Inflammatory T helper 17 cells in humans are distinguished by selective expression of MDR1 and are enriched in the gut of patients with Crohn’s disease.
Abstract
IL-17A–expressing CD4+ T cells (Th17 cells) are generally regarded as key effectors of autoimmune inflammation. However, not all Th17 cells are pro-inflammatory. Pathogenic Th17 cells that induce autoimmunity in mice are distinguished from nonpathogenic Th17 cells by a unique transcriptional signature, including high Il23r expression, and these cells require Il23r for their inflammatory function. In contrast, defining features of human pro-inflammatory Th17 cells are unknown. We show that pro-inflammatory human Th17 cells are restricted to a subset of CCR6+CXCR3hiCCR4loCCR10−CD161+ cells that transiently express c-Kit and stably express P-glycoprotein (P-gp)/multi-drug resistance type 1 (MDR1). In contrast to MDR1− Th1 or Th17 cells, MDR1+ Th17 cells produce both Th17 (IL-17A, IL-17F, and IL-22) and Th1 (IFN-γ) cytokines upon TCR stimulation and do not express IL-10 or other anti-inflammatory molecules. These cells also display a transcriptional signature akin to pathogenic mouse Th17 cells and show heightened functional responses to IL-23 stimulation. In vivo, MDR1+ Th17 cells are enriched and activated in the gut of Crohn’s disease patients. Furthermore, MDR1+ Th17 cells are refractory to several glucocorticoids used to treat clinical autoimmune disease. Thus, MDR1+ Th17 cells may be important mediators of chronic inflammation, particularly in clinical settings of steroid resistant inflammatory disease.
Like all CD4+ memory T cell subsets, Th17 cells are defined by the coordinate expression of select cytokines, lymphoid homing (e.g., chemokine) receptors, and transcription factors. Th17 cells express the cytokines IL-17A, IL-17F and IL-22, as well as the inflammatory chemokine receptor CCR6, which are all induced during Th17 cell development by the synergistic actions of STAT3 and the Th17-specific orphan nuclear receptor RORγt (Miossec et al., 2009). Human peripheral blood memory (CD45RO+) T cells that express IL-17A ex vivo are CCR6+, and are further enriched within cells that coexpress CCR4 (Acosta-Rodriguez et al., 2007). However, a second subset of CCR6+ IL-17A–expressing human memory T cells has been described that coexpresses the Th1-associated chemokine receptor CXCR3 (Sallusto et al., 2012). Whereas fewer CCR6+CXCR3+ memory T cells produce IL-17A upon ex vivo stimulation compared with CCR6+CCR4+ cells, more CCR6+CXCR3+ cells produce IFN-γ, and some are polyfunctional, displaying expression of both IL-17A and IFN-γ (Acosta-Rodriguez et al., 2007; Cohen et al., 2011), which is a hallmark of T cells isolated from inflamed tissue (Kebir et al., 2009; Ahern et al., 2010; Nistala et al., 2010; Hirota et al., 2011).
Recent studies in mice indicate that IL-17A expression is not sufficient to define Th17 cells with pathogenic activity. For example, in vitro differentiation of naive mouse CD4+ T cells using combinations of TGF-β1 plus IL-6, TGF-β3 plus IL-6, or IL-1β plus IL-6 plus IL-23 all induce equivalent proportions of IL-17A–expressing effector cells, yet only the latter two Th17 populations cause experimental autoimmune encephalomyelitis (EAE) when transferred into mice (Ghoreschi et al., 2010; Lee et al., 2012). Pathogenic mouse Th17 cells express a unique transcriptional signature compared with nonpathogenic Th17 cells, which includes elevated expression of the IL-23 receptor (Il23r). Accordingly, IL23r-deficient Th17 cells cannot induce autoimmune pathology in vivo, irrespective of how they are generated in vitro (Lee et al., 2012). IL-23 acts on effector/memory Th17 cells to induce proliferation and also augments production of the cytokines IL-17A, IL-17F, and IL-22 (Croxford et al., 2012). Furthermore, IL-23 has been reported to induce GM-CSF expression in Th17 cells, and GM-CSF–deficient Th17 cells fail to induce adoptively transferred EAE (Codarri et al., 2011). In contrast to pathogenic Th17 cells, nonpathogenic mouse Th17 cells: develop in vitro in the presence of TGF-β1 plus IL-6; develop in vivo in the absence of IL-23/IL-23R; produce IL-17A together with IL-10; and can have anti-inflammatory/immunoregulatory functions (McGeachy et al., 2007, 2009; Ghoreschi et al., 2010; Esplugues et al., 2011; Lee et al., 2012). These data begin to explain why Th17 cells that develop in the gut at steady-state do not induce pathological inflammation. Moreover, the distinctive features of pathogenic mouse Th17 cells indicate that cellular responses to IL-23, not just IL-17A per se, are a major factor underlying Th17-driven inflammation. These findings also suggest that IL23R expression levels may distinguish between pro- and anti-inflammatory Th17 cells in humans, independent of IL-17A.
Data from the clinic further support the notion that Th17 cells can promote inflammation independent of IL-17A. For example, in Crohn’s disease (CD), a chronic inflammatory bowel disease (IBD) in which affected gut tissue is infiltrated by IL-17A+ Th17 cells (Kleinschek et al., 2009), treatment of patients with Secukinumab, a fully human anti–IL-17A monoclonal antibody, actually exacerbates disease symptoms (Hueber et al., 2012). Collectively, these findings call for a reevaluation of human Th17 cells to better define specific subsets that associate with clinical disease and molecules that promote their pro-inflammatory function.
Killer cell lectin-like receptor B1 (KLRB1/CD161) is a reported marker of activated human Th17 cells, naive T cells that are Th17-biased, and ex-Th17 or non-classic Th1 cells, which are CCR6−IL-17A− Th1-like memory cells that retain expression of some Th17-lineage markers such as RORC and IL23R (Cosmi et al., 2008; Kleinschek et al., 2009; Maggi et al., 2012). Indeed, CD161+ human Th17 cells have been shown to infiltrate inflamed gut tissue of CD patients and synovial tissue of patients with active rheumatoid arthritis (Kleinschek et al., 2009; Nistala et al., 2010). However, it is unlikely that CD161 expression is sufficient to distinguish human pro-inflammatory Th17 cells, as it is also expressed, to varying degrees, on many other human CD4+ memory T cell subtypes, including Th2 and T regulatory (T reg) cells (Cosmi et al., 2010; Afzali et al., 2013).
Here, we report that high-level IL23R expression within human memory T cells is restricted to a subset of CCR6+CXCR3hiCCR4loCCR10−CD161+ cells that selectively expresses the multi-drug transporter MDR1 (also known as P-glycoprotein [P-gp] and ABCB1). MDR1 is an ATP-dependent membrane efflux pump with broad substrate specificity best known for its role in promoting tumor resistance to chemotherapy (Gottesman et al., 2002). In nonmalignant cells, MDR1 is expressed on intestinal epithelium, endothelial cells of the blood-brain-barrier, and hepatocytes, where it controls the accumulation of xenobiotic compounds and exogenous pharmacologic molecules (Schinkel, 1997). MDR1 is also expressed in progenitor cell types, and is thought to play a role in the survival and longevity of these cells (Chaudhary and Roninson, 1991; Sincock and Ashman, 1997). Consistent with this, we show that a sizeable proportion of human MDR1+ Th17 cells also express the stem cell marker c-Kit. All c-Kit+ memory T cells display robust MDR1 activity, and these cells give rise to c-Kit−MDR1+ progeny after inflammatory T cell activation in the presence of IL-23. Both c-Kit+ and c-Kit− MDR1+ Th17 cells display unique pro-inflammatory characteristics, including production of Th17 and Th1 cytokines, reduced expression of IL-10 and other anti-inflammatory molecules, and hypersensitivity to IL-23 stimulation, where they display enhanced activation of STAT3 and marked up-regulation of IL-17A compared with MDR1− memory T cell subsets.
Importantly, we show that c-Kit−CD161+MDR1+ Th17 cells are enriched and activated in CD patient lesions, and that MDR1+ Th17 cells are uniquely resistant to the anti-inflammatory actions of several glucocorticoids used to treat CD and other clinical autoimmune syndromes. Thus, MDR1 is a unique feature of pro-inflammatory and steroid-resistant Th17 cells in humans, which may be exploited to improve the diagnosis, characterization, and treatment of patients with chronic and steroid-resistant inflammatory diseases.
RESULTS
Th17 cytokines and IL23R are independently regulated in human memory T cell subsets
CCR6 expression broadly defines human CD4+ memory T cells with Th17 characteristics. Although not all CCR6+ memory cells produce IL-17A upon ex vivo activation, even those failing to produce IL-17A can express other Th17-signature genes, including IL23R (Nistala et al., 2010; Wan et al., 2011; Maggi et al., 2012). CCR6+ memory T cells are also heterogeneous with respect to expression of other cytokines and chemokine receptors. In particular, two human CCR6+ Th17 cell subsets have been described: (1) CCR6+CCR4+ T cells that express IL-17A but not IFN-γ; and (2) CCR6+CXCR3+ T cells that can express both IL-17A and IFN-γ (Acosta-Rodriguez et al., 2007; Cohen et al., 2011).
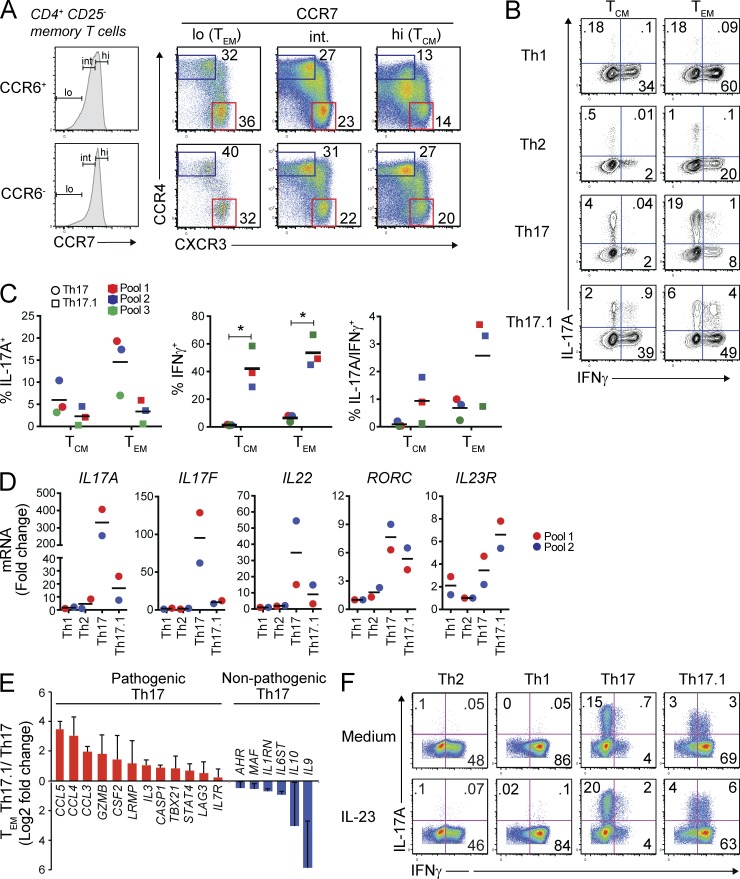
To more broadly define human memory T cell subsets with Th17 characteristics and relate them to the pathogenic and nonpathogenic Th17 subsets recently described in mice (Ghoreschi et al., 2010; Lee et al., 2012), we first examined whether CCR6+CCR4+ and CCR6+CXCR3+ cells are stable subsets or transient phenotypes. For this, expression of CCR4 and CXCR3 was monitored on CCR6+ or CCR6− memory (CD45RO+CD25−) T cells from healthy adult donor peripheral blood upon their maturation from CCR7hi central memory (TCM) to CCR7lo effector memory (TEM) cells. Memory T cell maturation, as defined by CCR7 expression, is particularly important to immune pathology given that human peripheral blood is comprised largely of TCM cells, whereas peripheral tissues, which serve as the sites for active inflammation, are highly enriched in TEM cells (Sallusto et al., 1999). We found that both CCR6+ and CCR6− memory cells were progressively polarized into two discreet subsets upon maturation from TCM to TEM based on reciprocal, high-level expression of CCR4 or CXCR3 (Fig. 1 A). Specifically, whereas 27–47% of either CCR6+ or CCR6− TCM cells from a representative donor were either CCR4hiCXCR3lo or CCR4loCXCR3hi, approximately twice as many TEM cells displayed these polarized phenotypes (Fig. 1 A). Thus, CCR6+CCR4+ and CCR6+CXCR3+ human memory cells are not only stable; they are actively selected for during memory T cell maturation in vivo. For simplicity and consistency with previous studies (Nistala et al., 2010), we refer to CCR6+CCR4hiCXCR3lo and CCR6+CCR4loCXCR3hi cells hereafter as Th17 and Th17.1 cells, respectively.
Figure 1.
Th17 cytokines and IL23R are independently regulated in human T cell subsets. (A) CD4+CD25− memory (CD45RO+) T cell subsets from healthy adult donor peripheral blood were analyzed by flow cytometry. CCR6+ or CCR6− cells were gated as CCR7hi (central memory; TCM), CCR7int (CCR7-intermediate), or CCR7lo (effector memory; TEM) cells, and CCR4 and CXCR3 expression was analyzed. Data represent >20 stains performed on individual donors or donor pools. (B) FACS-sorted TCM (CCR7hi) or TEM (CCR7lo) subsets were stimulated with PMA and ionomycin (P + I) and production of IFN-γ and IL-17A was determined by intracellular cytokine staining. Th1, CCR6−CCR4loCXCR3hi; Th2, CCR6−CCR4hiCXCR3lo; Th17, CCR6+CCR4hiCXCR3lo; Th17.1, CCR6+CCR4loCXCR3hi. Representative flow cytometry plots from 3 experiments performed on different pools of healthy adult donor blood are shown, and each donor pool contained blood from 2–4 individual donors. (C) IFN-γ and IL-17A production by FACS-sorted TCM or TEM cells was determined by intracellular cytokine staining as in B on 3 different pools of healthy adult donor peripheral blood. Each donor pool contained blood from 2–4 different donors. Individual and mean percentages of IL-17A+ (left), IFN-γ+ (middle), or IL-17A+/IFN-γ+ (right) T cells are shown, and data from each donor pool is color-coded. *, P < 0.05 by paired Student’s t test. (D) TEM Th1, Th2, Th17, or Th17.1 subsets were FACS-sorted as in B from two different pools of healthy donor peripheral blood. Both donor pools contained blood from 2–4 individual donors. Sorted cells were stimulated with anti-CD3/anti-CD28 for 72 h and expression of the indicated genes was measured by nanostring. Data are shown as fold change in gene expression within each donor pool; data from the two donor pools are color-coded. Horizontal bars in C and D represent the mean values. (E) Expression of pathogenic (red) or nonpathogenic (blue) murine Th17-signature genes (Lee et al., 2012) was determined in TEM Th17 and Th17.1 cells by nanostring as in D. Data are shown as mean relative (Log2 fold change) mRNA expression ± SD from 2 experiments on cells from different pools of healthy adult donor blood as in D. (F) TEM Th1, Th2, Th17, or Th17.1 cells (FACS-sorted as in B) were stimulated with anti-CD3/anti-CD28 and cultured for 6 d with or without IL-23. Cells were then restimulated with PMA and ionomycin and IFN-γ and IL-17A expression was determined by intracellular cytokine staining and FACS analysis. Data represent 3 experiments performed on independent donor pools, with each pool containing blood from 2–4 individual donors.
Consistent with previous studies (Acosta-Rodriguez et al., 2007; Cohen et al., 2011), a larger proportion of Th17 cells expressed intracellular IL-17A protein after ex vivo stimulation versus Th17.1 cells, and this increased consistently in Th17 cells, but not Th17.1 cells, as they transitioned from TCM to TEM (Fig. 1, B and C). In contrast to IL-17A, both IFN-γ–producing and IL-17A/IFN-γ–dual producing cells were largely restricted to the Th17.1 compartment, and both increased modestly but consistently upon Th17.1 maturation from TCM to TEM (Fig. 1, B and C). Neither Th17 nor Th17.1 cells produced IL-17A together with Th2 cytokines (IL-4, IL-13), although a fraction of IL-17A−IL-4+IL-13+ cells were observed within the Th17 population (unpublished data), consistent with earlier studies linking CCR4 expression to the Th2 phenotype (Sundrud et al., 2003; Rivino et al., 2004). In addition, small proportions (5–10%) of both CCR6+ (Th17) and CCR6− (Th2) CCR4hiCXCR3lo cells, but not Th1 or Th17.1 cells, expressed CRTH2 or CCR10, markers of terminally differentiated Th2 cells or IL-22–expressing Th22 cells, respectively (Duhen et al., 2009; Eyerich et al., 2009; Trifari et al., 2009; Mutalithas et al., 2010; not depicted). We confirmed that CCR6−CCR4hiCXCR3lo TEM cells were highly enriched for IL-4– and IL-13–producing Th2 cells, and that CCR6−CCR4loCXCR3hi TEM cells were predominantly IFN-γ+ Th1 cells that produced neither IL-17A nor Th2 cytokines (Fig. 1 B and not depicted).
Broader analyses of gene expression in FACS-sorted and ex vivo–stimulated TEM Th1, Th2, Th17, and Th17.1 cells revealed that, despite substantially (4- to 90-fold) higher expression of Th17 cytokine mRNAs (IL17A, IL17F, and IL22) in Th17 versus Th17.1 cells, both subsets expressed equivalent levels of RORC (Fig. 1 D) and CCL20 (not depicted), and Th17.1 cells consistently displayed twofold higher levels of IL23R mRNA compared with Th17 cells (Fig. 1 D). Expression of IL22, specifically within Th17 TEM cells from different donor pools correlated with the levels of contaminating Th22 cells that produced IL-22 but not IL-17A (not depicted; Duhen et al., 2009; Eyerich et al., 2009; Trifari et al., 2009). Th1 or Th2 TEM cells expressed low levels of all these Th17-associated transcripts (Fig. 1 D) but expressed high levels of TBX21 (T-bet) and IFNG, or IL5 and GATA3, respectively (not depicted). Thus, expression of Th17 cytokines and IL23R is independently regulated in human TEM subsets. In addition to elevated IL23R, we found that Th17.1 cells expressed higher levels of other pathogenic mouse Th17-signature mRNAs, including CCL3, CCL4, CCL5, CASP1, and GZMB, relative to Th17 cells, whereas Th17 cells expressed higher levels of nonpathogenic Th17-signature transcripts, including IL9, IL10, MAF, AHR, IL6ST (gp130), and IL1RN (Ghoreschi et al., 2010; Lee et al., 2012; Fig. 1 E). Together, these data indicate that despite reduced expression of canonical Th17 cytokines, human Th17.1 cells are enriched for expression of genes associated with pathogenic Th17 cell activity in mice.
Given their elevated expression of IL23R, we tested Th17.1 cells, and other TEM subsets, for their functional responses to IL-23 stimulation. Human TEM Th1, Th2, Th17, or Th17.1 cells were sorted by FACS, and then stimulated through the TCR with or without IL-23. After 1 wk, these cells were restimulated to assess the impact of IL-23 on IL-17A production. Consistent with our ex vivo analyses and independent of IL-23 stimulation, only Th17 and Th17.1 subsets produced detectable levels of IL-17A after in vitro activation and expansion; a larger subset of expanded Th17 cells produced IL-17A, whereas more Th17.1 cells produced IFN-γ, either alone or in combination with IL-17A (Fig. 1 F, top). Addition of IL-23 to the activated T cell cultures did not affect IFN-γ production by any subset and only modestly increased production of IL-17A by both Th17 and Th17.1 cells (Fig. 1 F). Thus, despite measurable differences in IL23R expression between human Th17 and Th17.1 cells, both subsets display similar, and very modest, responses to IL-23 stimulation in vitro. We therefore hypothesized that bona fide IL-23–responsive, pro-inflammatory human Th17 cells, although enriched within the Th17.1 compartment, comprise only a subset of these cells.
Transient c-Kit and stable MDR1 expression define a novel subset of human Th17.1 cells
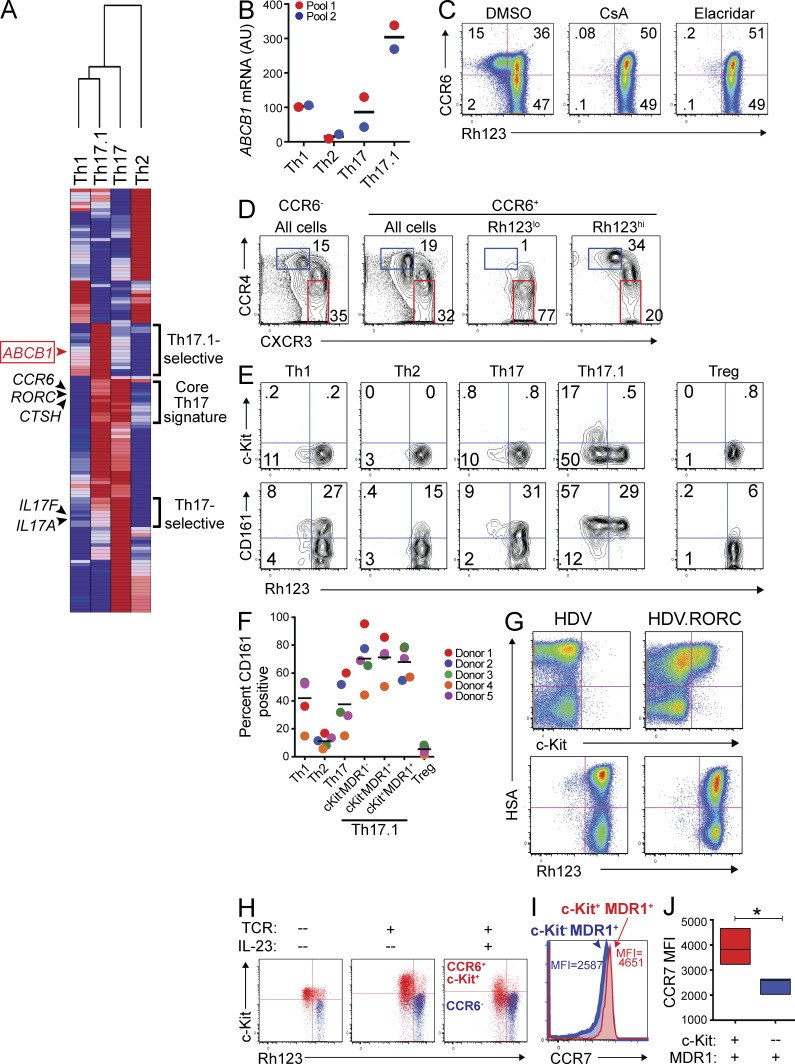
To identify additional markers selectively expressed by Th17.1 cells that further enrich for IL23R expression, we analyzed the transcriptional profiles of ex vivo–stimulated TEM human Th1, Th2, Th17, and Th17.1 cells by microarray. Analysis of all differentially expressed genes (1.8-fold cutoff) revealed the presence of both a core Th17 signature—genes expressed at high levels in both Th17 and Th17.1 cells, but low in Th1 and Th2 cells—and unique gene signatures present in only Th17 or Th17.1 cells (Fig. 2 A and Table S1). Genes within the core Th17 signature included well-established Th17-signature genes, such as RORC and CCR6, as well as those not previously associated with Th17 cells, such as CTSH and PTPN13 (Fig. 2 A and Table S1). As above, Th17 cells preferentially expressed IL17A and IL17F, whereas Th17.1 cells were unique in their expression of, among other genes, the multi-drug resistance type 1 membrane transporter ABCB1/ MDR1 (also known as P-gp; Fig. 2 A and Table S1). Selective ABCB1 gene expression in Th17.1 cells was confirmed by nanostring experiments performed on TEM subsets isolated from two additional pools of healthy adult donor blood (Fig. 2 B). Of the 50 known human ABC (ATP-binding cassette) transporter genes, only ABCB1 (MDR1) was differentially expressed across TEM subsets (Fig. S1).
Figure 2.
A novel subset of human Th17.1 cells is characterized by transient c-Kit and stable MDR1 expression. (A) TEM Th1, Th2, Th17, or Th17.1 cells were FACS-sorted from healthy adult donor peripheral blood as in Fig. 1 B. Sorted cells were stimulated with anti-CD3/anti-CD28 for 36 h and RNA was isolated for microarray analysis. Mean normalized raw gene expression values from two independent microarray experiments on cells sorted from different donor pools (each pool containing blood from 3–4 donors) were used to identify differentially expressed genes (1.8-fold cutoff). Data shown is a hierarchical clustering heatmap of all differentially expressed genes, with log2 transformation and row normalization. Red, high relative gene expression; dark blue, low relative gene expression. Representative gene symbols within each cluster are shown (a complete list of the genes and their absolute expression values within each cluster is provided in Table S1). (B) TEM Th1, Th2, Th17, or Th17.1 cells, FACS-sorted as in Fig. 1 B, were stimulated with anti-CD3/anti-CD28 for 72 h and expression of ABCB1 was determined by nanostring. Data are shown as ABCB1 mRNA expression (AU, arbitrary units) in two experiments performed on independent (color-coded) donor pools, with each pool containing blood from 2–4 individual donors. Horizontal bars represent the mean values. (C) Total CD4+CD25−CD45RO+ memory T cells isolated from healthy adult donor peripheral blood were labeled with rhodamine 123 (Rh123). After a 1-h efflux period at 37°C in the presence of vehicle (DMSO) or MDR1 inhibitors (CsA, cyclosporine A; Elacridar, selective MDR1 inhibitor), cells were stained with antibodies against CCR6, and Rh123 efflux and CCR6 expression was analyzed by FACS. Data shown are FACS plots from one experiment performed on cells from a healthy adult donor, and represent 3–4 independent experiments performed on cells isolated from different donors. (D) Rh123 efflux by CD4+ memory T cells was determined by FACS analysis as in C. After Rh123 efflux, cells were stained with antibodies against CCR6, CCR4, and CXCR3, and CCR4 and CXCR3 expression was analyzed on total CCR6− or CCR6+ cells, or on CCR6+ cells gated as Rh123lo (MDR1+) or Rh123hi (MDR1−). Data shown are representative FACS plots of >10 experiments performed on memory T cells isolated from independent donors. (E) CD4+ memory T cells were labeled with Rh123 and analyzed for Rh123 efflux as in C. After Rh123 efflux, cells were stained with antibodies against CD25, CCR6, CCR4, CXCR3, CCR7, c-Kit (CD117), and CD161. Cells were gated as TEM (CCR7lo) Th1 (CD25−CCR6−CCR4loCXCR3hi), Th2 (CD25−CCR6−CCR4hiCXCR3lo), Th17 (CD25−CCR6+CCR4hiCXCR3lo), Th17.1 (CD25−CCR6+CCR4loCXCR3hi), or T reg (CD25hi), and Rh123 efflux versus c-Kit (CD117; top) or CD161 (bottom) expression was analyzed in each subset. The FACS plots shown are representative of 5 independent experiments performed on memory T cells isolated from different donors. (F) The percentage of CD161+ cells within human TEM Th1, Th2, Th17, Th17.1 subsets and T reg cells was determined by FACS analysis as in E. Th17.1 cells were further gated into 3 subsets based on c-Kit expression and Rh123 efflux: c-Kit−MDR1−/Rh123hi, c-Kit−MDR1+/Rh123lo, and c-Kit+MDR1+/Rh123lo. Individual and mean percentages of CD161+ cells within each subset ± SD from 5 independent experiments performed on memory T cells isolated from individual (color-coded) donors is shown. (G) Naive CD4+ T cells were isolated from healthy adult donor peripheral blood, stimulated with anti-CD3/anti-CD28, and transduced with empty- (HDV) or RORC-containing (HDV.RORC) lentiviral particles that also contain a mouse HSA (heat stable antigen; a.k.a. CD24) expression cassette. Transduced T cells were expanded in IL-2–containing media for 7 d, and were then loaded with Rh123, incubated at 37°C for 1 h to allow for Rh123 efflux, and stained with antibodies against c-Kit or mouse HSA; Rh123 efflux and c-Kit expression was analyzed as a function of HSA expression in transduced T cells by FACS. FACS plots shown are representative of 3 independent experiments performed on naive T cells isolated from different donors. (H) CD4+ memory T cells isolated from healthy adult peripheral blood were FACS-sorted into CD25−CCR6+c-Kit+ (red) or CCR6− (blue) subsets. Cells were either left resting (no TCR), or were stimulated with anti-CD3/anti-CD28 and cultured for 6 d with or without IL-23. On day 6, cells were loaded with Rh123, stained with antibodies against c-Kit after Rh123 efflux, and analyzed by FACS. FACS plots shown are representative of 3 experiments using cells sorted from different donor pools, with each pool containing blood from 2–4 individual donors. (I) CD4+ memory T cells isolated from healthy adult peripheral blood were analyzed for Rh123 efflux as in E. CCR7 expression was assessed on c-Kit+ (red histogram) and c-Kit− (blue histogram) MDR1+/Rh123loCD25−CCR6+CCR4loCXCR3hi Th17.1 cells by FACS. The overlaid FACS histogram represents 4 experiments performed on memory T cells isolated from individual donors. CCR7 mean fluorescent intensity (MFI) is shown for c-Kit+ (red text) and c-Kit− (blue text) MDR1+ Th17.1 cells. (J) CCR7 MFI in c-Kit+ (red) and c-Kit− (blue) MDR1+ Th17.1 cells was determined by FACS analysis as in I. Data are shown as mean CCR7 MFI ± SD in c-Kit+ or c-Kit− MDR1+ Th17.1 cells from 4 individual donors. *, P < 0.05 by paired Student’s t test.
To assess specificity and function of MDR1 expression in human T cells, we labeled resting, ex vivo–isolated human CD4+ memory T cells with the fluorescent MDR1 substrate rhodamine 123 (Rh123; Ludescher et al., 1992) and analyzed the phenotype of cells capable of Rh123 efflux via FACS. As predicted by microarray, Rh123 efflux in human CD4+ memory T cells was restricted to a subset of CCR6+ cells (Fig. 2 C). Efflux of Rh123 was mediated by MDR1, as it was abolished by treatment of labeled cells with cyclosporine A, a known modulator of MDR1 activity, as well as by a selective MDR1 antagonist, elacridar (Fig. 2 C; Hyafil et al., 1993; Turtle et al., 2009). Further analysis revealed that nearly all CCR6+MDR1+ memory T cells were CCR4loCXCR3hi Th17.1 cells, whereas CCR4hiCXCR3lo Th17 cells were enriched in CCR6+ cells lacking MDR1 activity (Fig. 2 D).
MDR1 activity in nonmalignant cells is often associated with long-lived progenitor and stem cells, many of which also express the receptor for stem cell factor, c-Kit (Chaudhary and Roninson, 1991; Sincock and Ashman, 1997). In addition, Th17 cells in both mice and humans have been reported to display stem cell–like characteristics (Kryczek et al., 2011; Muranski et al., 2011). Consistent with these notions, we found that a sizeable fraction of MDR1+ (Rh123lo) Th17.1 cells also expressed c-Kit (Fig. 2 E). All c-Kit+ Th17.1 cells displayed robust MDR1 activity, as judged by Rh123 dye efflux, although MDR1 activity was also evident in c-Kit− Th17.1 cells (Fig. 2 E). We confirmed that Th17.1 memory cells expressing c-Kit and/or displaying MDR1 activity were CD3+CD4+ (unpublished data), ruling out the possibility that expression of these markers within our purified memory T cell preparations were coming from non–T cell contaminants. Like MDR1, c-Kit expression on CD4+ memory T cells was almost entirely restricted to the Th17.1 subset, and both c-Kit and MDR1 expression were either low or absent on Th1, Th2, Th17, T reg, or naive CD4+ T cells (Fig. 2 E and not depicted). In contrast to these tightly restricted expression patterns of c-Kit and MDR1, CD161 was more broadly expressed on effector and regulatory T cell lineages (Fig. 2, E and F). Although CD161 expression was enriched within the Th17.1 compartment, it was not differentially expressed among c-Kit+MDR1+, c-Kit−MDR1−, and c-Kit−MDR1− Th17.1 subsets (Fig. 2, E and F).
In accord with the selective expression of c-Kit and MDR1 on only a subset of CCR6+ Th17 cells, ectopic expression of the core Th17 transcription factor RORC in human naive CD4+ T cells induced c-Kit expression in only a portion of transduced cells, and it failed to promote any detectable MDR1 activity over control-transduced cells (Fig. 2 G). In contrast, RORC strongly induced IL-17A expression and uniformly up-regulated CCR6, as previously reported (not depicted; Manel et al., 2008). Thus, c-Kit and MDR1 are selectively expressed on a small subset of human Th17.1 cells, and RORC expression alone is not sufficient to fully induce their expression.
In stem cells, c-Kit signaling controls pluripotency and self-renewal; it is rapidly down-regulated as stem cells are mobilized or induced to differentiate (Simmons et al., 1994; Roberts et al., 1999). This knowledge, together with our microarray data indicating that TCR-stimulated TEM subsets do not express KIT mRNA (not depicted), suggested that c-Kit expression in mature CD4+ memory T cells is transient and is suppressed upon T cell activation/differentiation. To test this, we FACS-sorted CCR6+c-Kit+ or CCR6− memory T cells (which uniformly lack c-Kit expression), and either left them resting in media alone or activated them with anti-CD3/anti-CD28 beads with or without IL-23. After 6 d, c-Kit expression and MDR1 activity was assessed via FACS analysis. Unstimulated CCR6+c-Kit+ cells and those activated via TCR in the absence of IL-23 both maintained c-Kit expression (Fig. 2 H). In contrast, CCR6+c-Kit+ cells stimulated through TCR in the presence of IL-23 down-regulated c-Kit (Fig. 2 H). Despite loss of c-Kit, CCR6+c-Kit+ cells expanded by TCR plus IL-23 stimulation maintained stable MDR1 activity (Fig. 2 H). CCR6− memory T cells, which lacked c-Kit expression and MDR1 activity ex vivo, remained c-Kit−MDR1− after activation and expansion in vitro (Fig. 2 H). Further consistent with a model in which c-Kit+MDR1+ cells give rise to c-Kit−MDR1+ cells in vivo, we found that c-Kit+MDR1+ Th17.1 memory cells expressed higher levels of CCR7 ex vivo versus c-Kit−MDR1+ Th17.1 cells (Fig. 2, I and J). Collectively, these data suggest that MDR1 activity stably marks a novel lineage of human Th17 memory cells, comprised of both c-Kit+ and c-Kit− cells, and that c-Kit+MDR1+ cells give rise to c-Kit−MDR1+ cells after activation in pro-inflammatory settings.
MDR1+ Th17.1 cells display unique, pro-inflammatory characteristics
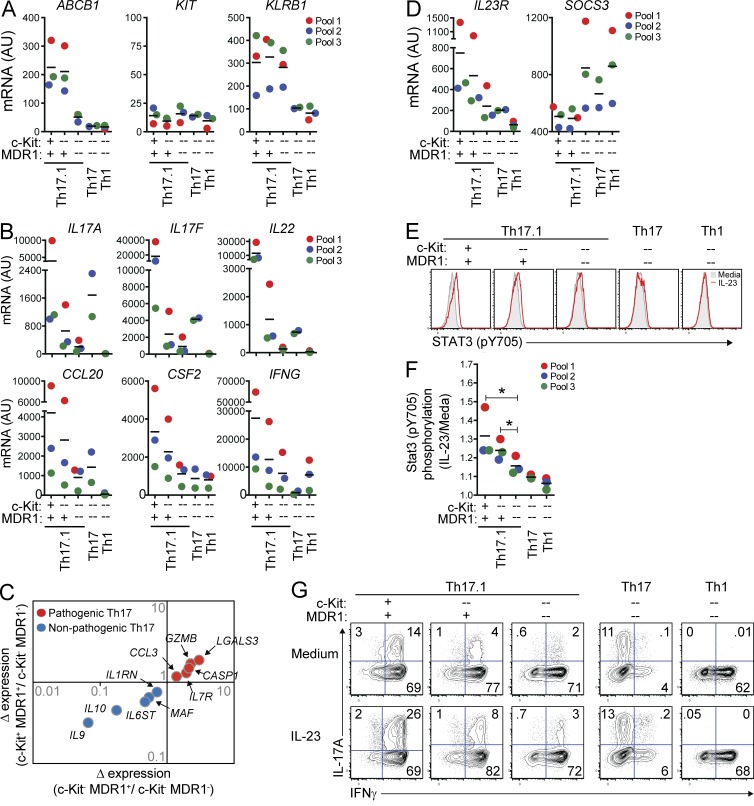
To investigate the putative function of human MDR1+ Th17.1 subsets, we analyzed gene expression in c-Kit+MDR1+, c-Kit−MDR1+, or c-Kit−MDR1− Th17.1 cells after FACS purification (Fig. S2) and ex vivo stimulation. For comparison, we also analyzed FACS-sorted c-Kit−MDR1− Th17 or Th1 (CCR6−CCR4loCXCR3hi) memory cells (Fig. S2). We confirmed that c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells sorted based on Rh123 efflux displayed selective, high-level ABCB1 (MDR1) gene expression, whereas KIT (c-Kit) expression, presumably as a function of ex vivo stimulation, was low in all subsets (Fig. 3 A). Consistent with our ex vivo FACS analyses, KLRB1 (CD161) mRNA expression was elevated in Th17.1 cells versus either Th17 or Th1 cells, and was high in Th17.1 cells independent of c-Kit expression or MDR1 activity (Fig. 3 A).
Figure 3.
Unique pro-inflammatory characteristics of human MDR1+ Th17.1 cells. (A and B) Human CD4+ memory T cells from pooled healthy adult donor peripheral blood were FACS-sorted into TEM (CCR7lo) Th17.1 (CCR6+CCR4loCXCR3hi), Th17 (CCR6+CCR4hiCXCR3lo), or Th1 (CCR6−CCR4loCXCR3hi) cells. Th17.1 cells were sub-sorted into c-Kit+ or c-Kit− MDR1+ (Rh123lo), or c-Kit−MDR1− (Rh123hi) cells as indicated; Th17 and Th1 cells were sorted as c-Kit−MDR1− cells (see Fig. S1 for gating/sorting strategy). All cells were stimulated with anti-CD3/anti-CD28 for 72 h and expression of ABCB1 (MDR1), KIT (c-Kit), and KLRB1 (CD161; A), or IL17A, IL17F, IL22, CCL20, CSF2 (GM-CSF), and IFNG (B) was determined by nanostring. Data are shown as individual (color-coded) and mean normalized raw expression values (AU, arbitrary units) in cells sorted from 2–3 independent donor pools, with each pool containing blood from 2–4 individual donors. (C) Expression of pathogenic (red dots) or nonpathogenic (blue dots) murine Th17-signature genes (Lee et al., 2012) in TEM c-Kit+MDR1+, c-Kit−MDR1+, or c-Kit−MDR1− Th17.1 cells was determined by nanostring as in A and B. Mean normalized raw expression values from 3 independent experiments performed on different donor pools containing blood from 2–4 individual donors were used for fold change calculations. (D) Expression of IL23R and SOCS3 mRNA was determined in human TEM subsets by nanostring as in A and B. (E) FACS-sorted TEM subsets from healthy donor peripheral blood (as in A and B) were stimulated with anti-CD3/anti-CD28 and cultured for 3 d with or without IL-23. Stat3 phosphorylation (pY705) was determined on day 3 by phospho-intracellular staining and FACS analysis. Gray filled histograms, media alone; red traced histograms, plus IL-23. Representative FACS plots are shown and represent 3 independent experiments using cells isolated from different donor pools, with each pool containing blood from 2–4 individual donors. (F) Stat3 phosphorylation was determined by flow cytometry in TEM subsets cultured with or without IL-23 for 3 d as in E. Data are shown as mean fold change in Stat3 pY705 MFI (IL-23/media alone) ± SD from 3 experiments using cells isolated from different (color-coded) donor pools. *, P < 0.05 by paired Student’s t test. (G) FACS-sorted TEM subsets from healthy donor peripheral blood were stimulated with anti-CD3/anti-CD28 and cultured for 6 d with or without IL-23. On day 6, cells were restimulated with PMA and ionomycin and IL-17A and IFN-γ expression was determined by intracellular cytokine staining and FACS analysis. FACS plots shown are representative of 3 experiments using cells sorted from different donor pools, with each pool containing blood from 2–4 individual donors. Horizontal bars represent the mean values.
c-Kit−MDR1− Th17 or Th1 cells displayed polarized expression of Th17-associated (IL17A, IL17F, IL22, CSF2, and CCL20) or Th1-associated (IFNG) transcripts, respectively, whereas c-Kit+MDR1+ Th17.1 cells, and to a lesser extent c-Kit−MDR1+ Th17.1 cells, were polyfunctional, expressing high levels of both Th17- and Th1-associated mRNAs (Fig. 3 B). In contrast, and in spite of high-level CD161 expression, c-Kit−MDR1− Th17.1 cells displayed reduced levels of both Th17- and Th1-associated transcripts versus either c-Kit+MDR1+ or c-Kit−MDR1+ Th17.1 cells, indicating that T cell polyfunctionality is associated with c-Kit and MDR1 expression specifically, and not the broader Th17.1 (CCR6+CCR4loCXCR3hiCD161+) phenotype (Fig. 3 B). c-Kit+ and c-Kit− MDR1+ Th17.1 subsets also displayed increased levels of transcripts associated with pathogenic mouse Th17 cells, including CCL3, GZMB, IL7R, CASP1, and LGALS3, relative to c-Kit−MDR1− Th17.1 counterparts, and showed decreased expression of mRNAs found in nonpathogenic mouse Th17 cells, most notably IL9, IL10, MAF, IL6ST, and IL1RN (Fig. 3 C; Ghoreschi et al., 2010; Lee et al., 2012). Along these lines, IL23R expression was consistently two- to threefold higher in both c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells relative to c-Kit−MDR1− Th17.1 or Th17 cells, and 5- to 10-fold higher than Il23R expression seen in c-Kit−MDR1− Th1 cells (Fig. 3 D). Pro-inflammatory signaling downstream of IL-23/IL-23R is thought to proceed through STAT3, and both MDR1+ Th17.1 cell subsets displayed consistently reduced expression of the STAT3 negative regulator SOCS3 (Chen et al., 2006) versus c-Kit−MDR1− Th17.1, Th17, or Th1 subsets (Fig. 3 D). Thus, human MDR1+ Th17.1 subsets are endowed with a unique, pro-inflammatory transcriptional signature that both mimics disease-inducing Th17 subsets in mice and predicts increased responsiveness to IL-23 stimulation.
To directly measure the responses of MDR1+ Th17.1 subsets to IL-23, we activated FACS-sorted c-Kit+MDR1+ or c-Kit−MDR1+ Th17.1 cells, as well as c-Kit−MDR1− Th17.1, Th17, or Th1 cells, through the TCR and expanded them with or without IL-23. IL-23 responses were determined by FACS analyses of STAT3 tyrosine phosphorylation (Y705) and IL-17A up-regulation. IL-23–induced STAT3 phosphorylation was indeed augmented in both c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells versus c-Kit−MDR1− Th17.1, Th17, or Th1 cells across three experiments in different donor pools (Fig. 3, E and F). IL-23 stimulation also strongly increased IL-17A production in c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells (two- to fourfold) compared with the same cells activated in the absence of IL-23 (Fig. 3 G). In contrast, IL-23 stimulation had little impact on IL-17A production by c-Kit−MDR1− Th17.1, Th17, or Th1 subsets (<1.5-fold over cells activated without IL-23). In addition to making more IL-17A and IFN-γ, greater percentages of c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells expressed TNF and GM-CSF versus c-Kit−MDR1− memory T cell subsets—irrespective of IL-23 stimulation—and both MDR1+ Th17.1 subsets displayed reduced production of IL-10 relative to MDR1− T cell subsets (unpublished data). Collectively, these findings establish that c-Kit+MDR1+ and c-Kit−MDR1+ Th17.1 cells display several hallmarks of a pro-inflammatory Th17 cell lineage, including: production of both Th17 and Th1 cytokines; a transcriptional signature akin to that of pathogenic mouse Th17 cells; and heightened functional responses to IL-23.
MDR1 distinguishes pro-inflammatory Th17.1 cells within clinically inflamed tissue
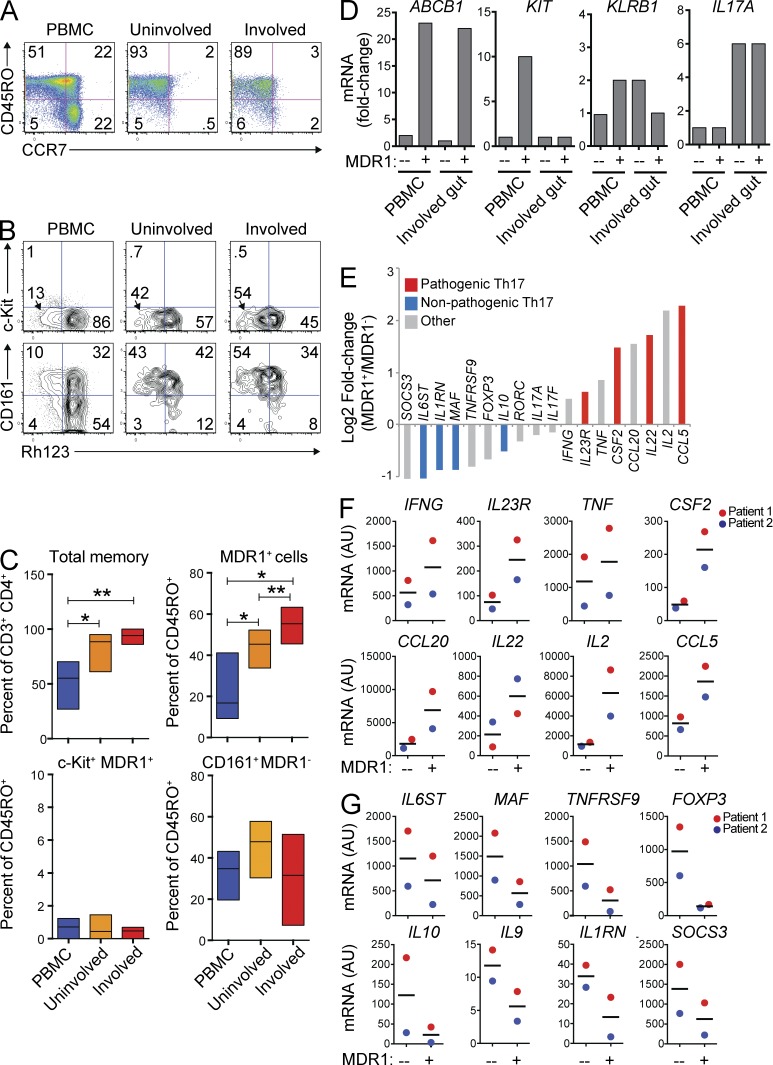
To determine their relevance in a clinical setting of inflammatory disease, we analyzed MDR1+ Th17.1 cells in the peripheral blood and gut of patients with active CD (Table 1). As expected, CD4+ T cells isolated from CD patient uninvolved or involved gut tissues were highly enriched for CD45RO+CCR7lo TEM cells compared with those from blood (Fig. 4 A). Furthermore, and consistent with previous studies (Kleinschek et al., 2009; Zorzi et al., 2013), ex vivo–stimulated memory T cells from CD patient gut produced more IL-17A and IFN-γ, but less IL-4, compared with memory cells from patient-matched peripheral blood (unpublished data). Despite these observations indicating the presence of Th17 and Th1 TEM cells, surface expression of CCR6 and CXCR3 was markedly decreased on CD4+ memory T cells after isolation from gut tissues, in contrast to cells from patient-matched peripheral blood (unpublished data). MDR1 activity, in contrast, was readily detectable in both blood- and gut-derived T cells from CD patients; in both cases MDR1 activity was restricted, within CD3+CD4+ T cells, to a subset of CD45RO+ memory T cells (Fig. 4 B). The proportions of both total and MDR1+ (Rh123lo) memory T cells were enriched in uninvolved and involved gut, relative to peripheral blood, and MDR1+ memory cells were further increased in involved versus uninvolved gut tissue (Fig. 4, B and C). MDR1+ memory T cells from either uninvolved or involved gut tissue were almost entirely c-Kit− and CD161+ (Fig. 4 B), and CD161 expression was high in both MDR1+ and MDR1− memory T cells from CD patient gut tissues (Fig. 4 B). Unlike MDR1+ T cells, however, MDR1−CD161+ memory T cells were not enriched in involved tissue (Fig. 4 C). Thus, as in healthy donor peripheral blood, MDR1 expression in the gut of CD patients distinguishes a subset of CD161+ Th17.1 cells, and these cells are increased in actively inflamed tissue.
Table 1.
CD patient data
| Characteristic | Number |
| Total patients | 6 |
| Gender | |
| Male | 2 |
| Female | 4 |
| Ethnicity | |
| White non-Hispanic | 3 |
| Hispanic/white | 2 |
| Hispanic | 1 |
| Age | |
| Range | 26–59 |
| Mean ± SD | 39.8 ± 13.8 |
| Diagnosis | |
| CD | 5 |
| IBD, unspecified | 1 |
Demographic data on the cohort of CD patients used in this study is shown.
Figure 4.
MDR1+ Th17.1 cells are enriched and activated in clinically inflamed tissue. (A) Mononuclear cells were isolated from CD patient peripheral blood (PBMC; left), uninvolved gut (middle), or involved gut (right), and were analyzed for expression of CD45RO and CCR7. Data shown are on CD3+CD4+CD25− gated T cells and represent 5 experiments on cells from different patients. (B) Mononuclear cells from CD patient PBMC, uninvolved gut, or involved gut tissue (as in A) were loaded with Rh123, stained with antibodies against CD3, CD4, CD25, CD45RO, c-Kit, and CD161 after a 1-h Rh123 efflux period at 37°C, and analyzed by FACS. Data shown are on CD3+CD4+CD25−CD45RO+ gated memory T cells. Rh123 efflux versus c-Kit (top) or CD161 (bottom) expression is shown. Data represent 3 (CD161 staining) or 4 (c-Kit staining) experiments on cells isolated from different patients. (C) Percentages of total memory (CD45RO+) cells (top left), MDR1+/Rh123lo memory cells (top right), c-Kit+MDR1+/Rh123lo memory T cells (bottom left), or CD161+MDR1+/Rh123lo memory cells (bottom right) were determined in CD patient PBMC, uninvolved gut, or involved gut tissue by FACS analysis as in B. Data are shown as mean percentages ± SD from 3–5 individual patients. *, P < 0.05; **, P < 0.01 by paired Student’s t test. (D) MDR1+ (Rh123lo) or MDR1− (Rh123hi) CD3+CD4+CD25−CD45RO+ memory T cells were FACS-sorted from the PBMC of one HC donor or from mononuclear cells isolated from the involved gut of one CD patient. Sorted cells were lysed directly ex vivo, and RNA was isolated for microarray analysis. Relative (fold change) expression of ABCB1 (MDR1), KIT (c-Kit), KLRB1 (CD161), or IL17A is shown for the T cell subsets as indicated. (E) Relative ex vivo expression (Log2 fold change) of pathogenic mouse Th17-signature genes (red; Lee et al., 2012), nonpathogenic Th17-signature genes (blue; Lee et al., 2012), or other notable (gray) genes was determined by microarray analysis of MDR1+ or MDR1− memory T cells sorted from involved CD patient gut tissue as in D. (F and G) Mononuclear cells isolated from involved CD patient gut tissue were FACS-sorted into MDR1+ or MDR1− CD3+CD4+CD25−CD45RO+ memory T cells, and expression of pathogenic (F) or nonpathogenic (G) mouse Th17-signature genes (Lee et al., 2012) was analyzed by nanostring. Data are shown as individual (color coded) and mean expression values (AU – arbitrary units) from 2 independent patients. Horizontal bars represent the mean values.
To assess whether MDR1+ memory T cells in involved CD tissue display hallmarks of pro-inflammatory Th17 cells, we defined the ex vivo transcriptional profiles of MDR1+ and MDR1− CD4+ memory T cells isolated from surgically resected involved gut tissue of a CD patient. MDR1+ or MDR1− CD4+ memory T cells were FACS-sorted based on Rh123 efflux, and, as controls, we also analyzed gene expression profiles of MDR1+ or MDR1− memory T cells similarly sorted from the peripheral blood of an unmatched healthy adult donor (healthy control [HC]). Both blood- and involved gut-derived Rh123lo CD4+ memory T cells expressed ∼20-fold higher levels of ABCB1 mRNA compared with Rh123hi-sorted memory cells, whereas only blood-derived MDR1+ (Rh123lo) T cells expressed appreciable levels of KIT mRNA (Fig. 4 D). These latter results were consistent with the c-Kit− surface phenotype of gut-derived MDR1+ T cells observed by FACS analysis. In contrast to either MDR1+ or MDR1− memory T cells from peripheral blood, both subsets isolated from involved gut tissue showed evidence of in vivo activation (e.g., elevated CD69 expression; not depicted), and both expressed equivalent levels of KLRB1 (CD161) and IL17A (Fig. 4 D). Despite these similarities, MDR1+ T cells from involved gut displayed selective expression of numerous pro-inflammatory cytokine and chemokine transcripts, as well as many pathogenic murine Th17-signature mRNAs, including IL23R, CSF2, IL22, TNF, CCL20, and CCL5, relative to local MDR1− T cells (Fig. 4 E; Lee et al., 2012). In contrast, MDR1− cells from involved gut displayed increased expression of nonpathogenic murine Th17-signature genes, including MAF, IL10, IL6ST, IL1RN, and IKZF3 (Lee et al., 2012), as well as other genes generally associated with T reg cells, such as FOXP3 and TNFRSF9 (4-1BB; Fig. 4 E; So et al., 2008; Josefowicz et al., 2012). This selective, pro-inflammatory transcriptional profile of MDR1+ memory T cells was confirmed in nanostring experiments of MDR1+ and MDR1− memory T cells isolated from involved gut tissue of two additional CD patients (Fig. 4, F and G). As a whole, these data indicate that pro-inflammatory MDR1+ Th17.1 cells are enriched and activated in involved gut tissue of CD patients.
MDR1+ Th17.1 cells are resistant to glucocorticoid-mediated T cell suppression
MDR1 is best known for mediating tumor resistance to chemotherapy (Gottesman et al., 2002). In T cells, MDR1 reportedly modulates activity of anti-retroviral compounds (Weiss and Haefeli, 2010), though little is known about its physiological substrates and functions. Several natural and synthetic glucocorticoids (i.e., steroids), including dexamethasone, prednisolone, and 6α-prednisolone, have been shown to be substrates for MDR1 efflux (Crowe and Tan, 2012). Because steroids are first-line therapy for many autoimmune diseases, including CD (Flammer and Rogatsky, 2011), we sought to determine whether MDR1+ Th17.1 cells are differentially responsive to steroids, and whether MDR1 activity directly modulates T cell responses to steroids.
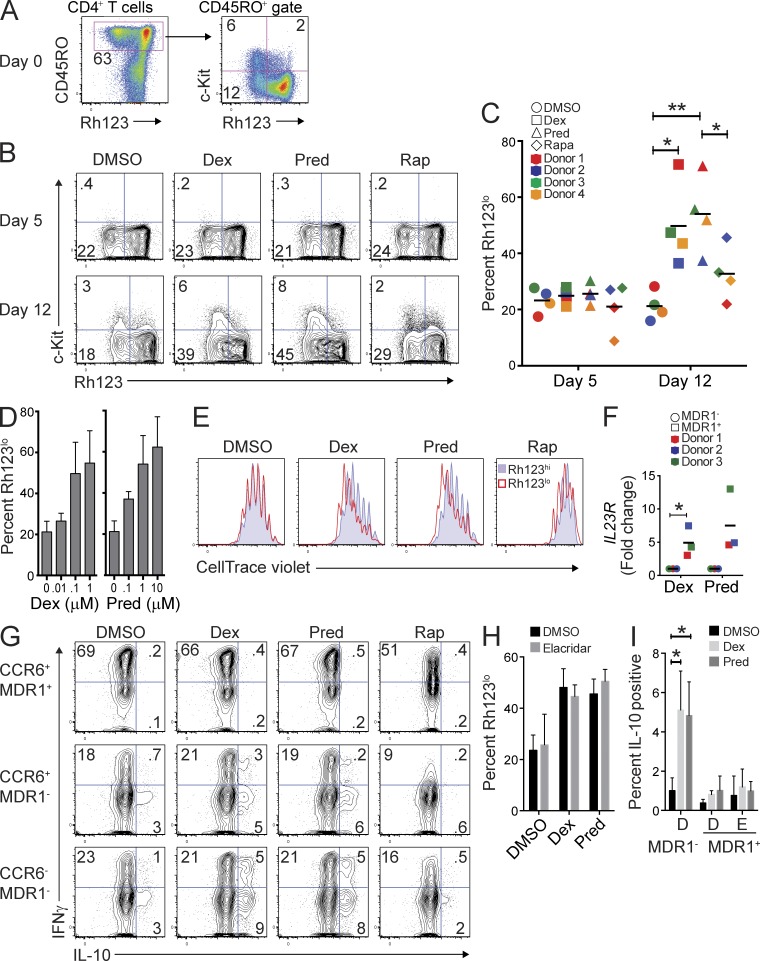
To address these questions, we cultured CD4+ T cells (from healthy donor peripheral blood) stimulated through the TCR with or without glucocorticoids. To distinguish between steroid-specific effects and those associated with other types of immunosuppression, some cultures were treated with the nonsteroidal immunosuppressant rapamycin (Araki et al., 2011). Before culturing cells, we determined the baseline percentages of c-Kit+ and c-Kit− MDR1+ memory T cells within individual donor CD4+ T cell compartments by ex vivo FACS analysis (Fig. 5 A). We then analyzed the proportion of MDR1+ cells within control- or steroid-treated CD4+ T cell cultures at various time points after activation. With the exception of activation-dependent c-Kit down-regulation, which we noted previously, no changes were observed in the proportions of MDR1+ (Rh123lo) T cells within control- or steroid-treated T cell cultures during the first 5 d (Fig. 5, B and C). In contrast, proportions of both c-Kit− and c-Kit+ MDR1+ T cells increased markedly within steroid-treated cultures by day 12, in most experiments increasing two- to threefold over baseline (day 0; Fig. 5, B and C). Selection for MDR1+ T cells was steroid-specific, as neither DMSO- nor rapamycin-treated cultures showed similar increases in MDR1+ T cell proportions, although smaller increases in MDR1+ T cell frequencies were observed in the presence of rapamycin in most experiments (Fig. 5, B and C). The prednisolone derivative, 6α-methylprednisolone, also prompted expansion of MDR1+ T cells within mixed T cell cultures (unpublished data).
Figure 5.
MDR1+ Th17.1 cells are refractory to glucocorticoid-mediated T cell suppression. (A) Total CD4+ T cells from healthy donor peripheral blood were labeled with Rh123, stained for CD45RO and c-Kit expression after Rh123 efflux, and analyzed for the frequency of c-Kit+ and c-Kit− MDR1+ (Rh123lo) memory T cells ex vivo (day 0) by flow cytometry. FACS plots show MDR1 (Rh123 efflux) activity versus CD45RO (left) or c-Kit (right) expression. Data represent 4 experiments on individual donors. (B) Total CD4+ T cells were stimulated with anti-CD3/anti-CD28 and were cultured for 5 d (top) or 12 d (bottom) in the presence of DMSO (vehicle), dexamethasone (Dex; 0.1 µM), prednisolone (Pred; 1 µM), or rapamycin (Rap; 0.1 µM). At days 5 and 12, cells were analyzed by FACS for Rh123 efflux and c-Kit expression as in A. FACS plots shown are representative of 4 experiments on individual donors. (C) Percentage of Rh123lo (MDR1+) cells within total CD4+ T cells cultured for 5 or 12 d with DMSO, Dex, Pred, or Rap was determined by FACS analysis as in B. Data are shown as individual (color coded) and mean percentages of Rh123lo cells from 4 independent donors. *, P < 0.05; **, P < 0.01 by paired Student’s t test. Horizontal bars represent the mean values. (D) Total CD4+ T cells were stimulated with anti-CD3/anti-CD28 and treated with titrating concentrations of dexamethasone (Dex; left) or prednisolone (Pred; right). At day 12, the frequency of Rh123lo cells was determined by Rh123 efflux and FACS analysis. Data are shown as mean percentages of Rh123lo (MDR1+) T cells ± SD from 4 independent experiments performed on cells from different donors. (E) Total CD4+ T cells were labeled with CellTrace violet, stimulated with anti-CD3/anti-CD28, and treated with DMSO, Dex, Pred, or Rap as in B. On day 5, cells were further labeled with Rh123 and were analyzed for Rh123 efflux and CellTrace violet dilution by FACS analysis. Overlaid histograms show CellTrace violet dilution in T cells gated as MDR1+/Rh123lo (red histogram) or MDR1−/Rh123hi (blue histogram); data represent 4 experiments performed on T cells isolated from individual donors. (F) Total CD4+ T cells were stimulated with anti-CD3/anti-CD28, treated with dexamethasone (Dex; 0.1 µM) or prednisolone (Pred; 1 µM), and cultured for 12–14 d. Cells were then loaded with Rh123, FACS-sorted into MDR1+/Rh123lo and MDR1−/Rh123hi cells, and RNA was isolated to determine IL23R gene expression by qPCR. Expression of IL23R was normalized to ACTB (β-actin). Data are shown as relative (fold change) IL23R expression in MDR1+ (Rh123lo) and MDR1− (Rh123hi) T cells sorted from Dex- or Pred-treated cultures. Individual (color-coded) and mean values are shown for 3 experiments performed on cells from different donors. *, P < 0.05 by paired Student’s t test. Horizontal bars represent the mean values. (G) FACS-sorted CCR6+MDR1+/Rh123lo, CCR6+MDR1−/Rh123hi, or CCR6−MDR1−/Rh123hi cells were stimulated with anti-CD3/anti-CD28, treated with DMSO, dexamethasone (Dex; 0.1 µM), prednisolone (Pred; 1 µM), or rapamycin (Rap; 0.1 µM), and cultured for 5 d. Cells were restimulated with PMA and ionomycin, and IFN-γ and IL-10 expression was determined by intracellular staining and flow cytometry. FACS plots are representative of 4 experiments using cells sorted from individual donors. (H) Total CD4+ T cells were stimulated with anti-CD3/anti-CD28, treated with DMSO, dexamethasone (Dex; 0.1 µM), or prednisolone (Pred; 1 µM), in the absence (DMSO) or presence of elacridar (0.1 µM). Cells were cultured for 12 d and were then analyzed for Rh123 efflux by FACS as in A, B, and D. Data are shown as percentage of MDR1+/Rh123lo T cells ± SD from 3 independent experiments using cells from different donors. (I) FACS-sorted MDR1− or MDR1+ (CCR6+) cells were stimulated with anti-CD3/anti-CD28 and cultured in the absence (DMSO) or presence of dexamethasone (Dex; 0.1 µM), or prednisolone (Pred; 1 µM) plus vehicle (DMSO; D) or 0.1 µM elacridar (E) for 5 d. Cells were restimulated with PMA and ionomycin, and IL-10 expression was determined by intracellular cytokine staining and FACS analysis. Data are shown as mean percentages of IL-10–expressing cells ± SD from 3 donors. *, P < 0.05 by paired Student’s t test.
Expansion of MDR1+ T cells in the presence of dexamethasone or prednisolone was dose-dependent (Fig. 5 D) and was associated with increased proliferation of MDR1+ (Rh123lo) versus MDR1− (Rh123hi) T cells (Fig. 5 E), as judged by CellTrace dye dilution experiments. In contrast, MDR1+ and MDR1− T cells proliferated equally in vehicle (DMSO)-treated cultures or cultures treated with rapamycin (Fig. 5 E), indicating that differences in MDR1+ T cell proliferation were context (steroid)-dependent, not intrinsic. Furthermore, MDR1+ (Rh123lo) T cells expanded in the presence of dexamethasone or prednisolone for up to 2 wk remained high for ABCB1 gene expression (not depicted), and these cells maintained a pro-inflammatory transcriptional signature relative to bystander MDR1− T cells, as exemplified by elevated IL23R expression (Fig. 5 F).
Increased expression of anti-inflammatory cytokines, particularly IL-10, is another hallmark of steroid action on T cells (Barrat et al., 2002). Therefore, to examine the impact of steroids on T cell cytokine production, we FACS-sorted CCR6+MDR1+, CCR6+MDR1−, and CCR6−MDR1− memory T cell subsets, activated them through the TCR in the presence or absence of steroids, and then restimulated the cells after 5 d to measure cytokine production via intracellular staining. As before, more MDR1+ T cells expressed pro-inflammatory cytokines, including IFN-γ, and fewer of these cells produced IL-10 relative to MDR1− T cell subsets (Fig. 5 G). Addition of either dexamethasone or prednisolone, but not rapamycin, to CCR6+ or CCR6−MDR1− memory T cells increased IL-10 production between two- and fourfold, and led some cells to produce both IFN-γ and IL-10 (Fig. 5 G). In contrast, neither dexamethasone nor prednisolone increased IL-10 production by MDR1+ T cells (Fig. 5 G). Expression of most pro-inflammatory cytokines, including IFN-γ and IL-17A, were not responsive to steroid treatment in any of the subsets analyzed (Fig. 5 G and not depicted). Thus, in addition to their unique, pro-inflammatory transcriptional signature and their enrichment within involved tissue from CD patients, human MDR1+ Th17.1 cells are broadly resistant to glucocorticoids.
To address the contribution of MDR1 efflux activity to glucocorticoid resistance in MDR1+ Th17.1 cells, we asked whether perturbing efflux activity via the selective MDR1 inhibitor elacridar could sensitize MDR1+ Th17.1 cells to the anti-inflammatory effects of glucocorticoids. However, using concentrations of Elacridar (0.1 µM) that block 99% of Rh123 efflux activity, we observed that elacridar did not reduce MDR1+ T cell expansion within mixed T cell cultures treated with dexamethasone or prednisolone, and elacridar did not facilitate IL-10 up-regulation in steroid-treated MDR1+ T cells (Fig. 5, H and I). Thus, whereas MDR1 expression marks steroid-resistant Th17.1 cells, its function, at least as manipulated by elacridar, is not a major determinant of this phenotype.
DISCUSSION
IL-23 is a central regulator of pathogenic Th17 cell function in humans and mice (Croxford et al., 2012). Using IL23R expression levels as a surrogate to define the pro-inflammatory potential of human effector/memory T cell subsets (Ghoreschi et al., 2010; Lee et al., 2012), we show that IL23R expression does not track with that of canonical Th17 cytokines (e.g., IL17A and IL17F) but rather is increased in CCR6+ Th17 subsets that display Th1-like properties (i.e., Th17.1 cells), namely expression of IFN-γ and CXCR3. Previous studies in mice suggest that IL-23 promotes inflammation, in part though induction of (or selection for) IFN-γ expression in Th17 cells, for example in EAE and T cell–induced colitis (Ahern et al., 2010; Codarri et al., 2011; Hirota et al., 2011). Consistent with these data, we find that an increased percentage of effector/memory T cells from involved gut tissue of CD patients produce both IL-17A and IFN-γ, compared with those from patient-matched peripheral blood; a previous study indicates that gut-infiltrating T cells in CD patients are highly sensitive to IL-23 stimulation (Kleinschek et al., 2009). Furthermore, we show that elevated expression of IL23R in human Th17.1 cells coincides with expression of a broader pathogenic Th17 transcriptional signature defined previously in mice (Ghoreschi et al., 2010; Lee et al., 2012).
Th22 cells are another effector/memory T cell subset that has both similar and nonoverlapping features relative to Th17 cells. Like Th17 cells, Th22 cells are CCR6+CCR4hiCXCR3lo and they produce IL-22 (Duhen et al., 2009; Eyerich et al., 2009; Trifari et al., 2009). Unlike Th17 cells, however, Th22 cells express additional skin-homing receptors, including CCR10 and CLA (cutaneous lymphocyte antigen), and they do not express IL-17A or IL-17F (Duhen et al., 2009; Eyerich et al., 2009; Trifari et al., 2009). Not surprisingly, we observed that IL22 mRNA expression in CCR6+CCR4hiCXCR3lo (Th17) cells sorted from independent donors (or donor pools) tracked with the level of contaminating Th22 cells, which varied between 4 and 16% of Th17 cells in our experiments. Despite this variable, however, expression of both IL23R and pathogenic murine Th17-signature genes was consistently increased in Th17.1 versus Th17 cells. Similar expression profiling of purified Th22 cells and other recently identified TEM lineages (e.g., Tfh cells, Th9 cells; Cosmi et al., 2013) will be important for a broader picture of pathogenic gene expression in the human immune system.
Despite elevated IL23R expression in human Th17.1 cells, neither they nor Th17 cells display robust functional responses to IL-23 stimulation, as judged by IL-17A up-regulation. Rather, both high-level IL23R expression and marked functional responses to IL-23 stimulation (i.e., STAT3 phosphorylation, IL-17A induction) are restricted to a subset of Th17.1 cells that also expresses the multi-drug transporter MDR1/P-gp. MDR1+ Th17.1 cells also show increased expression of Th17 and Th1 cytokines upon activation, and they selectively express pathogenic mouse Th17-signature genes (Ghoreschi et al., 2010; Lee et al., 2012), relative to either MDR1− Th17 or Th1 cells. Notably, these pro-inflammatory characteristics of MDR1+ Th17.1 cells are independent of another putative marker of activated human Th17 cells, CD161, which is generally enriched within Th17.1 cells but is not different between MDR1+ and MDR1− Th17.1 subsets.
Importantly, we show that MDR1+ Th17.1 cells are prevalent in actively inflamed gut tissue from CD patients, and that these cells display a marked pro-inflammatory transcriptional signature compared with local MDR1− memory T cells. MDR1+ memory T cells are proportionately increased in both uninvolved and involved gut tissue from CD patients, relative to patient-matched peripheral blood, and are further enriched in involved versus uninvolved gut tissue. Although the cohort analyzed in this study is small and it is difficult to draw general conclusions about MDR1+ Th17.1 cells in broader CD patient populations, these data suggest that MDR1+ T cell prevalence in involved CD patient gut tissue is a function of both preferential gut homing and active inflammation. As with Th17.1 cells from peripheral blood, both MDR1+ and MDR1− memory T cells from CD patient gut tissue are largely CD161+, consistent with a previous study describing CD161-expressing T cells in CD patient gut biopsies (Kleinschek et al., 2009). However, unlike MDR1+ memory T cells, MDR1−CD161+ memory cells are not enriched in involved versus uninvolved gut tissue of CD patients. Thus, MDR1 expression discriminates human pro-inflammatory Th17 cells in both blood and clinically inflamed tissue beyond that identified by current makers, including CD161.
MDR1 activity assays have been leveraged previously to characterize human tumors and to anticipate patient responses to therapy (Ludescher et al., 1992). Our experiments serve as a proof-of-principal that similar assays could be used for diagnostic or patient stratification purposes in CD patients. It will also be important to determine how the current findings extend to other inflammatory diseases, and to ask whether MDR1+ Th17 cell numbers change in autoimmune patients proportionate with disease remission/progression or upon treatment with specific therapies.
Of further relevance to clinical inflammation and therapy, we show that MDR1+ Th17.1 cells are resistant to the immunosuppressive effects of several natural and synthetic glucocorticoids. Glucocorticoids, including some tested in this study (dexamethasone, prednisolone, and 6α-methylprednisolone), are widely used in the treatment of CD, other IBDs, and other, non-IBD autoimmune disorders (Flammer and Rogatsky, 2011). However, patient responses to steroid therapy are unpredictable and are often plagued by the development of steroid-resistant disease. The reasons underlying clinical steroid resistance remain uncertain (Barnes and Adcock, 2009). Our data show not only that MDR1+ Th17.1 cells are refractory to the anti-proliferative and IL-10–inducing actions of steroids but also that these cells are actively and specifically selected for within mixed T cell cultures upon exposure to steroids. On one hand, these results are consistent with previous studies linking Th17 cells to steroid-resistant airway inflammation in mice and humans (McKinley et al., 2008; Alcorn et al., 2010; Ano et al., 2013). On the other hand, these data extend current notions of steroid resistance in Th17 cells by identifying a distinctive subset of Th17 cells that may underlie steroid hyporesponsiveness. It is also interesting to speculate, based on our findings, that steroid treatment of autoimmune patients may directly enrich for precisely the pro-inflammatory T cell subsets linked with inflammation and drug resistance. Of course, larger studies will be required to determine if, indeed, MDR1+ Th17.1 cell selection occurs in autoimmune patients upon steroid treatment, and whether such selection is tied to (or is predictive of) clinical outcomes.
It is unclear from our studies whether MDR1 contributes directly to pro-inflammatory T cell function or steroid resistance. In spite of the fact that MDR1+ Th17.1 cells display resistance to several glucocorticoids that are also known MDR1 substrates (Crowe and Tan, 2012), perturbing MDR1 efflux activity with the small molecule inhibitor elacridar neither influences MDR1+ Th17.1 cell function at steady-state nor sensitizes these cells to the immunosuppressive effects of glucocorticoids. Consistent with this, a previous study suggests that steroid resistance in Th17 cells is, at least partly, regulated by epigenetic repression of glucocorticoid target genes (McKinley et al., 2008). Although our results suggest that MDR1 function is not involved in the regulation of T cell responses to steroids, it remains an open question whether MDR1 contributes to T cell steroid resistance in a manner that is not sensitive to perturbation by elacridar. A recent study by Strouse et al. (2013) demonstrated that individual MDR1 inhibitors do not uniformly regulate efflux of all MDR1 substrates. Furthermore, structural studies indicate that MDR1 has two discreet substrate-binding pockets and is capable of accommodating multiple substrates/inhibitors simultaneously (Aller et al., 2009). Thus, whether elacridar or other MDR1 inhibitors block glucocorticoid efflux will need to be determined empirically. MDR1 may also regulate the intracellular concentrations of other endogenous small molecules that influence Th17 differentiation, Th17 cell effector function, and/or IL-23R signaling. Alternatively, MDR1 may act in T cells as a survival factor, for example by effluxing toxins or other xenobiotic compounds in the gut.
Drug transporter/dye efflux activity is widely considered a feature of long-lived stem and progenitor cells (Chaudhary and Roninson, 1991; Sincock and Ashman, 1997). Furthermore, Th17 cells in both mice and humans have been shown to harbor stem cell–like properties (Kryczek et al., 2011; Muranski et al., 2011). Consistent with these notions, we show that a sizeable proportion of MDR1+ Th17.1 cells from healthy donor peripheral blood express the stem cell marker c-Kit. We show several results supporting a model wherein c-Kit+MDR1+ memory T cells act as precursors to seed pathogenic c-Kit−MDR1+ progeny cells after activation under inflammatory conditions: c-Kit+MDR1+ Th17.1 cells can give rise to c-Kit−MDR1+ cells in vitro, after T cell activation in the presence of IL-23; c-Kit+MDR1+ Th17.1 cells are less mature than c-Kit−MDR1+ Th17.1 cells in vivo, as judged by increased levels of CCR7; and MDR1+ Th17.1 from CD lesions are uniformly c-Kit−. If c-Kit+MDR1+ Th17.1 cells indeed represent a long-lived human memory T cell compartment, further insight into their development and function may also prove important to understanding mechanisms of long-term immune memory and responses to vaccines.
In summary, we have identified a novel lineage of pro-inflammatory human Th17 cells that is defined by transient c-Kit expression and stable MDR1 activity. The facts that c-Kit+ and c-Kit− MDR1+ human Th17 cells express a unique pro-inflammatory transcriptional signature, produce both Th17 and Th1 cytokines, are highly sensitive to IL-23 stimulation, are enriched and activated within CD patient gut tissue, and are refractory to glucocorticoids speaks to the pathogenic potential of these cells in settings of clinical inflammation. We suggest that MDR1+ Th17 cells are important in the pathogenesis and drug resistance of autoimmune diseases, and that approaches to quantify or manipulate these cells in patients may prove useful in the diagnosis, molecular characterization, or treatment of immune-mediated inflammatory diseases.
MATERIALS AND METHODS
Human blood and tissue samples.
All experiments using human blood and tissue samples were conducted in accordance with IRB protocols approved by committees at Research Blood Components, New York Blood Center, OneBlood, Conversant Bio., and the University of Miami. Peripheral blood from healthy adult volunteers was purchased from Research Blood Components (Boston, MA), New York Blood Center (New York, NY), and OneBlood (Lauderhill, FL). Peripheral blood and biopsied gut tissues were collected under informed consent at the University of Miami Hospital from patients with active CD in accordance with the approved IRB protocol 20081100. Additional CD patient samples (peripheral blood, involved/uninvolved gut) collected during surgery under informed consent were purchased from Conversant Bio (Huntsville, AL) in accord with the approved IRB protocol Alabama Biobank Research Trial 001 V2 (ABRT_001-V2). Inflammation in tissue samples was confirmed by histopathology.
Media and cell culture.
Human mononuclear or purified T cells were cultured in complete T cell medium prepared as follows: DMEM or IMDM media (Gibco) supplemented with 10% FBS (Hyclone, Thermo Fisher Scientific), 1% l-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, 1% Hepes, and 1% Pen-Strep (all from Gibco). Purified T cells were activated using anti-CD3–/anti-CD28–coated beads (Dynabeads; Invitrogen) with or without recombinant human IL-23 (R&D Systems). For T cell activation and expansion, anti-CD3–/anti-CD28–coated beads were removed using Dynabead magnets (Invitrogen) on day 3, and cells were maintained with or without IL-23 for an additional 3–4 d. In some experiments, total CD4+ T cells, total memory T cells (CD45RO+CD25−), or FACS-sorted memory T cell subsets (CCR6+MDR1+, CCR6+MDR1−, and CCR6−MDR1−) were activated in the presence or absence (vehicle control; DMSO) of glucocorticoids—dexamethasone, prednisolone, or 6α-methylprednisolone (all from Sigma-Aldrich)—or rapamycin (EMD Chemicals). In some experiments, cells treated with or without glucocorticoids or rapamycin were further cultured with 0.1 µM Elacridar (Tocris Bioscience) or an equal volume of DMSO. For assessing T cell proliferation, cells were labeled with CellTrace violet (Invitrogen) before activation with or without glucocorticoids. For experiments involving expansion, activated T cells treated with or without glucocorticoids were split on day 4 and maintained for up to 10 more days in media supplemented with 20 ng/ml IL-2.
Cell isolation.
Mononuclear cells were isolated from peripheral blood by Ficoll density centrifugation. In brief, diluted blood (1:1 in PBS; Gibco) was overlaid onto lymphocyte separation media containing Ficoll (Ficoll Paque; GE Healthcare) and centrifuged at ambient temperature for 20 min at 2,000 rpm. Mononuclear cells were collected from the interface and washed two to three times with complete T cell medium for further analysis. For mononuclear cell isolation from clinical gut sections, biopsied tissue (∼0.5 × 1.5 in) was minced with scissors and rinsed thoroughly in DMEM (Gibco). Sections were placed in 50-ml conical vials containing DMEM supplemented with 0.15% DTT (Sigma-Aldrich) and agitated at room temperature for 30 min in a bacterial shaker (Environ Shaker; Labline) to remove residual mucus and debris. Sections were rinsed with DMEM medium and transferred to fresh 50 ml conical vials containing DMEM plus 1 mM EDTA and agitated at room temperature for 30 min to release epithelial cells. Sections were washed three times to remove EDTA, transferred to a fresh 50-ml conical vial, and resuspended in 20 ml DMEM containing 250 µg/ml Liberase and 10 U/ml DNase I (both from Roche). Samples were agitated at 37°C for 25 min, and digested tissue was filtered with a 70-µm filter (BD). Mononuclear cells from the flow through were isolated by 70/30% percoll (Sigma-Aldrich) gradient centrifugation. Total or memory CD4+ T cells were isolated using a Dynabeads CD4 positive T cell isolation kit (Invitrogen) or a human memory CD4+ T cell negative selection kit (STEMCELL Technologies; EasySep). Purified cells were 95–99% pure as determined by FACS analysis.
FACS sorting.
All FACS sorting was performed using a FACS Aria II (BD). For characterization of human memory T cell subsets from healthy adult donor peripheral blood, magnetically isolated memory CD4+ T cells were stained using the following antibodies in combination: anti-CD4, anti-CD45RO, anti-CD25, anti-CRTH2, anti-CCR6, anti-CCR4, anti-CXCR3, and anti-CCR7 (all from BioLegend). Stained cells were resuspended in IMDM medium without serum, filtered through a 70-µm filter, and sorted. In some experiments, anti–TCR-α/β or anti-CD3 antibodies (BioLegend) were included. For isolation of memory T cell subsets based on c-Kit expression and Rhodamine 123 (Rh123) efflux from healthy donor peripheral blood, negatively selected memory CD4+ T cells were labeled with Rh123 (Sigma-Aldrich), and, after efflux assay (see below), cells were stained with surface antibodies against anti-CD25, anti-CD45RO, anti-CCR7, anti-CRTH2, anti-CCR6, anti-CCR4, anti-CXCR3, and anti-c-Kit (CD117; BioLegend). Stained cells were resuspended, filtered, and sorted as above. For isolation of T cell subsets from biopsied patient tissues, total CD4+ T cells were first isolated from mononuclear cell preparations using positive selection (Miltenyi Biotec). CD4+ cells were then labeled with Rh123, and, after efflux assay (see below), stained with surface antibodies against CD3, CD45RO, CD25, and CD4 (BioLegend) before cell sorting.
FACS analysis.
Cell surface FACS staining was performed in FACS buffer (PBS + 2% FBS; HyClone) by incubating cells with fluorochrome-conjugated antibodies for 20 min at room temperature. After staining, cells were washed with PBS and stored in PBS plus 1% PFA. Intracellular cytokine and phospho-Stat3 staining were performed as previously described (Sundrud et al., 2009). In brief, cells used for intracellular cytokine staining were stimulated for 3–5 h at 37°C with 10 nM PMA and 1 µM ionomycin in the presence of 5 µg/ml Brefeldin A (all from Sigma-Aldrich), and stained after fixation and permeabilization using antibodies against IL-17A, IFN-γ, TNF, GM-CSF, and IL-10 (all from BioLegend). Phospho-Stat3 staining was determined according to manufacturer’s instructions using an anti-Stat3 (pY705) antibody (BD).
MDR1 activity assay.
Purified T cells in complete medium were loaded with Rh123 (Sigma-Aldrich) at a final concentration of 1 µg/ml for 30 min on ice. Cells were then washed and moved to a 37°C incubator for 2 h. After efflux period, cells were washed once in PBS, stained with surface markers, washed again in PBS, and stained cells were kept on ice before FACS analysis or FACS sorting. For some experiments 1 µM cyclosporine A (CsA; Sigma-Aldrich), 1 µM Elacridar (GlaxoSmithKline), or equal volume of DMSO (vehicle) was added to cells immediately before 37°C incubation step.
Microarrays.
Total RNA was processed and hybridized by the Boston University Microarray Resource Facility. All procedures were performed as described in the GeneChip Whole Transcript (WT) Sense Target Labeling Assay Manual (Affymetrix). RNA was hybridized to human 1.0 ST gene chips. Data were analyzed using GenePattern software (Broad Institute). Microarray data on FACS-sorted TEM Th1, Th2, Th17, and Th17.1 cells isolated from HC PBMC (accession ID: GSE49703), and on FACS-sorted MDR1+ and MDR1− memory CD4+ T cells isolated from healthy donor blood and CD tissue lesion (accession ID: GSE49702), are available at the gene expression omnibus (GEO).
Nanostring.
Gene expression was quantified using a custom nCounter probeset containing 92 probe pairs with additional probes for positive/negative controls and housekeeping genes (Nanostring Technologies). In brief, 30,000 T cells were lysed in 5 µl RLT buffer containing β-ME (QIAGEN). Biotin-conjugated capture probes and fluorescent-barcoded reporter probes were hybridized to cell lysates by overnight incubation at 65°C in a thermocycler (MJ Research). The next day, flow cell preparation and scanning were performed using an nCounter instrument (Nanostring Technologies) according to the manufacturer’s instructions. Raw data were normalized using nSolver software (Nanostring Technologies) and exported as raw transcript counts for presentation.
qPCR.
Messenger RNA was isolated from frozen cell pellets using the RNeasy kit (QIAGEN). cDNA was synthesized using a high capacity cDNA reverse transcription kit (Life Technologies/Applied Biosystems) per the manufacturer’s instructions. Detection of IL23R and ACTB mRNA was achieved using TaqMan qPCR performed on a StepOnePlus real time PCR instrument (Life Technologies/Applied Biosystems). TaqMan primer/probe sets for human IL23R (assay ID: Hs00332759_m1) and ACTB (assay ID: Hs01060665_g1) were purchased from Life Technologies/Applied Biosystems.
Lentivirus production and T cell transduction.
Human RORC (RORγ) cDNA was synthesized, cloned into pUC57, and shuttled into an HIV-1–derived lentiviral (HDV) plasmid containing IRES-murine CD24 (HSA) downstream of multi-cloning site (cloning done at Genscript; plasmid gift of D. Unutmaz; Sundrud et al., 2003). VSV-G pseudotyped empty or RORC-containing HDV particles with were produced in HEK293T cells by calcium phosphate transfection according to the manufacturer’s protocol (Profection kit; Promega). Lentiviral supernatants were concentrated by centrifugation (Sundrud et al., 2009), resuspended in complete T cell medium, and stored at −80°C. For T cell transduction experiments, purified naive CD4+ T cells (isolated using naive CD4 T-Cell isolation Kit; Miltenyi Biotec) were activated with anti-CD3–/anti-CD28–coated beads and infected with lentiviral supernatants (MOI = 50). Viral supernatants were removed on day 3 and infected T cells were expanded in media containing 10 U/ml recombinant human IL-2 (BD).
Online supplemental material.
Fig. S1 shows expression of ABC (ATP-binding cassette) family drug transporters in human TEM subsets. Table S1 shows gene expression signatures of human Th17 effector memory subsets. Table S2 shows ex vivo transcriptional signature of MDR1+ Th17.1 cells isolated from involved CD patient tissue. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130301/DC1.
Supplementary Material
Acknowledgments
We thank Drs. John Cleveland, Victor Torres, Sergei Koralov, and Christoph Rader for helpful discussions and critical review of the manuscript. We thank Scott Davis for bioinformatics support, and the Biorepository in the Center for Genome Technology at the Hussman Institute for Human Genomics for biospecimen processing, storage, and shipment. Tempero Pharmaceuticals, Inc. is majority-owned and funded by GlaxoSmithKline.
A portion of this work was supported through National Institutes of Health (NIH) grants R01AI065303 and NIH R21AI087973 (to D. Unutmaz), 1R01CA137869 (to M.T. Abreu), and a senior investigator award from the Crohn’s and Colitis Foundation of America (to M.T. Abreu). Additional support was provided by The Micky & Madeleine Arison Family Foundation Crohn’s & Colitis Discovery Laboratory at the University of Miami, and by The Scripps Research Institute, Florida from The State of Florida (to M.S. Sundrud)
The authors declare no competing financial interests.
Author contributions: R. Ramesh designed experiments, analyzed data, and developed Rh123 efflux assay. L. Kozhaya designed and performed glucocorticoid assays, analyzed data, and edited the manuscript. K. McKevitt performed experiments on clinical samples from CD patients and analyzed data. I.M. Djuretic developed staining for TCM and TEM subsets, performed microarray experiments, and edited the manuscript. T.J. Carlson provided technical support and key discussion, and edited the manuscript. M.A. Quintero and J.L. McCauley coordinated and supervised collection and shipment of CD patient samples, and edited the manuscript. M.T. Abreu performed procedures to collect blood and biopsied tissues from CD patients, supervised the study, provided key discussion, and edited the manuscript. D. Unutmaz supervised glucocorticoid assays, provided key discussion, and edited the manuscript. M.S. Sundrud supervised the study, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used:
- EAE
- experimental autoimmune encephalomyelitis
- IBD
- inflammatory bowel disease
- P-gp
- P-glycoprotein
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Afzali B., Mitchell P.J., Edozie F.C., Povoleri G.A., Dowson S.E., Demandt L., Walter G., Canavan J.B., Scotta C., Menon B., et al. 2013. CD161 expression characterizes a subpopulation of human regulatory T cells that produces IL-17 in a STAT3-dependent manner. Eur. J. Immunol. 43:2043–2054 10.1002/eji.201243296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., Powrie F. 2010. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 33:279–288 10.1016/j.immuni.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcorn J.F., Crowe C.R., Kolls J.K. 2010. TH17 cells in asthma and COPD. Annu. Rev. Physiol. 72:495–516 10.1146/annurev-physiol-021909-135926 [DOI] [PubMed] [Google Scholar]
- Aller S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P.M., Trinh Y.T., Zhang Q., Urbatsch I.L., Chang G. 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 323:1718–1722 10.1126/science.1168750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano S., Morishima Y., Ishii Y., Yoh K., Yageta Y., Ohtsuka S., Matsuyama M., Kawaguchi M., Takahashi S., Hizawa N. 2013. Transcription factors GATA-3 and RORγt are important for determining the phenotype of allergic airway inflammation in a murine model of asthma. J. Immunol. 190:1056–1065 10.4049/jimmunol.1202386 [DOI] [PubMed] [Google Scholar]
- Araki K., Ellebedy A.H., Ahmed R. 2011. TOR in the immune system. Curr. Opin. Cell Biol. 23:707–715 10.1016/j.ceb.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P.J., Adcock I.M. 2009. Glucocorticoid resistance in inflammatory diseases. Lancet. 373:1905–1917 10.1016/S0140-6736(09)60326-3 [DOI] [PubMed] [Google Scholar]
- Barrat F.J., Cua D.J., Boonstra A., Richards D.F., Crain C., Savelkoul H.F., de Waal-Malefyt R., Coffman R.L., Hawrylowicz C.M., O’Garra A. 2002. In vitro generation of interleukin 10-producing regulatory CD4+ T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J. Exp. Med. 195:603–616 10.1084/jem.20011629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary P.M., Roninson I.B. 1991. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell. 66:85–94 10.1016/0092-8674(91)90141-K [DOI] [PubMed] [Google Scholar]
- Chen Z., Laurence A., Kanno Y., Pacher-Zavisin M., Zhu B.M., Tato C., Yoshimura A., Hennighausen L., O’Shea J.J. 2006. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl. Acad. Sci. USA. 103:8137–8142 10.1073/pnas.0600666103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., Becher B. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12:560–567 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Cohen C.J., Crome S.Q., MacDonald K.G., Dai E.L., Mager D.L., Levings M.K. 2011. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. J. Immunol. 187:5615–5626 10.4049/jimmunol.1101058 [DOI] [PubMed] [Google Scholar]
- Cosmi L., De Palma R., Santarlasci V., Maggi L., Capone M., Frosali F., Rodolico G., Querci V., Abbate G., Angeli R., et al. 2008. Human interleukin 17–producing cells originate from a CD161+CD4+ T cell precursor. J. Exp. Med. 205:1903–1916 10.1084/jem.20080397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmi L., Maggi L., Santarlasci V., Capone M., Cardilicchia E., Frosali F., Querci V., Angeli R., Matucci A., Fambrini M., et al. 2010. Identification of a novel subset of human circulating memory CD4(+) T cells that produce both IL-17A and IL-4. J. Allergy Clin. Immunol. 125:222–230: e1–e4 10.1016/j.jaci.2009.10.012 [DOI] [PubMed] [Google Scholar]
- Cosmi L., Maggi L., Santarlasci V., Liotta F., Annunziato F. 2013. T helper cells plasticity in inflammation. Cytometry A.:n/a 10.1002/cyto.a.22348 [DOI] [PubMed] [Google Scholar]
- Crowe A., Tan A.M. 2012. Oral and inhaled corticosteroids: differences in P-glycoprotein (ABCB1) mediated efflux. Toxicol. Appl. Pharmacol. 260:294–302 10.1016/j.taap.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Croxford A.L., Mair F., Becher B. 2012. IL-23: one cytokine in control of autoimmunity. Eur. J. Immunol. 42:2263–2273 10.1002/eji.201242598 [DOI] [PubMed] [Google Scholar]
- Duhen T., Geiger R., Jarrossay D., Lanzavecchia A., Sallusto F. 2009. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10:857–863 10.1038/ni.1767 [DOI] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W., Jr, Rongvaux A., Van Rooijen N., Haberman A.M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S., Eyerich K., Pennino D., Carbone T., Nasorri F., Pallotta S., Cianfarani F., Odorisio T., Traidl-Hoffmann C., Behrendt H., et al. 2009. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Invest. 119:3573–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flammer J.R., Rogatsky I. 2011. Minireview: Glucocorticoids in autoimmunity: unexpected targets and mechanisms. Mol. Endocrinol. 25:1075–1086 10.1210/me.2011-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M.M., Fojo T., Bates S.E. 2002. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer. 2:48–58 10.1038/nrc706 [DOI] [PubMed] [Google Scholar]
- Hirota K., Duarte J.H., Veldhoen M., Hornsby E., Li Y., Cua D.J., Ahlfors H., Wilhelm C., Tolaini M., Menzel U., et al. 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12:255–263 10.1038/ni.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber W., Sands B.E., Lewitzky S., Vandemeulebroecke M., Reinisch W., Higgins P.D., Wehkamp J., Feagan B.G., Yao M.D., Karczewski M., et al. , for the Secukinumab in Crohn’s Disease Study Group 2012. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 61:1693–1700 10.1136/gutjnl-2011-301668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyafil F., Vergely C., Du Vignaud P., Grand-Perret T. 1993. In vitro and in vivo reversal of multidrug resistance by GF120918, an acridonecarboxamide derivative. Cancer Res. 53:4595–4602 [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., Rudensky A.Y. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebir H., Ifergan I., Alvarez J.I., Bernard M., Poirier J., Arbour N., Duquette P., Prat A. 2009. Preferential recruitment of interferon-gamma-expressing TH17 cells in multiple sclerosis. Ann. Neurol. 66:390–402 10.1002/ana.21748 [DOI] [PubMed] [Google Scholar]
- Kleinschek M.A., Boniface K., Sadekova S., Grein J., Murphy E.E., Turner S.P., Raskin L., Desai B., Faubion W.A., de Waal Malefyt R., et al. 2009. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 206:525–534 10.1084/jem.20081712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Zhao E., Liu Y., Wang Y., Vatan L., Szeliga W., Moyer J., Klimczak A., Lange A., Zou W. 2011. Human TH17 cells are long-lived effector memory cells. Sci. Transl. Med. 3:104ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. 2012. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13:991–999 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludescher C., Thaler J., Drach D., Drach J., Spitaler M., Gattringer C., Huber H., Hofmann J. 1992. Detection of activity of P-glycoprotein in human tumour samples using rhodamine 123. Br. J. Haematol. 82:161–168 10.1111/j.1365-2141.1992.tb04608.x [DOI] [PubMed] [Google Scholar]
- Maggi L., Santarlasci V., Capone M., Rossi M.C., Querci V., Mazzoni A., Cimaz R., De Palma R., Liotta F., Maggi E., et al. 2012. Distinctive features of classic and nonclassic (Th17 derived) human Th1 cells. Eur. J. Immunol. 42:3180–3188 10.1002/eji.201242648 [DOI] [PubMed] [Google Scholar]
- Manel N., Unutmaz D., Littman D.R. 2008. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9:641–649 10.1038/ni.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M.J., Bak-Jensen K.S., Chen Y., Tato C.M., Blumenschein W., McClanahan T., Cua D.J. 2007. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8:1390–1397 10.1038/ni1539 [DOI] [PubMed] [Google Scholar]
- McGeachy M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., Cua D.J. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley L., Alcorn J.F., Peterson A., Dupont R.B., Kapadia S., Logar A., Henry A., Irvin C.G., Piganelli J.D., Ray A., Kolls J.K. 2008. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J. Immunol. 181:4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miossec P., Korn T., Kuchroo V.K. 2009. Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361:888–898 10.1056/NEJMra0707449 [DOI] [PubMed] [Google Scholar]
- Muranski P., Borman Z.A., Kerkar S.P., Klebanoff C.A., Ji Y., Sanchez-Perez L., Sukumar M., Reger R.N., Yu Z., Kern S.J., et al. 2011. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 35:972–985 10.1016/j.immuni.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalithas K., Guillen C., Day C., Brightling C.E., Pavord I.D., Wardlaw A.J. 2010. CRTH2 expression on T cells in asthma. Clin. Exp. Immunol. 161:34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala K., Adams S., Cambrook H., Ursu S., Olivito B., de Jager W., Evans J.G., Cimaz R., Bajaj-Elliott M., Wedderburn L.R. 2010. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc. Natl. Acad. Sci. USA. 107:14751–14756 10.1073/pnas.1003852107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivino L., Messi M., Jarrossay D., Lanzavecchia A., Sallusto F., Geginat J. 2004. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J. Exp. Med. 200:725–735 10.1084/jem.20040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.M., Swart B.W., Simmons P.J., Basser R.L., Begley C.G., To L.B. 1999. Prolonged release and c-kit expression of haemopoietic precursor cells mobilized by stem cell factor and granulocyte colony stimulating factor. Br. J. Haematol. 104:778–784 10.1046/j.1365-2141.1999.01231.x [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Sallusto F., Zielinski C.E., Lanzavecchia A. 2012. Human Th17 subsets. Eur. J. Immunol. 42:2215–2220 10.1002/eji.201242741 [DOI] [PubMed] [Google Scholar]
- Schinkel A.H. 1997. The physiological function of drug-transporting P-glycoproteins. Semin. Cancer Biol. 8:161–170 10.1006/scbi.1997.0068 [DOI] [PubMed] [Google Scholar]
- Simmons P.J., Leavesley D.I., Levesque J.P., Swart B.W., Haylock D.N., To L.B., Ashman L.K., Juttner C.A. 1994. The mobilization of primitive hemopoietic progenitors into the peripheral blood. Stem Cells. 12:187–201, discussion :201–202 [PubMed] [Google Scholar]
- Sincock P.M., Ashman L.K. 1997. Expression of c-Kit and functional drug efflux are correlated in de novo acute myeloid leukaemia. Leukemia. 11:1850–1857 10.1038/sj.leu.2400823 [DOI] [PubMed] [Google Scholar]
- So T., Lee S.W., Croft M. 2008. Immune regulation and control of regulatory T cells by OX40 and 4-1BB. Cytokine Growth Factor Rev. 19:253–262 10.1016/j.cytogfr.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouse J.J., Ivnitski-Steele I., Waller A., Young S.M., Perez D., Evangelisti A.M., Ursu O., Bologa C.G., Carter M.B., Salas V.M., et al. 2013. Fluorescent substrates for flow cytometric evaluation of efflux inhibition in ABCB1, ABCC1, and ABCG2 transporters. Anal. Biochem. 437:77–87 10.1016/j.ab.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundrud M.S., Grill S.M., Ni D., Nagata K., Alkan S.S., Subramaniam A., Unutmaz D. 2003. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J. Immunol. 171:3542–3549 [DOI] [PubMed] [Google Scholar]
- Sundrud M.S., Koralov S.B., Feuerer M., Calado D.P., Kozhaya A.E., Rhule-Smith A., Lefebvre R.E., Unutmaz D., Mazitschek R., Waldner H., et al. 2009. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 324:1334–1338 10.1126/science.1172638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S., Kaplan C.D., Tran E.H., Crellin N.K., Spits H. 2009. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 10:864–871 10.1038/ni.1770 [DOI] [PubMed] [Google Scholar]
- Turtle C.J., Swanson H.M., Fujii N., Estey E.H., Riddell S.R. 2009. A distinct subset of self-renewing human memory CD8+ T cells survives cytotoxic chemotherapy. Immunity. 31:834–844 10.1016/j.immuni.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Kozhaya L., ElHed A., Ramesh R., Carlson T.J., Djuretic I.M., Sundrud M.S., Unutmaz D. 2011. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J. Exp. Med. 208:1875–1887 10.1084/jem.20102516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J., Haefeli W.E. 2010. Impact of ATP-binding cassette transporters on human immunodeficiency virus therapy. Int Rev Cell Mol Biol. 280:219–279 10.1016/S1937-6448(10)80005-X [DOI] [PubMed] [Google Scholar]
- Zorzi F., Monteleone I., Sarra M., Calabrese E., Marafini I., Cretella M., Sedda S., Biancone L., Pallone F., Monteleone G. 2013. Distinct profiles of effector cytokines mark the different phases of Crohn’s disease. PLoS ONE. 8:e54562 10.1371/journal.pone.0054562 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.