Roe and Vakoc discuss the role of C/EBPα in the initiation of acute myeloid leukemia.
Abstract
Acute myeloid leukemia (AML) is a hematopoietic malignancy characterized by clonal expansion of myeloid progenitor cells. A major mechanistic theme in AML biology is the extensive collaboration among fusion oncoproteins, transcription factors, and chromatin regulators to initiate and sustain a transformed cellular state. A new study in this issue describes how the C/EBPα transcription factor is crucial for the initiation of AML induced by MLL fusion oncoproteins, but is entirely dispensable for the maintenance of established disease. These observations provide a unique glimpse into the pioneer round of regulatory events that are critical at the origin of AML formation. Furthermore, this study implies the existence of oncogene-induced positive feedback loops capable of bypassing the continuous need for certain regulators to propagate disease.
Acute myeloid leukemia (AML) is a blood cancer characterized by uncontrolled proliferation of myeloid progenitors that exhibit a severe block in their ability to differentiate into mature granulocytes or macrophages. A critical event in AML initiation is the acquisition of aberrant self-renewal characteristics in leukemia stem cells (LSCs), a cell population that has been associated with disease propagation and the evolution of therapy-resistance. The estimated 5 yr survival rate among AML patients is ∼30%, with mortality being largely attributed to the emergence of chemotherapy resistance. Deregulated transcriptional pathways feature prominently in the genetic etiology of AML, which can occur in the form of gain- or loss-of-function mutations in lineage-specific transcription factors, histone modifying enzymes, and DNA methylation machineries (Rosenbauer and Tenen, 2007; Shih et al., 2012). Of clinical relevance, pharmacological targeting of transcriptional regulatory proteins represents a validated approach for undermining the driver oncogenes in certain subtypes of AML, as exemplified by the clinical success of using all-trans retinoic acid and arsenic trioxide to inactivate fusion oncoproteins involving the retinoic acid receptor (Wang and Chen, 2008; Ablain et al., 2013).
One important genetic subtype of AML is defined by rearrangements of the mixed lineage leukemia (MLL) gene, which is found in ∼5% of patients (Krivtsov and Armstrong, 2007). MLL encodes a histone-modifying enzyme belonging to the SET1 family of H3 lysine 4 (H3K4) methyltransferases, which often function as transcriptional coactivators (Shilatifard, 2012). In AML, chromosomal translocations can involve MLL and various other genes, of which >50 partner loci have been described to date (Krivtsov and Armstrong, 2007). Some of the most common translocation partners of MLL include AF9, ENL, AF4, AF6, AF10, and ELL. The in-frame fusion of the N terminus of MLL with the C terminus of its fusion partner leads to the formation of a potent oncoprotein (e.g., MLL-ENL) that can efficiently transform hematopoietic stem and progenitor cells. This genetic driver that has been widely used to generate animal models of leukemia for studying all aspects of disease biology, including LSC specification and chemotherapy resistance (Krivtsov et al., 2006; Somervaille and Cleary, 2006; Zuber et al., 2009). MLL-rearranged leukemia is considered a poor/intermediate prognosis subtype of leukemia for which targeted therapies have yet to make any clinical impact (Krivtsov and Armstrong, 2007).
Mechanistically, MLL fusion proteins form hybrid multisubunit protein complexes that include factors associated with the MLL N-terminal fragment (Menin, LEDGF, and PAF1) and factors associated with the fusion partner C-terminal fragment (Mohan et al., 2010). Remarkably, many of the common C-terminal fusion partners associate with one another in a complex that includes the protein kinase pTEFb and the histone methyltransferase DOT1L, which act as effectors to promote transcription elongation (Okada et al., 2005; Lin et al., 2010; Yokoyama et al., 2010). Because MLL fusion proteins lack the domains required for catalysis of H3K4 methylation, they instead rely on the C-terminal fusion partner to recruit pTEFb and DOT1L to promote transcriptional activation of their target genes (Okada et al., 2005). Among the well-validated direct targets of MLL fusion proteins are genes encoding homeodomain-containing transcription factors, including Hoxa9 and Meis1 (Armstrong et al., 2002). Forced co-overexpression of Hoxa9 and Meis1 can itself efficiently transform hematopoietic cells and form leukemias that resemble those initiated by MLL fusion proteins (Kroon et al., 1998). In addition to Hoxa9/Meis1, MLL fusion proteins also activate the expression of numerous additional transcription factor–encoding genes, such as Myc, Myb, and Mef2c, which are crucial for disease maintenance (Krivtsov et al., 2006; Zuber et al., 2011a). Furthermore, MLL fusion leukemia cells are also dependent on chromatin regulatory activities to maintain a transformed cellular state, such as the histone demethylase LSD1 and the BET protein BRD4 (Zuber et al., 2011b; Harris et al., 2012). Thus, MLL-rearranged leukemia is a paradigm for understanding how a vast network of transcriptional regulators cooperates to sustain an aberrant lineage program. A new study in this issue of the JEM, Ohlsson et al., demonstrates an essential collaboration between MLL fusion proteins and the transcription factor C/EBPα during leukemogenesis. Interestingly, C/EBPα is critical for the initiation of MLL fusion leukemia but becomes dispensable once the disease is fully established.
Function of C/EBPα in normal and malignant myelopoiesis
The CCAAT/enhancer-binding protein-α (C/EBPα) is a lineage-specific transcription factor in the hematopoietic system that is required for the formation of committed myeloid progenitors from multipotent precursor cells (Rosenbauer and Tenen, 2007). C/EBPα executes this function by coupling the direct transcriptional activation of myeloid-specific genes with the arrest of cell proliferation (Nerlov, 2004). C/EBPα belongs to a subfamily of basic-region leucine zipper (BR-LZ) transcription factors, several of which are also expressed in the myeloid lineage (e.g., C/EBPβ and C/EBPε; Rosenbauer and Tenen, 2007). C/EBPα is an intronless gene whose mRNA can be translated from two different AUG codons to give rise to two distinct isoforms (p42 and p30). p30 lacks two N-terminal trans-activation domains that are only present on p42. Notably, this unique N-terminal region of p42 can interact with and inhibit E2F transcription factors (Slomiany et al., 2000; Porse et al., 2001). Hence, only the p42 isoform of C/EBPα can promote proliferation arrest. p30 retains the BR-LZ and a third trans-activation domain and can promote myeloid lineage specification, but lacks the capacity to promote terminal differentiation and proliferation-arrest (Kirstetter et al., 2008). The relative levels of p42 and p30 in the cell can be regulated at the level of translation initiation through mTOR signaling to control cell fate transitions (Calkhoven et al., 2000). Notably, a genetic knockout C/EBPα (p30 and p42) results in a complete block in the transition from common myeloid progenitor (CMP) to the granulocyte/monocyte progenitor (GMP) stage of differentiation and leads to enhanced self-renewal of hematopoietic stem cells, highlighting the essential function of C/EBPα for normal hematopoiesis (Zhang et al., 2004).
Genetic alteration of C/EBPα is a major etiology of human AML, with a mutational frequency ∼10% across this disease (Pabst et al., 2001b). C/EBPα mutations fall into two major categories: those that abolish p42 protein expression while retaining p30 and insertions/deletions near the BR-LZ region that compromise DNA binding, which may result in dominant-negative homodimers or heterodimers with other C/EBP family members (Nerlov, 2004). Interestingly, these two classes of mutations often co-occur in a biallelic manner in AML patients. C/EBPα expression can also be down-regulated through direct transcriptional repression by fusion oncoproteins, such as AML1-ETO (Pabst et al., 2001a). A selective knockout of p42 in mice (while preserving p30 expression) leads to AML formation with complete penetrance, confirming the p42 isoform of C/EBPα as a myeloid tumor suppressor gene (Kirstetter et al., 2008). Null mutations of C/EBPα that abolish both p42 and p30 are not observed in human AML, and mice with a complete knockout of p42/p30 fail to develop AML, suggesting that residual C/EBPα function (presumably via the p30 isoform) is necessary to form myeloid lineage leukemias (Zhang et al., 2004). Consistent with this possibility, C/EBPα knockout bone marrow will not form myeloid leukemias when transduced with BCR-ABL, but instead will only form leukemias of the erythroid lineage in this genetic background (Wagner et al., 2006). These results highlight a critical dosage-dependent role of C/EBPα in AML pathogenesis: partial loss of function (e.g., through selective loss of p42) can drive AML progression whereas complete loss (e.g., through complete loss of p30/42) might be incompatible with leukemias of the myeloid lineage.
C/EBPα is essential for the initiation but not the maintenance of MLL fusion leukemia
In an effort to discover novel transcription factors that support MLL fusion-mediated leukemogenesis, Ohlsson et al. (2013) noted that C/EBPα target genes tended to be more highly expressed in MLL-rearranged human AML samples when compared with normal hematopoietic progenitors. To determine the functional relevance of this observation, the authors evaluated whether C/EBPα knockout hematopoietic stem and progenitor cells were capable of being transformed by the MLL-ENL oncogene. Importantly, in this experimental design they first inactivated C/EBPα in vivo with conditional alleles, followed by retroviral transduction of MLL-ENL into the hematopoietic stem and progenitor cells isolated from these animals. The authors found that C/EBPα was absolutely essential for MLL-ENL–mediated transformation in colony formation assays, liquid culture, and after transplantation in vivo. Based on prior studies (such as the aforementioned BCR-ABL study), this result was not entirely surprising. C/EBPα is critical in the formation of normal GMPs, and thus a failure to form myeloid lineage leukemias may simply reflect the normal lineage requirement for C/EBPα in myeloid progenitors. To address this, the authors first sorted GMPs from C/EBPαfloxed/floxed mice and subsequently inactivated C/EBPα, since upon reaching the GMP stage C/EBPα has been shown to be dispensable for further myeloid maturation (Zhang et al., 2004). Importantly, C/EBPα-null GMP cells were still unable to be transformed by MLL-ENL, suggesting an intrinsic requirement for C/EBPα that is independent of lineage establishment (Ohlsson et al., 2013). C/EBPα-knockout cells also failed to be transformed by co-transduction with Hoxa9/Meis1, indicating that a defect in MLL-ENL–mediated activation of these two downstream target genes was not the primary cause of impaired leukemogenesis. This finding also suggests that the C/EBPα requirement is not restricted to MLL fusion AML, but applies to other oncogenes that activate related pathways. However, C/EBPα is not required for transformation induced by the E2A-HLF oncogene, demonstrating that C/EBPα is only required for leukemogenesis in specific genetic contexts.
Extending these observations to the setting of disease maintenance, Ohlsson et al. (2013) went on to evaluate whether conditional inactivation of C/EBPα after MLL-ENL transduction also impaired cell expansion. Remarkably, conditional inactivation of C/EBPα in MLL-ENL–transformed cells had no effect on colony formation in serial replating assays or under in vivo conditions. C/EBPα-null AML could also be readily transplanted into tertiary recipients without any delay in disease progression, highlighting the dispensability of C/EBPα in leukemia stem cell subpopulations. These findings provide a dramatic example where a gene can be required exclusively at the stage of cancer initiation but is rendered superfluous in the setting of a fully transformed state.
What could be the mechanistic basis for the C/EBPα-requirement for leukemia initiation? Ohlsson et al. (2013) found that the majority of genes induced immediately after MLL-ENL transduction were not properly induced in C/EBPα-null cells. Furthermore, ChIP-seq analysis revealed that C/EBPα binding to DNA correlates with MLL-ENL target genes. These observations suggest, although do not prove, that C/EBPα binding to DNA might be a prerequisite for MLL fusion proteins to efficiently occupy chromatin during the initial stages of leukemogenesis. Such a model, however, cannot explain why C/EBPα is also required for initiation of AML by Hoxa9/Meis1 overexpression, since these cells lack an MLL fusion protein. It is interesting to note that a prior ChIP-seq analysis performed in leukemia cells found a close correlation between C/EBPα and Hoxa9/Meis1-binding sites at a large number of distal enhancer elements (Huang et al., 2012). This observation raises the interesting possibility that C/EBPα might also prime chromatin for subsequent binding of Hoxa9/Meis1 proteins at regulatory elements. Such a function would be reminiscent of pioneer transcription factors, described previously as a class of DNA-binding proteins that render chromatin competent for other factors to bind (Zaret and Carroll, 2011).
Open questions
Perhaps the most provocative question raised by this study is why C/EBPα becomes dispensable after full leukemic transformation. Although this issue was not directly addressed in this study, one can envision several regulatory scenarios that could explain this result. MLL fusion proteins are known to induce the expression of numerous additional transcription factors (e.g., Myb, Mef2c, Myc; Zuber et al., 2011a). These transcription factors, once achieving high levels of expression in transformed cells, might themselves be capable of stabilizing MLL fusion protein recruitment to chromatin, thereby establishing an oncogene-induced positive feedback loop (Fig. 1). Myb has previously been shown to physically associate with MLL fusion proteins and would be a logical candidate for bypassing the C/EBPα requirement for oncoprotein recruitment to DNA (Jin et al., 2010). Other C/EBP family homologues (e.g., C/EBPβ) expressed in fully transformed MLL fusion leukemia cells would be additional candidates that could alleviate the C/EBPα requirement after leukemic transformation. An alternative explanation for a transient C/EBPα requirement could be that MLL fusion proteins regulate distinct genes at different temporal stages of leukemic transformation. In this situation, C/EBPα is critical for the primary wave of regulatory events that occur upon leukemia initiation, but becomes dispensable as disease progresses; as MLL-ENL occupancy on chromatin shifts to a secondary set of target genes that are regulated in a C/EBPα-independent manner. Elucidating the mechanistic basis for the transient role of C/EBPα during AML progression will be a worthwhile endeavor and may redefine our understanding of the stepwise progression of leukemogenesis.
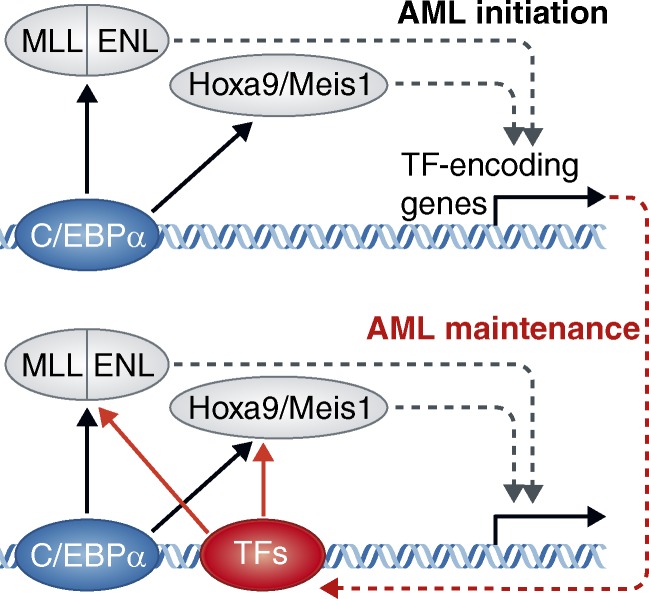
Figure 1.
A speculative model to account for a transient C/EBPα requirement during initiation of MLL-ENL– or Hoxa9/Meis1-induced acute myeloid leukemia. (top) At the onset of oncoprotein expression, C/EBPα is critical for initial recruitment of MLL-ENL or Hoxa9/Meis1 to their target sites on DNA, as depicted by arrows pointing from C/EBPα to these factors. Such an effect could be mediated through direct physical interactions or indirectly by increasing DNA accessibility. Many of the known target genes of MLL-ENL and Hoxa9/Meis1 are other transcription factors (TFs), hence C/EBPα would be critical to drive the initial burst in expression of these factors. (bottom) Upon reaching a fully transformed state, these additional TFs (encoded by genes up-regulated by the oncoproteins) would now have achieved sufficient levels of expression to participate in stabilizing oncoprotein recruitment. In this situation, they might act in parallel with (or replace) C/EBPα at MLL-ENL– or Hoxa9/Meis1-binding sites. In this fully transformed state, the C/EBPα requirement would be alleviated by these alternative TFs.
References
- Ablain J., Leiva M., Peres L., Fonsart J., Anthony E., de Thé H. 2013. Uncoupling RARA transcriptional activation and degradation clarifies the bases for APL response to therapies. J. Exp. Med. 210:647–653 10.1084/jem.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30:41–47 10.1038/ng765 [DOI] [PubMed] [Google Scholar]
- Calkhoven C.F., Müller C., Leutz A. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920–1932 [PMC free article] [PubMed] [Google Scholar]
- Harris W.J., Huang X., Lynch J.T., Spencer G.J., Hitchin J.R., Li Y., Ciceri F., Blaser J.G., Greystoke B.F., Jordan A.M., et al. 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 21:473–487 10.1016/j.ccr.2012.03.014 [DOI] [PubMed] [Google Scholar]
- Huang Y., Sitwala K., Bronstein J., Sanders D., Dandekar M., Collins C., Robertson G., MacDonald J., Cezard T., Bilenky M., et al. 2012. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 119:388–398 10.1182/blood-2011-03-341081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhao H., Yi Y., Nakata Y., Kalota A., Gewirtz A.M. 2010. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J. Clin. Invest. 120:593–606 10.1172/JCI38030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstetter P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T.A., Pabst T., et al. 2008. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 13:299–310 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Armstrong S.A. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 7:823–833 10.1038/nrc2253 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714–3725 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Smith E.R., Takahashi H., Lai K.C., Martin-Brown S., Florens L., Washburn M.P., Conaway J.W., Conaway R.C., Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 37:429–437 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M., Lin C., Guest E., Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer. 10:721–728 10.1038/nrc2915 [DOI] [PubMed] [Google Scholar]
- Nerlov C. 2004. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer. 4:394–400 10.1038/nrc1363 [DOI] [PubMed] [Google Scholar]
- Ohlsson E., Hasemann M.S., Willer A., Lauridsen F.K.B., Rapin N., Jendholm J., Porse B.T. 2013. Initiation of MLL-rearranged AML is dependent on C/EBPa. J. Exp. Med. 211:xxx–xxx [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell. 121:167–178 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Pabst T., Mueller B.U., Harakawa N., Schoch C., Haferlach T., Behre G., Hiddemann W., Zhang D.E., Tenen D.G. 2001a. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat. Med. 7:444–451 10.1038/86515 [DOI] [PubMed] [Google Scholar]
- Pabst T., Mueller B.U., Zhang P., Radomska H.S., Narravula S., Schnittger S., Behre G., Hiddemann W., Tenen D.G. 2001b. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 27:263–270 10.1038/85820 [DOI] [PubMed] [Google Scholar]
- Porse B.T., Pederson T.A., Xu X., Lindberg B., Wewer U.M., Friis-Hansen L., Nerlov C., Nerlov C. 2001. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 107:247–258 10.1016/S0092-8674(01)00516-5 [DOI] [PubMed] [Google Scholar]
- Rosenbauer F., Tenen D.G. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117 10.1038/nri2024 [DOI] [PubMed] [Google Scholar]
- Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 12:599–612 10.1038/nrc3343 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81:65–95 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomiany B.A., D’Arigo K.L., Kelly M.M., Kurtz D.T. 2000. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986–5997 10.1128/MCB.20.16.5986-5997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille T.C., Cleary M.L. 2006. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10:257–268 10.1016/j.ccr.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Wagner K., Zhang P., Rosenbauer F., Drescher B., Kobayashi S., Radomska H.S., Kutok J.L., Gilliland D.G., Krauter J., Tenen D.G. 2006. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc. Natl. Acad. Sci. USA. 103:6338–6343 10.1073/pnas.0508143103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Chen Z. 2008. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 111:2505–2515 10.1182/blood-2007-07-102798 [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Lin M., Naresh A., Kitabayashi I., Cleary M.L. 2010. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 17:198–212 10.1016/j.ccr.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S., Carroll J.S. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25:2227–2241 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A., et al. 2004. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 21:853–863 10.1016/j.immuni.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Zuber J., Radtke I., Pardee T.S., Zhao Z., Rappaport A.R., Luo W., McCurrach M.E., Yang M.M., Dolan M.E., Kogan S.C., et al. 2009. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 23:877–889 10.1101/gad.1771409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Rappaport A.R., Luo W., Wang E., Chen C., Vaseva A.V., Shi J., Weissmueller S., Fellmann C., Taylor M.J., et al. 2011a. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 25:1628–1640 10.1101/gad.17269211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. 2011b. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478:524–528 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]