procedure and equation deductions are described in detail in the supplementary material).32 In this model, heating and cooling curves at each scan rate were analyzed independently with a two-state irreversible model to obtain the temperature dependence of the forward and reverse rate constants for unfolding and refolding. The equilibrium constant was calculated from the ratio of these kinetic rate constants to obtain the thermodynamic stability at equilibrium.
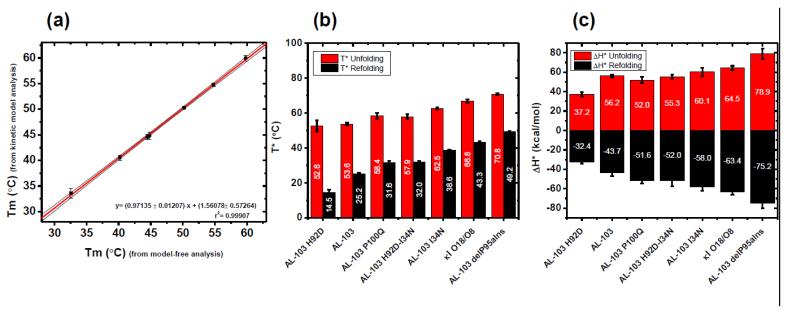
The goodness of fit for the unfolding/refolding transitions showed an average R2=0.9932 (values range from 0.9772-0.9992). Kinetic parameters were the activation enthalpy (ΔH‡) and the temperature (T*) where the rate constant for folding and refolding=1 min−1(see supplemental material). However, the quality of the fits does not, themselves, constitute a proof of the adequacy of the model. An additional test of consistency is based on the effect of the thermal scanning rate on the apparent melting temperature (apparent Tm) values. Figure 3 show a comparison between the thermal scan rate dependence of the apparent Tm values extrapolated to zero scan rate equilibrium (model-free analysis) and the Tm values obtained by fitting all thermal transitions using the kinetic two-state model. Data obtained from both procedures are in agreement (Fig. 4a) for all proteins tested. Such agreement constitutes strong evidence for the applicability of the kinetic model proposed, as it implies that the model accounts, not only for the transition fitting, but also for the hysteresis in unfolding/refolding transitions and the scan rate effect on them.
Fig. 4. Contribution of restorative mutants to the kinetic stability.
(a) Correlation between Tm estimate from the model-free analysis and the Tm values obtained from kinetic two-state model. Continuous lines represent the best fitting. The 95% confidence interval is shown in black thin lines. (b) T* and (c) ΔH‡ values for each protein characterized in this study. Labels in the bars show the average value (n=30). Values derived from unfolding experiments are shown in red, values from refolding experiments are shown in black.
The individual contributions of the restorative mutations on the kinetic stability, as a function of T* and ΔH‡ values are compared in Fig. 4b and 4c respectively. We observe that the values of T* for unfolding and refolding follows a pattern that is consistent with the stability of the proteins. The ΔH‡ values are identical in value and of opposite sign for the two different processes (folding and unfolding) as expected for a reversible process where unfolding is endothermic and refolding is exothermic. This suggests that the model is in agreement with a reversible folding process that is kinetically controlled. To the best of our knowledge, this is the first time that a reversible folding process that is kinetically controlled has been described for amyloidogenic light chains.
The T* and ΔH‡ values for unfolding and refolding from figure 4 are also provided on supplementary table 1. Tm values were obtained from both the model-free analysis and from the kinetic model analysis (supplementary Table 2). These results show that, although the thermal unfolding is not at equilibrium at the scan rate of 0.5°C/min, the Tm derived from the model-free analysis is equal to the Tm obtained from the kinetic model.
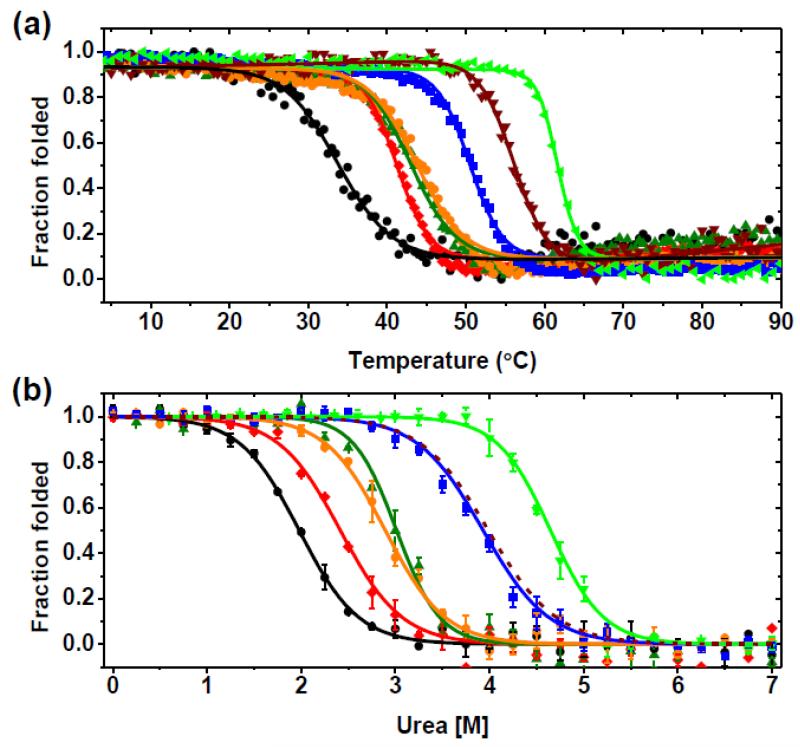
The most striking results were found for AL-103 H92D and AL-103 del95aIns. For AL-103 H92D, the change from Histidine to Aspartic acid at position 92 decreased the Tm value by 6.9°C (from 40.5°C to 33.6°C) compared to AL-103’s Tm value, while the deletion of the extra Proline after residue 95 in AL-103 del95aIns mutant showed a 19.4°C increment in the Tm value (from 40.5°C to 59.9°C) (Fig. 5a), resulting in a Tm value higher than the one for κI O18/O8. We previously reported that restoring the Asparagine at residue 34 on AL-09 has an halfway stabilizing effect with respect to κI O18/O8.4 The same effect was found on AL-103 I34N, with a Tm value of 50.3°C, very close to the Tm value of 48.6 °C obtained for AL-09 I34N. The double restorative mutant AL-103 H92D-I34N tested the combined effects of the destabilizing mutation H92D and the stabilizing mutation I34N. This double restorative mutant shows an increase in the Tm value slightly above the Tm for AL-103. The AL-103 P100Q restorative mutant also has a Tm value slightly above the Tm for AL-103 (Table 1). It should be noted that H92D, I34N, and P100Q are all located within the dimer interface (Fig. 1a). These results indicate that the differences in stability between AL-103 light chain and the non-amyloidogenic κI O18/O8 germline are a result of a combination of destabilizing (N34I, Q100P, delP95aIns) and protective somatic mutations (D92H). This is in contrast with the somatic mutations found in AL-09 where the conservative mutations were destabilizing or neutral.4 In our previous work, we reported that equilibrium chemical denaturation of AL-103 did not follow a two-state unfolding transition using either urea or guanidinium hydrochloride.28 In this study, we are using a slightly modified urea-induced unfolding protocol (see Materials and Methods for details) and have been able to demonstrate that the urea-induced unfolding followed by fluorescence showed monophasic reversible sigmoidal unfolding curves for κI O18/O8, AL-103, and all AL-103 mutant proteins (Fig. 5b).
Fig. 5. Thermal and urea-induced unfolding transitions.
(a) Thermal unfolding curves following molar ellipticity change at 217 nm. (b) Urea unfolding curves at 4°C following change in fluorescence intensity at 350nm. Experimental data from AL-103 (red) and AL-103 restorative mutants (AL-103 H92D, black; AL-103 H92D-I34N, olive green; AL-103 P100Q, orange; AL-103 I34N, blue; AL-103 del95aIns, green) (n=3). Continuous lines represent the best fit to a two-state model; see Table 1 for thermodynamic parameters. Data from κI O18/O8 (dark red) was calculated using thermodynamic parameters from Baden et al. 2008. 29
Table 1.
Thermodynamic parameters of kl O18/O8, AL-103 and AL-103 restorative mutants
| Protein | aTm (°C) (from kinetic bCm (M) model analysis) |
m (kcal/mol M) | dDGunfold (kcal/mol) |
|
|---|---|---|---|---|
| AL-103 H92D | 33.6 ± 1.0 | 1.98 | 1.75 ± 0.10 | 3.4 ± 0.2 |
| AL-103 | 40.5 ± 0.6 | 2.41 | 1.68 ± 0.13 | 4.0 ± 0.3 |
| AL-103 P100Q | 44.6 ± 0.5 | 2.99 | 1.29 ± 0.11 | 3.9 ± 0.3 |
| AL-103 H92D- I34N |
44.8 ± 0.7 | 3.14 | 2.21 ± 0.39 | 6.9 ± 1.5 |
| AL-103 I34N | 50.3 ± 0.3 | 3.93 | 1.59 ± 0.09 | 6.2 ± 0.3 |
| kI O18/O8c | 54.7 ± 0.3 | 3.98c | 1.53c | 6.1 ± 0.2 |
| AL-103 delP95aIns |
59.9 ± 0.5 | 4.62 | 1.73 ± 0.13 | 8.6 ± 0.3 |
| a n=30, error is the S.D |
c Values for kI O18/O8 are from Baden et at. (2008). The m-value for tis variant was calculated from those values. |
|||
| b The Cm values were calculate as the ratio DG/m |
d From chemical denaturation data at 4 °C |
|||
Urea-induced unfolding and refolding curves were adequately fitted using a two-state process, and thermodynamic parameters were obtained from fitting parameters (Fig. 5b and Table 1). Thermodynamic parameters obtained from urea unfolding experiments showed a similar trend like those derived from thermal unfolding experiments, except for AL-103 H92D-I34N. Even though the urea concentration at the midpoint of the transition (Cm) value for AL-103 H92D-I34N was similar to the one obtained for AL-103 P100Q, and the ΔGunfolding is within error of the one obtained for AL-103 I34N and κI O18/O8, there is a larger cooperativity (m value) observed in the AL-103 H92D-I34N unfolding transition (Table 1).
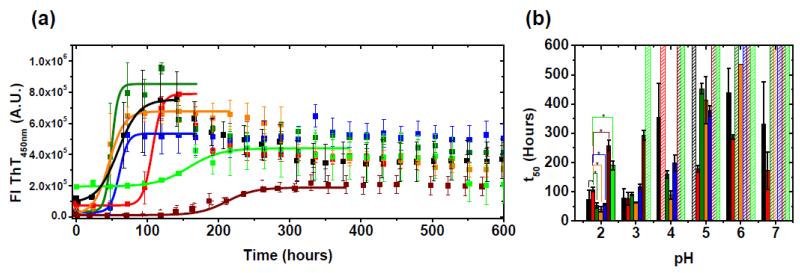
Figure 6 summarizes the results of the amyloid fibril formation reactions followed by Thioflavin T (ThT) fluorescence. Fibril formation was considered completed when ThT fluorescence enhancement reached a plateau. The kinetic traces of fibril formation were sigmoidal, with all proteins showing amyloid formation at pH 2 (Fig. 6a). All proteins, except AL-103 delP95aIns and κI O18/O8, form fibrils significantly faster than AL-103 (Fig. 6b). The correlation between low thermodynamic stability (measured at pH 7.4) and fast amyloid formation seems to start occurring at pH 3, although there is no observable amyloid formation for AL-103 delP95aIns from pH 3-11 after monitoring the reaction for 600 hours. The amyloid formation reactions for all proteins become slower and more stochastic as the pH of the reaction increases, until only the most destabilized proteins form amyloid fibrils at pH 7.0. None of the proteins form fibrils from pH 8-11.
Fig. 6. In vitro fibril formation assay.
(a) Average fibril formation kinetic traces of κI O18/O8 (dark red), AL-103 (red) and AL-103 restorative mutants (AL-103 H92D, black; AL-103 H92D-I34N, green olive; AL-103 P100Q, orange; AL-103 I34N, blue; AL-103 del95aIns, green) at pH 2. Average t50 values (mean ± SE) were obtained by fitting each triplicate to Boltzmann equation. Continuous lines represent the best fitting. (b) pH dependence of fibril formation kinetics. Absence of error bar indicates that only one triplicate reaction increased to three-fold ThT fluorescence (>200,000 A.U). *p-value<0.005 with respect to AL-103.
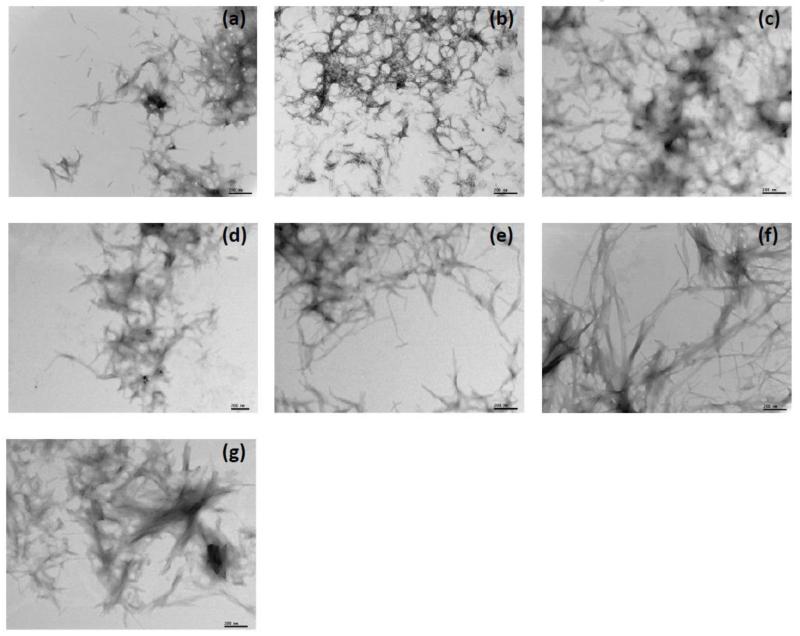
In vitro fibril formation at pH 2 was confirmed by electron microscopy (Fig. 7). Restorative mutants with the lowest thermodynamic stability (Fig. 7, a-d) formed short rods, associated in large tangles, while the most stable mutants (AL-103 I34N, κI O18/O8 and AL-103 del95aIns) formed long and unbranched fibrils with significant lateral association, similar to other AL protein fibrils previously characterized in our laboratory and others.6,25,29 In some cases, individual fibrils were difficult to discern (Fig. 7, e-g). Interestingly, fibril formation reactions for restorative mutants AL-103 P100Q and AL-103 del95aIns, mutants where proline residues were replaced or deleted, present spherical structures along with the mature fibrils (supplementary Fig. S4). Similar structures were found when AL-09 seeded reactions were performed in the presence of chondroitin sulfate-A.20 These spherical structures resemble the pre-amyloid species found in early stages of α-synuclein fibril formation.33 These results suggest that the spherical species were stabilized by the proline mutation/deletion and could be on-pathway intermediates that nucleate and assemble into amyloid fibrils.
Fig. 7. Electron microscopy images of fibrils formed at pH 2.
(a) AL-103 H92D. (b) AL-103. (c) AL-103 P100Q. (d) AL-103 H92D-I34N. (e) AL-103 I34N. (f) κI O18/O8 and (g) AL-103 del95aIns. Scale bars represent 200 nm.
One of the most outstanding structural features of AL-103 is the insertion of Proline 95a. As described previously,27,28 this insertion creates a di-proline motif with Proline 95 in trans conformation and Proline 95a in cis conformation. This insertion induces a structural deviation in the CDR3 region and the beginning of β-strand G in AL-103. Because AL-103 del95aIns exhibited the highest thermodynamic stability and presented delayed fibril formation compared to κI O18/O8 (Fig. 8), we wanted to know if the deletion of Proline 95a has a significant effect on the protein structure. We solved the structure of AL-103 delP95Ins at 2.87 Å resolution (Fig. 9). There are 10 molecules in the unit cell with a dimer as a relevant biological assembly. The structural alignment of the crystal structures of κI O18/O8, AL-103, and the new structure obtained in this study, AL-103 delP95Ins showed that all three proteins have the characteristic immunoglobulin fold and crystallized with the same canonical dimer geometry, with no differences in the dimer interface. As we published previously 28, the main structural deviations found with the AL-103 structure center around the CDR3 region, where the Pro95aIns mutation is located. RMS comparison via Cα backbone superposition shows that AL-103 shares more structural similarities with κI O18/O8 than AL-09. We had previously reported that the AL-103 CDR3 region appears to be a dynamic region based on the poor electron density data. Our current crystallographic data on AL-103 delP95Ins also presents with poor supporting density on that region, suggesting that this mutant presents the same behavior as its parent protein AL-103.The remaining Proline 95 adopts a cis conformation as in the κI O18/O8 structure.
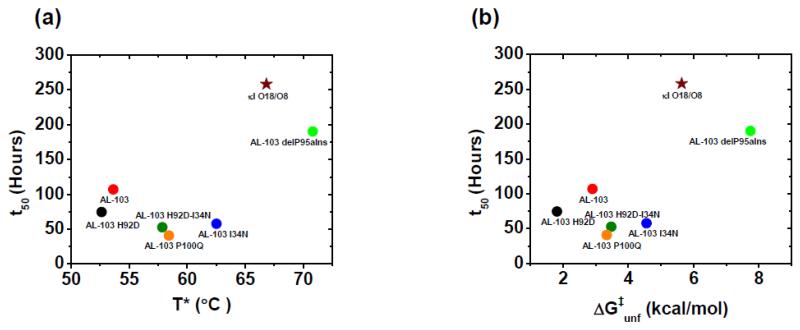
Fig. 8. Correlation of fibril formation vs. protein stability of AL-103 restorative mutants.
(a) T* value vs t50 value (hours). (b) ΔG‡unf (kcal/mol) at 37°C vs rates of fibril formation or t50 value (hours). t50 value where obtained from fibril formation kinetics at pH 2, 37°C.
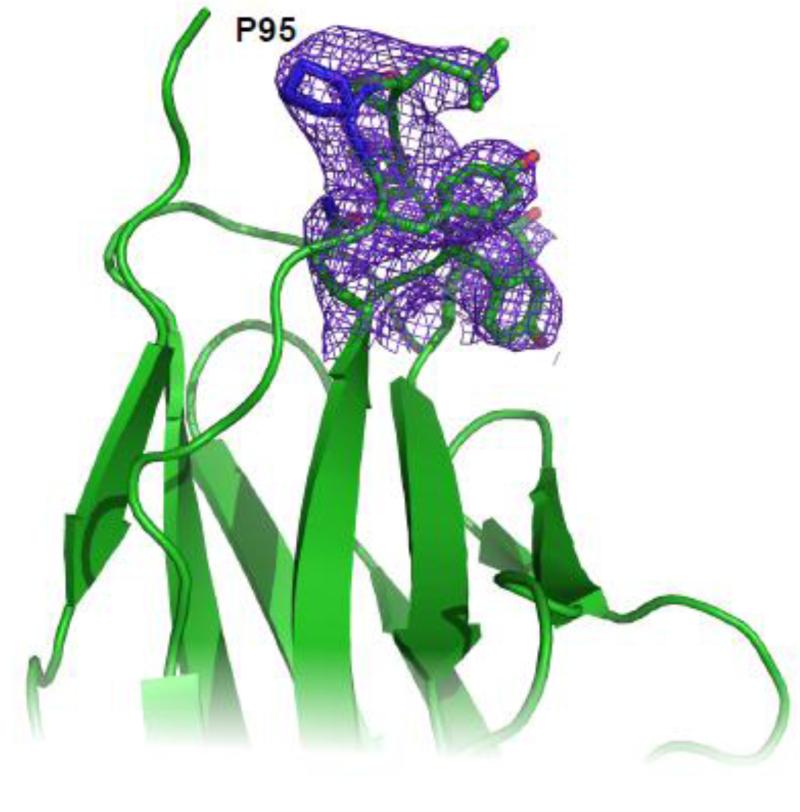
Fig. 9. Electron density for AL-103 del P95aIns CDR3 region.
Electron density map (2Fo-Fc at 1σ contouring) for the Proline 95 region of AL-103 del P95aIns. P95 is labeled.
Discussion
We previously reported a systematic mutational analysis of the non-conservative mutations of AL-09,4 where a correlation between thermodynamic protein stability and the kinetics of amyloid fibril formation among the non-conservative mutations of AL-09 was found. To understand if this correlation between protein stability and amyloid formation kinetics is a general feature in AL amyloidosis, here we characterized four single and one double AL-103 restorative mutants.
Our CD thermal unfolding data revealed that the seven proteins presented hysteresis for the unfolding and refolding transitions (a signature of kinetically controlled folding process). The refolding processes were completely reversible.
Usually it is assumed that the protein stability equals to thermodynamic stability - related to the energetic difference between the native and unfolded states-. However, if the folding reaction is under kinetic control, kinetic stability -related to a high free-energy barrier “separating” the native state from the non-functional forms (unfolded states, irreversibly-denatured protein)- is more relevant than thermodynamic stability. 34 Our study with AL-103 restorative mutants showed that rather than an equilibrium two-state model, thermal transitions for these proteins could be adequately described by a fully kinetic two-state model; where the rate constant for unfolding and refolding reactions are explicitly considered as first-order reactions. This approach has a strong theoretical foundation. 32,35 A similar procedure has been successfully applied to obtain kinetic and thermodynamic parameters from scan rate dependent experimental data on collagen. 36 Hence it is possible to obtain the true (thermodynamic) Tm values, as well as meaningful associated thermodynamic parameters, as ΔH‡, from scan rate-dependent unfolding transitions.
According to our model, to obtain equilibrium transitions curves, unfolding experiments should be carried out at a scan rate of ≤ 0.1°C/min (Fig. 3). Tm values obtained using the two-state kinetic model (40.5±0.6°C for AL-103; 54.7±0.3°C for κI O18/O8) agree within error with Tm values previously reported (41.6±0.5°C for AL-103; 56.1±0.2°C for κI O18/O8) 28,29 and indicate that, although thermal transitions obtained at 0.5°C/min are not at equilibrium, the scan rate is slow enough to obtain reliable thermodynamic Tm values. Such an agreement implies that the model accounts, not only to fit individual unfolding/refolding transitions, but also for the hysteresis and the scan rate effect on them.
Urea-induced unfolding experiments were performed with samples incubated for 24 h at 4°C with specific concentrations of urea, allowing each sample to reach equilibrium. This would explain why we did not observe any hysteresis in our chemical denaturation results.
Analysis of the thermodynamic parameters from chemical and thermal unfolding experiments of AL-103 restorative mutants show that the presence of Histidine in position 92 has a stabilizing effect on AL-103 with respect to κI O18/O8, while Asparagine in position 34 shows the same destabilizing effect observed in AL-09. 4
Deletion of Proline 95a (AL-103 delP95aIns), which is located in the CDR3 loop, causes a dramatic increment of thermodynamic stability, even beyond the κI O18/O8 value. This is an unexpected result because the CDR3 loop is known for its hypervariability in both length and sequence, commonly associated with a high structural plasticity. This striking result suggests that the presence of the di-proline motif, due to the Proline 95a insertion, results in an unfavorable contribution to the thermodynamic stability of AL-103, which leads us to think that this could be a key region in κI light chain stability.
The AL-103 crystal structure reveals that the insertion of Proline 95a (P95aIns) creates a diproline motif with proline 95 in trans conformation and Proline 95a in cis conformation.27,28 Some studies have proposed that the cis Proline residue at position 95 within the variable domain plays a rate limiting step role for the folding of variable fragments and single chain variable fragments.37-40
Rather than affecting the equilibrium thermodynamic stability, Proline residues are widely recognized for playing a special role in the kinetic folding and unfolding pathway of globular proteins. However, it is currently unclear if the presence of a particular cis/trans Proline conformation in the native state has a favorable or unfavorable effect on thermodynamic stability.
Fibril formation is pH dependent for AL-103 and its restorative mutants; lowering the pH in the solution accelerates the fibril formation reaction for all proteins with fairly similar time at which the fibril formation reaction is 50% complete (t50) values. Figure 8 shows two plots of thermodynamic parameters (T* and ΔG‡unf) as a function of the t50 values of fibril formation at pH 2. It is worth noting that although AL-103 restorative mutants show some correlation between thermodynamic stability with amyloid fibril formation kinetics from pH 3-7, fibril formation reactions at these conditions become more stochastic as the pH increases and only the most destabilizing proteins form fibrils at pH 7, which hampers the ability to make quantitative comparisons at physiological pH. In addition, AL-103 delP95aIns is unable to form fibrils above pH 2 and κI O18/O8 is unable to form fibrils above pH 3.
The results do not show the same correlation found for AL-09 30. We propose that in addition to the overall thermodynamic stability (clearly playing a dominant role with the most stable proteins such as κI O18/O8), kinetic stability (possibly influenced by the presence of a di-proline motif) may influence the AL-103 amyloid fibril formation process. The rates of fibril formation of these light chains are much slower occurring over many hours and days than the rates of denaturation obtained from thermal scans. So, although the kinetic stability against unfolding is correlated to the kinetics of fibril formation as shown in Figure 8, these two processes are separate reactions and whether they are linked functions remains to be determined.
As we discussed before, the scan rate dependency and hysteresis observed in the thermal unfolding/refolding transitions of AL-103 and its restorative mutants indicate that the process is under kinetic control.31 This result leads to the speculation that the presence of cis Proline (either in the di-proline motif in AL-103 and restorative mutants or as a cis Proline in the deletion mutant delPro95aIns) could enhance the population of a transient kinetic intermediate prone to fibril formation, as has been proposed previously.17,24,40
As was mentioned above, AL-09 and AL-103 have more than 90% sequence identity, and they share the N34I mutation and have similar thermodynamic stability, however, the clinical phenotypes of these proteins differed significantly. Patient AL-09 suffered an aggressive cardiac amyloidosis, surviving just months after diagnosis whereas patient AL-103, which showed a prominent cardiac and tongue involvement, survived almost 3 years.
In previous work, we reported that the differences observed between AL-09 (fast fibril formation kinetics as measured with the t50 values) and AL-103 (slow, very stringent conditions that allow fibril formation) correlates very well with the relative aggressiveness of the disease related with these two proteins in vivo, when monomeric species are enriched in the protein solution at the beginning of the fibril formation reaction.25 However, in a more recent work from our laboratory,27 we found that if the protein solutions are enriched with dimeric species; the differences in the t50 values of AL-09 and AL-103 are minimal and not statistically significant. Results reported here display fibril formation behavior similar to the results reported by Martin et al. previously.25 In this study, we removed preformed aggregates using the ultracentrifugation protocol reported by DiCostanzo et al. 27 enriching for dimeric species. The only difference is that we incubated the samples at 4°C for 24 hours prior to the initiation of the fibril formation reactions, while the protein was used immediately after the ultracentrifugation step in the study by DiCostanzo et al. In this study, we identified that protein folding is kinetically controlled for AL-103 and its mutants. It is possible that ultracentrifugation prior to fibril formation caused a disturbance in the equilibrium conditions that were not restored in the DiCostanzo study but were allowed to be restored in this study. What is very clear with this study and our previous reports is that the initial state of the species present at the beginning of the reaction plays a major role in the kinetics of fibril formation for these proteins. Further characterization of the species found after these steps will be conducted in order to understand how the protein is affected by these processes.
Collectively, our results lead us to conclude that not only thermodynamic, but also kinetic instability plays a major role in AL-103 amyloid fibril formation.
AL-103 and its restorative mutants are therefore, excellent candidates for protein folding kinetics studies aiming to determine their kinetic folding pathway. We hope that future kinetic studies will help us elucidate additional information that will shed light into the exact molecular mechanism of light chain amyloid formation that together with cytotoxicity studies will help us identify species that can be possible therapeutic targets for AL amyloidosis.
Materials and Methods
Chemicals
Water was Milli-Q grade. Yeast extract and tryptone were from Difco. Other reagents were from Sigma-Aldrich.
Cloning, Expression, Extraction and purification of recombinant κI O18/O8, AL-103 and AL-103 mutant proteins
The AL-103 variable domain sequence was previously obtained 28 from a patient who presented heart, liver, and tongue AL deposits (sequence deposited in GenBank with the accession code AY701640). Because the κI O18/O8 (IGKV 1-33) protein is not expressed naturally, the κI O18/O8 germline DNA was previously generated 4 by mutating the cDNA of AL-103 (sequence deposited under GenBank accession number EF640313), which differs from the germline by only 4 codons. Restorative AL-103 mutants were generated by using the QuikChange® Multi Site-directed Mutagenesis kit (Stratagene, La Jolla, CA). The Mayo Clinic DNA Sequencing Core facility confirmed the mutagenesis. Recombinant κI O18/O8, AL-103, and restorative AL-103 mutants proteins were expressed in Escherichia coli and purified as described previously.4,28 Briefly, all plasmids were transformed into Escherichia coli BL21 (DE3) Gold competent cells (Stratagene, La Jolla, CA) and protein expression was induced with 0.8 mM IPTG (isopropyl-β-D-thiogalactopyranoside) at an A600 nm (O.D.)= 0.7. After 17-20 h of post induction growth, the bacteria were collected, pelleted, and frozen.
The over expressed κI O18/O8 protein was extracted from the periplasmic space of the bacteria by breaking the cells through one freeze-thaw cycle using phosphate-buffered saline (PBS). The periplasmic fraction was then dialyzed against 10mM Tris HCl 0.02% NaN3, pH 7.4. AL-103, AL-103 H92D, AL-103 P100Q, and AL-103 del95aIns were found in inclusion bodies. After overnight growth, cells were harvested, most of the time; the pellets were frozen at −20°C. The thawed cell pellet was resuspended into 10 mM Tris buffer, and lysed by ultrasonication. Proteins were extracted from solubilized inclusion bodies using 6.0 M urea and immediately dialyzed overnight against 10 mM Tris-HCl (pH 7.4) to remove the urea. AL-103 I34N and AL-103AL-103 H92D/I34N proteins were obtained from both inclusion bodies and the periplasmic space fraction.
All protein extracts were injected into a HiLoad 16/60 Superdex® 75 column on an AKTA FPLC (GE Healthcare, Piscataway, NJ) system. Protein purity was determined by SDS-PAGE. Protein concentration was determined by UV absorption using an extinction coefficient ε=14,890 M−1 cm−1 calculated from the amino acid sequence. Pure fractions were combined, concentrated, flash frozen, and stored at −80°C. Proteins were thawed at 4°C, filtered and/or ultracentrifuged before they were used for each study.
Analytical size-exclusion chromatography
In order to determine the oligomeric state of the purified protein, analytic size-exclusion chromatography was carried out at 4°C using a BioSil® 125-5 HPLC (Bio-Rad,) size exclusion column on an AKTA FPLC (GE Healthcare). The column was equilibrated with 50 mM Na2HPO4, 50 mM NaH2PO4, 150 mM NaCl, and 0.02% NaN3 at pH 6.8. Chromatographic analyses were carried out at 0.2 mL/min using the same buffer as elution buffer. 200 μL of purified proteins at 20 μM were injected onto the column, and chromatographic peaks were detected by UV absorbance at 280 nm. Molecular weight and oligomeric states were estimated from elution volumes using a molecular weight calibration curve as reported before.24,28 All proteins studied under these experimental conditions were predominantly monomeric at 20 μM. This allowed us to use analytical size-exclusion chromatography as a quality control measure before spectroscopic and fibril formation experiments (see supplementary Fig. S3).
Spectroscopic characterization of restorative mutants
Circular dichroism (CD) spectroscopy was used to determine the global secondary structure after protein extraction/purification as described previously.27 Briefly, far UV-CD spectra from 260-200 nm (1 nm bandwidth) were acquired at 4°C, on a Jasco Spectropolarimeter 810 (JASCO, Inc., Easton, MD) using a 0.2 cm path-length quartz cuvette. All samples (20 μM) were prepared in 10 mM Tris pH 7.4. Temperature was regulated within ±0.002 °C using a Peltier system. Immunoglobulin light chains share many structural features, including a disulfide bridge, which is buried in the hydrophobic core and often in close proximity to a conserved Tryptophan residue2,3,40. The fluorescence of the conserved Tryptophan residue 35 is quenched in the native state by the adjacent disulfide bond, so it can be used as a ‘reporter group’ for the changes in conformational state of immunoglobulin light chains41. Intrinsic Tryptophan fluorescence measurements were carried out using a PTI spectrofluorometer (Photon Technology International, Lawrenceville, NJ) using a quartz 1 cm path-length micro-cuvette (150 μL). Samples were excited at 295 nm (0.5 mm slit); the emission spectra were collected in the 310- to 410-nm range (1 mm slit). Temperature was regulated within ±0.2 °C using a Peltier system.
Thermal unfolding
Thermal unfolding/refolding experiments were carried out following the ellipticity at 217 nm over a temperature range of 4-90°C to determine the melting temperature (Tm) as reported previously 27. All proteins exhibited hysteresis in the thermal unfolding/refolding transitions. In order to evaluate the scan rate dependency of the Tm values, thermal unfolding experiments were carried out by employing thermal scan rates from 0.5 to 2.5 °C min−1.
The Tm values were calculated by two different procedures:
- Model-free analysis of the apparent Tm values (as shown on supplementary table 1): Thermal unfolding data from CD were converted to FN taking into account pre- and post-transition linear baselines. The apparent Tm values from spectroscopic data at each scan rate were calculated by nonlinear least-squares fitting to a two-state model between native (N) and unfolded (U) states:
where FN is the observed signal; yN and yU are the values for native and unfolded states, respectively; R is the ideal gas constant, T is temperature, Tm is the apparent temperature of the midpoint and ΔHm is the apparent enthalpy of the transition at the apparent Tm. The pre- and post-transition baselines are described by the linear slopes mN and mU, respectively. 24,42 The equilibrium values of Tm were determined empirically from linear extrapolation of the thermal scan rate dependence of the apparent Tm to zero scan rate.(Eq.1) - Kinetic reversible two-state analysis: For a reversible two-state thermal unfolding transition with hysteresis, the unfolding and refolding reactions are kinetically controlled by a first-order rate constant which varies with temperature according to the Arrhenius equation. We treated both heating and cooling transitions separately as an independent irreversible kinetic two-state model
(Eq.2) (Eq.3)
where N is the native state, U the unfolded state and k1 and k−1are the rate constants for the unfolding and refolding reactions respectively. For a complete description of this model and fitting procedure, please see the supplementary material.(Eq.4)
Urea-induced unfolding
In order to determine the thermodynamic stability of κI O18/O8 germline AL-103, and AL-103 restorative mutant proteins, chemical unfolding/refolding experiments using urea were carried out.
We previously reported that chemical denaturation of AL-103 did not follow a two-state unfolding transition using either urea or guanidinium hydrochloride28 using the protocol described in Baden et al., 2008 4. For that reason, in this work, 20 μM protein samples containing variable amounts of urea were incubated in 10 mM Tris pH 7.4 at 4°C for 24 hours before measurement. For the refolding experiment, a 400 μM protein stock was pre-incubated in 5 M urea, 10 mM Tris buffer, pH 7.4 at 4°C for 24 hours. Samples were refolded by dilution (20 μM) to different urea concentrations and were incubated at 4°C for 24 hours before measurement. Fluorescence measurements were made using a PTI spectrofluorometer, and temperature was regulated within ±0.02 °C using a Peltier system. Samples were excited at 295 nm (0.5 mm slit); the emission spectra were collected in the 310 to 410 nm range (1.0 mm slit). Fluorescence intensity data at 350 nm were plotted versus urea concentration.
Thermodynamic parameters from spectroscopic data were calculated by nonlinear least-squares fitting to a two-state model between native (N) and unfolded (U) states as reported before.24,43
| (Eq. 5) |
Where FN is the observed signal; yN and yU are the values for native and unfolded states, respectively; ΔG° is the free energy of unfolding in the absence of denaturant; [D] is urea concentration; m is δΔG/δ[D], R is the ideal gas constant and T is temperature. The pre- and post-transition baselines are described by the linear slopes mN and mU, respectively. Cm values were calculated as the ΔG/m ratio. When the chemical denaturation was followed by CD, we found out that unfortunately the change in far-UV CD signal at 217 nm was very small for the urea denaturation experiments to track changes in secondary structure (data not shown), in part because the urea contribution to the CD signal in far-UV region is too high at urea concentrations higher than 1.0 M urea. The data reported in this manuscript are derived from fluorescence experiments.
In vitro fibril formation assays
Proteins were thawed at 4°C and filtered using 0.45 μm membranes prior to these reactions. Once thawed, all proteins were ultracentrifuged at a speed of 90,000 rpm for 3.3 h (enrichment for dimeric species) in a NVT-90 rotor on an Optima L-100 XP centrifuge (Beckman Coulter). This step was carried out before fibril formation assays started to remove any preformed aggregates from soluble protein, as reported previously.27 Post-ultracentrifugation CD spectra were collected and thermal unfolding as well as size-exclusion chromatography were carried out to ensure the integrity and oligomeric state of the proteins before the initiation of the fibril formation reaction. In all cases, the major species found were monomeric In vitro fibrillogenesis reactions were carried out by monitoring the fluorescence intensity of ThT that is enhanced when ThT binds to amyloid fibrils. Samples of each filtered and ultracentrifuged protein (20 μM) were prepared in 10 mM sodium acetate, boric acid, and sodium citrate buffer containing 150 mM NaCl, 10 μM ThT, 0.02% NaN3 (260 mL final volume), at the appropriate pH value. All fibril formation assays were performed in triplicate using black 96-well polystyrene plates (Greiner, Monroe, NC) sealed with plate sealers (Nunc, Roskilde, Denmark), covered with a black polysterene cover and incubated at 37°C with continuous orbital shaking (300 rpm) in a New Brunswick Scientific Innova40 incubator shaker. Fibril formation was monitored daily for 1 month (~750 h) following fluorescence on a plate reader (Analyst AD, Molecular Devices, Sunnyvale, CA). The excitation wavelength used was 440 nm and the emission wavelength was 480 nm.27 The t50 value, or the time at which the fibril formation reaction is 50% complete was obtained by fitting each kinetic trace to a Boltzmann function as defined by the Origin software package (http://www.originlab.com/www/helponline/Origin/en/UserGuide/Boltzmann.html).
| (Eq. 6) |
Where A1 is the initial fluorescence value, A2 is the final fluorescence value, x0 is the center (t50 value), dx is defined as the time constant. A larger t50 value indicates a longer time required to form fibrils.
Electron Microscopy
A 3-μl fibril sample was placed on a 300 mesh copper formvar/carbon grid (Electron Microscopy Science, Hatfield, Pa), and excess liquid was removed. The samples were negatively stained with 2% uranyl acetate, washed twice with H2O, and air-dried. Grids were analyzed on a Philips Tecnai T12 transmission electron microscope at 80 kV (FEI, Hillsboro, OR).
X-ray Crystallography
Purified AL-103 del95aIns was concentrated to 13.4 mg/mL. AL-103 del95aIns crystals were obtained in hanging drops using vapor diffusion against 15% (w/v) polyethylene glycol 8000 and 443 mM ammonium nitrate in 1.0 M sodium Acetate buffer (pH 5.5) at 22 °C. Crystals were cryoprotected with a 10% glycerol solution in liquid N2. Diffraction data were collected at beamline 19BM (Structural Biology Consortium, Advanced Photon Source (APS) Argonne National Laboratory). The data sets were collected at 70 K. Diffraction patterns were processed with Crystal Clear and XDS. All structures were solved by molecular replacement with PHASER as implemented by the Phenix software package using the κI O18/O8 structure (Protein Data Bank code 2Q20) as a probe molecule. Phenix and Coot were used for structure refinement and model building. Structure was deposited into the Protein Data Bank with the code: 4K07 (AL-103 del95aIns).
Supplementary Material
Highlights.
Light chain amyloidosis presents light chain amyloid deposits in vital organs
Protein AL-103 shows kinetic control on the folding/unfolding process
Insertion of Proline in position 95a affects the folding and aggregation in AL-103
Kinetic stability plays a role in AL-103 amyloid fibril formation process
Table 2.
Data collection and refinement statistics (molecular replacement)
| Data collection | |
|---|---|
| Space group | P 21 21 2 |
| Cell dimensions a, b, c (Å) |
159.91, 200.33, 40.06 |
| a, b, g (°) | 90, 90, 90 |
| Resolution (Å) | 37.1-2.83 (3.05-2.83) |
| Unique reflections | 31753 (5672) |
| Wilson B-factor | 80.8 |
| Rmerge (%) | 7.8 (22.9) |
| I / sI | 21.9 (3.0) |
| Completeness (%) | 98 (90) |
| Redundancy | 6.2 (1.8) |
| Refinement | |
| Resolution (Å) | 37.1-2.83 (3.05-2.83) |
| Reflections used | 30878 |
| Rwork / Rfree (%) | 20.3/23.6 (27.2/29.5) |
| Atom Number Protein |
9351 |
| SO4 ion | 5 |
| Water | 33 |
| Ave. B-factors Protein |
91.6 |
| SO4 ion | 90.6 |
| Water | 62.5 |
| RMSD, Bond lengths (Å) |
0.006 |
| Bond angles (°) | 1.236 |
| MolProbity Clashscore |
14 |
| % Ramachandran Outliers |
0.09 |
| % Rotamer Outliers |
4.8 |
| * Highest resolution shell shown in parentheses. |
Acknowledgements
This work was supported by NIH grant R01 GM071514, the Mayo Foundation, and the generous contributions of amyloidosis patients and their families. We thank Diana Rommelfanger-Konkol for the initial thermodynamic characterization of the AL-103 mutants and Eric W. Mahlum for preliminary crystallographic work on AL-103 del95aIns.
Glossary
- t50 value
time at which the fibril formation reaction is 50% complete
- Amyloid fibrils
protein aggregates distinguished by their cross-β structure and their characteristic binding to Thioflavin T (in vitro)
- CDR
complementarity determining region; hypervariable region that determine specific antibody binding
- FRs
framework regions; The FR regions form a beta-sheet structure which serves as a scaffold to hold the HV regions in position to contact antigen.
- Restorative mutants
Mutant proteins which the one or more of the somatic mutations are restored to the amino acid residues found in the corresponding germline sequence.
Abbreviations
- AL amyloidosis
Light chain amyloidosis
- FRs
framework regions
- CDR
complementarity determining region
- ΔH‡
activation enthalpy
- T*
temperature where the rate constant for folding and refolding= 1 min−1
- t50 value
time at which the fibril formation reaction is 50% complete
- ΔG‡unf
activation energy of unfolding reaction
- k1
rate constant for the unfolding
- k−1
rate constant for the refolding
- ThT
Thioflavin T
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kumar SK, Gertz MA, Lacy MQ, Dingli D, Hayman SR, Buadi FK, et al. Recent improvements in survival in primary systemic amyloidosis and the importance of an early mortality risk score. Mayo Clin. Proc. 2011;86:12–18. doi: 10.4065/mcp.2010.0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke J, Cota E, Fowler SB, Hamill SJ. Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway. Structure. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Bork P, Holm L, Sander C. The Immunoglobulin Fold: Structural Classification, Sequence Patterns and Common Core. J. Mol. Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 4.Baden EM, Randles EG, Aboagye AK, Thompson JR, Ramirez-Alvarado M. Structural insights into the role of mutations in amyloidogenesis. J. Biol. Chem. 2008;283:30950–30956. doi: 10.1074/jbc.M804822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung CM, Chiu JD, Connors LH, Gursky O, Lim A, Dykstra AB, et al. Thermodynamic stability of a kappaI immunoglobulin light chain: relevance to multiple myeloma. Biophys. J. 2005;88:4232–4242. doi: 10.1529/biophysj.105.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Pozo Yauner L, Ortiz E, Sanchez R, Sanchez-Lopez R, Guereca L, Murphy CL, et al. Influence of the germline sequence on the thermodynamic stability and fibrillogenicity of human lambda 6 light chains. Proteins. 2008;72:684–692. doi: 10.1002/prot.21934. [DOI] [PubMed] [Google Scholar]
- 7.Helms LR, Wetzel R. Specificity of abnormal assembly in immunoglobulin light chain deposition disease and amyloidosis. J. Mol. Biol. 1996;257:77–86. doi: 10.1006/jmbi.1996.0148. [DOI] [PubMed] [Google Scholar]
- 8.Hurle MR, Helms LR, Li L, Chan W, Wetzel R. A role for destabilizing amino acid replacements in light-chain amyloidosis. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5446–5450. doi: 10.1073/pnas.91.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens PW, Raffen R, Hanson DK, Deng Y-L, Berrios-Hammond M, Westholm FA, et al. Recombinant immunoglobulin variable domains generated from synthetic genes provide a system for in vitro characterization of light-chain amyloid proteins. Protein Sci. 1995;4:421–432. doi: 10.1002/pro.5560040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall J, Schell M, Murphy C, Hrncic R, Stevens FJ, Solomon A. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 11.Wall JS, Gupta V, Wilkerson M, Schell M, Loris R, Adams P, et al. Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four Vlambda6 proteins. J. Mol. Recognit. 2004;17:323–331. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- 12.Wetzel R. Domain stability in immunoglobulin light chain deposition disorders. Adv. Protein Chem. 1997;50:183–242. doi: 10.1016/s0065-3233(08)60322-8. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Wall JS, Meyer J, Murphy C, Randolph TW, Manning MC, et al. Thermodynamic modulation of light chain amyloid fibril formation. J. Biol. Chem. 2000;275:1570–1574. doi: 10.1074/jbc.275.3.1570. [DOI] [PubMed] [Google Scholar]
- 14.Raffen R, Dieckman LJ, Szpunar M, Wunschl C, Pokkuluri PR, Dave P, et al. Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci. 1999;8:509–517. doi: 10.1110/ps.8.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YS, Cape SP, Chi E, Raffen R, Wilkins-Stevens P, Stevens FJ, et al. Counteracting effects of renal solutes on amyloid fibril formation by immunoglobulin light chains. J. Biol. Chem. 2001;276:1626–1633. doi: 10.1074/jbc.M007766200. [DOI] [PubMed] [Google Scholar]
- 17.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, et al. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 18.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Effect of association state and conformational stability on the kinetics of immunoglobulin light chain amyloid fibril formation at physiological pH. J. Biol. Chem. 2002;277:12657–12665. doi: 10.1074/jbc.M109230200. [DOI] [PubMed] [Google Scholar]
- 19.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Elucidation of the molecular mechanism during the early events in immunoglobulin light chain amyloid fibrillation. Evidence for an off- pathway oligomer at acidic pH. J. Biol. Chem. 2002;277:12666–12679. doi: 10.1074/jbc.M109229200. [DOI] [PubMed] [Google Scholar]
- 20.McLaughlin RW, De Stigter JK, Sikkink LA, Baden EM, Ramirez-Alvarado M. The effects of sodium sulfate, glycosaminoglycans, and Congo red on the structure, stability, and amyloid formation of an immunoglobulin light-chain protein. Protein Sci. 2006;15:1710–1722. doi: 10.1110/ps.051997606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikkink LA, Ramirez-Alvarado M. Biochemical and aggregation analysis of Bence Jones proteins from different light chain diseases. Amyloid. 2008;15:29–39. doi: 10.1080/13506120701815324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys. Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng X, Fink AL, Uversky VN. The effect of membranes on the in vitro fibrillation of an amyloidogenic light-chain variable-domain SMA. J. Mol. Biol. 2008;381:989–999. doi: 10.1016/j.jmb.2008.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blancas-Mejia LM, Tellez LA, del Pozo-Yauner L, Becerril B, Sanchez-Ruiz JM, Fernandez-Velasco DA. Thermodynamic and Kinetic Characterization of a Germ Line Human λ6 Light-Chain Protein: The Relation between Unfolding and Fibrillogenesis. J. Mol. Biol. 2009;386:1153–1166. doi: 10.1016/j.jmb.2008.12.069. [DOI] [PubMed] [Google Scholar]
- 25.Martin DJ, Ramirez-Alvarado M. Comparison of amyloid fibril formation by two closely related immunoglobulin light chain variable domains. Amyloid. 2010;17:129–136. doi: 10.3109/13506129.2010.530081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin DJ, Ramirez-Alvarado M. Glycosaminoglycans promote fibril formation by amyloidogenic immunoglobulin light chains through a transient interaction. Biophys. Chem. 2011;158:81–89. doi: 10.1016/j.bpc.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiCostanzo AC, Thompson JR, Peterson FC, Volkman BF, Ramirez-Alvarado M. Tyrosine residues mediate fibril formation in a dynamic light chain dimer interface. The Journal of biological chemistry. 2012;287:27997–28006. doi: 10.1074/jbc.M112.362921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randles EG, Thompson JR, Martin DJ, Ramirez-Alvarado M. Structural alterations within native amyloidogenic immunoglobulin light chains. J. Mol. Biol. 2009;389:199–210. doi: 10.1016/j.jmb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. Altered dimer interface decreases stability in an amyloidogenic protein. J. Biol. Chem. 2008;283:15853–15860. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson FC, Baden EM, Owen BA, Volkman BF, Ramirez-Alvarado M. A single mutation promotes amyloidogenicity through a highly promiscuous dimer interface. Structure. 2010;18:563–570. doi: 10.1016/j.str.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasshauer D, Antonin W, Subramaniam V, Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 32.Tischer A, Cruz MA, Auton M. The linker between the D3 and A1 domains of vWF suppresses A1-GPIbα catch bonds by site specific binding to the A1 domain. Protein Sci. 2013;22:1049–1059. doi: 10.1002/pro.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fauerbach Jonathan A., Yushchenko Dmytro A., Shahmoradian Sarah H., Chiu W, Jovin Thomas M., Jares-Erijman Elizabeth A. Supramolecular Non-Amyloid Intermediates in the Early Stages of α-Synuclein Aggregation. Biophys. J. 2012;102:1127–1136. doi: 10.1016/j.bpj.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Ruiz JM. Protein kinetic stability. Biophys. Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Lepock JR, Ritchie KP, Kolios MC, Rodahl AM, Heinz KA, Kruuv J. Influence of transition rates and scan rate on kinetic simulations of differential scanning calorimetry profiles of reversible and irreversible protein denaturation. Biochemistry. 1992;31:12706–12712. doi: 10.1021/bi00165a023. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno K, Boudko SP, Engel J, Bachinger HP. Kinetic hysteresis in collagen folding. Biophys. J. 2010;98:3004–3014. doi: 10.1016/j.bpj.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freund C, Honegger A, Hunziker P, Holak TA, Plückthun A. Folding Nuclei of the scFv Fragment of an Antibody†. Biochemistry. 1996;35:8457–8464. doi: 10.1021/bi952764a. [DOI] [PubMed] [Google Scholar]
- 38.Jäger M, Plückthun A. The rate-limiting steps for the folding of an antibody scFv fragment. FEBS Lett. 1997;418:106–110. doi: 10.1016/s0014-5793(97)01350-1. [DOI] [PubMed] [Google Scholar]
- 39.Jager M, Pluckthun A. Folding and assembly of an antibody Fv fragment, a heterodimer stabilized by antigen. J. Mol. Biol. 1999;285:2005–2019. doi: 10.1006/jmbi.1998.2425. [DOI] [PubMed] [Google Scholar]
- 40.Simpson ER, Herold EM, Buchner J. The folding pathway of the antibody V(L) domain. J. Mol. Biol. 2009;392:1326–1338. doi: 10.1016/j.jmb.2009.07.075. [DOI] [PubMed] [Google Scholar]
- 41.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem. Sci. 2010;35:189–198. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eftink MR. Use of multiple spectroscopic methods to monitor equilibrium unfolding of proteins. Methods Enzymol. 1995;259:487–512. doi: 10.1016/0076-6879(95)59058-7. [DOI] [PubMed] [Google Scholar]
- 43.Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988;27:8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.