EndoU is a critical regulator of a novel RNA-dependent pathway controlling peripheral B cell survival and antigen responsiveness.
Abstract
Balanced transmembrane signals maintain a competent peripheral B cell pool limited in self-reactive B cells that may produce pathogenic autoantibodies. To identify molecules regulating peripheral B cell survival and tolerance to self-antigens (Ags), a gene modifier screen was performed with B cells from CD22-deficient C57BL/6 (CD22−/−[B6]) mice that undergo activation-induced cell death (AICD) and fail to up-regulate c-Myc expression after B cell Ag receptor ligation. Likewise, lysozyme auto-Ag–specific B cells in IgTg hen egg lysozyme (HEL) transgenic mice inhabit the spleen but undergo AICD after auto-Ag encounter. This gene modifier screen identified EndoU, a single-stranded RNA-binding protein of ancient origin, as a major regulator of B cell survival in both models. EndoU gene disruption prevents AICD and normalizes c-Myc expression. These findings reveal that EndoU is a critical regulator of an unexpected and novel RNA-dependent pathway controlling peripheral B cell survival and Ag responsiveness that may contribute to peripheral B cell tolerance.
Molecules that regulate lymphocyte homeostasis, proliferation, and survival operate in concert to enable robust adaptive immune responses to foreign antigens (Ags). For the B cell lineage, the optimal outcome of these processes is a diverse antibody (Ab) repertoire purged of pathological (self-reactive) B cells. The elimination of pathological B cells occurs either through clonal deletion or receptor editing during B lymphopoiesis in the bone marrow, or in the periphery through the induction of anergy (Goodnow et al., 1988; Nemazee and Bürki, 1989; Gay et al., 1993; Tiegs et al., 1993). Anergic B cells primarily inhabit the spleen, are short-lived, and undergo activation-induced cell death (AICD) in response to B cell Ag receptor (BCR) stimulation (Goodnow et al., 1995; Shlomchik, 2008).
BCR ligation by agonistic anti-IgM Abs induces 30–50% of spleen B cells from WT mice to blast and undergo proliferation ex vivo (DeFranco et al., 1982). However, the threshold for B cell AICD can be influenced by genetically altering the stimulatory and inhibitory pathways that regulate BCR-induced activation (Inaoki et al., 1997). The B cell–restricted surface protein CD22 is generally considered to negatively regulate BCR signaling by recruiting potent intracellular phosphatases after BCR ligation (Doody et al., 1995; O’Keefe et al., 1996; Otipoby et al., 1996; Sato et al., 1996; Nitschke et al., 1997; Tedder et al., 1997; Poe et al., 2000), and CD22−/− mice produce augmented levels of isotype-switched auto-Abs against DNA and some protein Ags (O’Keefe et al., 1999; Poe et al., 2011). Nevertheless, B cells from inbred CD22−/− mice with a B6/129 genetic background (CD22−/−[inbr]) are phenotypically and functionally normal ex vivo (Poe et al., 2004). In contrast, spleen B cells from C57BL/6 (B6) mice genetically deficient in CD22 (CD22−/−[B6]) undergo AICD after BCR stimulation (Poe et al., 2004), which is likely to be a result of their inability to induce c-Myc transcription factor expression that balances B cell proliferation versus AICD (Donjerković and Scott, 2000; Poe et al., 2004). These striking phenotypic differences in B cells between mouse lines with a common deletion of Cd22 indicate that important B cell signaling events that promote AICD are influenced differently by the B6 and 129 genetic backgrounds. These two CD22−/− mouse lines were therefore used to identify genetic and molecular factors regulating B cell AICD.
In these studies, a forward genetic screen was used to identify an evolutionarily conserved single-stranded RNA (ssRNA) binding protein, EndoU, as a novel regulator of AICD in CD22−/−[B6] mice. EndoU was also overexpressed by anergic peripheral B cells from double-transgenic mice expressing BCRs specific for hen egg lysozyme (HEL) along with soluble HEL (sHEL) as the cognate auto-Ag (IgTgsHEL mice; Goodnow et al., 1989; Hippen et al., 2000; Shlomchik, 2008). EndoU deficiency in IgTgsHEL mice also reversed AICD ex vivo and led to augmented anti-HEL auto-Ab responses in vivo. Thus, EndoU defines a new posttranscriptional regulatory pathway that controls B cell AICD, particularly in response to auto-Ag.
RESULTS
A genetic modifier locus/loci regulates BCR-induced AICD and CD5 expression
Spleen B cells from an inbred B6/129 founder line (CD22−/−[inbr]), their WT littermates (WT[inbr]), and WT B6 (WT[B6]) mice developed into blasts at normal frequencies and proliferated similarly after ex vivo BCR ligation using agonistic anti-IgM Abs (Fig. 1, A and B). In contrast, B cells from CD22−/− mice that were extensively backcrossed onto the B6 genetic background (CD22−/−[B6]) underwent AICD after BCR ligation. CD22−/−[B6] B cells also expressed CD5 after BCR stimulation but failed to up-regulate c-Myc transcript expression, whereas B cells from CD22−/−[inbr] had normal CD5 and c-Myc expression (Fig. 1, C and D). Similarly, B cells from IgTgsHEL mice with a B6 background underwent AICD, expressed CD5, and failed to up-regulate c-Myc after ex vivo BCR stimulation (Fig. 1, E–G). In contrast, B cells from IgTg mice (lacking the sHEL auto-Ag) blasted robustly, expressed CD5 at normal levels, and expressed high c-Myc levels after BCR ligation. These striking phenotypic similarities between CD22−/−[B6] and IgTgsHEL mice suggested that shared genetic and signaling programs regulated B cell AICD.
Figure 1.
CD22−/−[B6] B cells are phenotypically similar to anergic IgTgsHEL B cells. (A) Representative B cell blast assay in which WT or CD22−/− spleen B cells purified from B6 (WT[B6], CD22−/−[B6]) or inbred B6:129 (WT[inbr], CD22−/−[inbr]) mice were cultured in medium alone or containing F(ab’)2 anti-IgM Abs. After 48 h, the cells were analyzed for viability (7-amino-actinomycin d [7AAD] exclusion) and cell size (forward scatter [FSC]) by flow cytometry. Percentages indicate viable B cell blasts (7AAD−FSChigh) within the gate shown. Results are representative of >10 mice of each genotype producing similar results. (B) The proliferation of purified spleen B cells from CD22−/−[B6] or CD22−/−[inbr] mice, or their WT littermates, was measured by [3H]-thymidine incorporation. Values represent mean (±SEM) cpm for triplicate wells from one of three independent experiments producing similar results. **, P < 0.01, Student’s t test. (C) Representative CD5 immunofluorescence staining of freshly isolated B220+ splenocytes from ≥5 mice/group of the indicated genotypes. Background staining using an isotype-matched control mAb is shown (Neg Ctl). (D) Spleen B cells from WT[B6] or CD22−/−[B6] mice were cultured with F(ab’)2 anti-IgM Abs for 18 h. [32P]-radiolabeled cDNA was generated from total RNA, followed by hybridization to GEArray signaling pathway gene arrays. Regions of the arrays containing the c-Myc gene (arrowheads) and other representative genes expressed in B cells, including the c-Myc binding partner Max, are shown. (E) Mean (±SEM) frequencies of B cell blasts in untreated or F(ab’)2 anti-IgM Ab-stimulated cultures (48 h) from four each IgTg and IgTgsHEL mice assessed in independent experiments. **, P < 0.01, Student’s t test. (F) Representative CD5 expression by spleen B cells from IgTgsHEL mice and IgTg mice as assessed by flow cytometry. A nonreactive isotype-matched Ab was used as a negative control (Neg Ctl). (G) Purified spleen B cells from IgTg or IgTgsHEL mice were cultured alone or with F(ab’)2 anti-IgM Abs. After 18 h, the cells were analyzed for c-Myc expression by intracellular staining, with flow cytometry analysis. Percentages of c-Myc+ B cells (those above the dashed line) are indicated in each plot. Bar graphs represent the mean (±SEM) percentage of c-Myc+ B cells in BCR-stimulated cultures compared with unstimulated cultures from three mice of each genotype assessed in independent experiments. **, P < 0.01, Student’s t test.
CD22−/−[inbr] mice were maintained as syngeneic brother/sister pairings for >8 yr, with genetic loci homozygous for either B6 germline DNA (B6:B6) or 129 germline DNA (129:129). To identify the genetic basis for the CD22−/−[B6] B cell phenotype, CD22−/−[B6] and CD22−/−[inbr] mice were first crossed to produce an F1 generation (CD22−/−[F1], Fig. 2 A) with either homozygous B6 (B6:B6) or heterozygous B6 and 129 (B6:129) genetic loci. CD22−/−[F1] B cell blast development after BCR stimulation was intermediate between B cells from CD22−/−[B6] and CD22−/−[inbr] mice (unpublished data), indicating that maximal AICD of CD22−/−[B6] B cells required B6:B6 homozygosity at a critical locus (or loci). CD22−/−[F1] and CD22−/−[B6] mice were subsequently crossed to generate genetically distinct CD22−/−[N1] littermates due to independent chromosomal segregation and crossovers between regions of heterozygous B6:129 genomic DNA during gametogenesis. Remarkably, B cells from CD22−/−[N1] mice underwent AICD (Fig. 2 B, top dot plots) or developed into blasts at substantial levels (Fig. 2 B, bottom dot plots), which correlated directly with increased or normal CD5 expression, respectively. These results confirmed the existence of a genetic modifier locus/loci that regulates BCR-induced AICD and CD5 expression.
Figure 2.
A Chr 15 locus segregates relative to CD22−/− B cell survival after BCR ligation. (A) Crossing CD22−/−[B6] mice with CD22−/−[inbr] mice generated genetically similar F1 progeny (CD22−/−[F1]). CD22−/−[F1] mice were then backcrossed to CD22−/−[B6] mice to generate genetically dissimilar N1 littermates (CD22−/−[N1]) that were used for QTL mapping. (B) Spleen B cells from CD22−/−[N1] mice were assessed for CD5 expression before BCR stimulation and for blast development after BCR stimulation as in Fig. 1. B cells from some CD22−/−[N1] mice were anergic (were CD5high and underwent AICD [7AAD+FSClow] after BCR stimulation; top), whereas others had a normal phenotype (were CD5low and developed into blasts [7AAD−FSChigh], bottom). Results are representative of those obtained from >40 CD22−/−[N1] littermates assessed in 10 independent experiments. (C) Survival analysis of BCR-stimulated B cells from CD22−/−[N1] littermates used for genomic SNP genotyping. Results are pooled from 10 independent CD22−/−[N1] mouse B cell blast assays (open squares), as described in B. Each value represents a single mouse, with dashed boxes distinguishing between CD22−/−[N1] littermates having B cells sufficient in blast development (top box), or insufficient in blast development (bottom box). Shown for comparison are the parental mouse B cells assessed in each of the assays as controls: CD22−/−[B6] B cells (apoptotic, black diamonds) and CD22−/−[inbr] B cells (viable, gray circles). (D) QTL mapping results from the 44 CD22−/−[N1] experimental mice shown in C, revealing a Chr 15 locus with a LOD score peaking at >12 (P < 0.000001, χ2 statistic, MapManager QTX permutation test). The Cd22 gene location (Chr 7) is indicated. (E) Detailed heat map display of the Chr 15 mapping data shown in D. The locus of significance was defined by four consecutive SNPs positioned at 68, 71, 78, and 91 Mbp with LOD scores of 8.3–12.6. Columns represent individual CD22−/−[N1] mice, with blue squares indicating homozygous B6:B6 SNP loci and red squares indicating heterozygous B6:129 SNP loci. Representative genotypes of parental CD22−/−[B6] and CD22−/−[inbr] mice are shown at the right, with green squares indicating regions of 129 homozygosity. (F) Refined PCR-based SNP mapping of the Chr 15 locus of CD22−/−[N1] mice at 1 Mbp intervals from 78 to 102 Mbp. Open squares represent SNPs that were not assessed. (G) B cells from an additional 59 CD22−/−[N1] mice were identified as viable or apoptotic in addition to the 44 CD22−/−[N1] mice shown in C–F. All 103 mice (55 with viable B cells and 48 with apoptotic B cells) were then assessed at 0.25–1 Mbp resolution by PCR-based SNP genotyping within the proximal (93.3 Mbp) to distal (98.3 Mbp) region of Chr 15.
A genetic locus regulates AICD of B cells from CD22−/−[B6] mice
Segregation of the B cell survival versus AICD phenotypes in CD22−/−[N1] mice indicated that the genomes of the parental CD22−/− mouse lines were stable and amenable to genetic analyses. Therefore, a forward genetic linkage analysis was used to identify the gene locus/loci responsible for CD22−/−[B6] B cell AICD. Genome-wide genotyping of 250 informative single nucleotide polymorphisms (SNPs) between the B6 and 129 mouse strains used genomic DNA from 44 CD22−/−[N1] mice; 22 mice had viable B cells after BCR ligation, whereas 22 mice had B cells that underwent AICD (Fig. 2 C). Quantitative trait loci (QTL) mapping regression analysis revealed a single locus with high logarithm of odds (LOD) scores (P < 10−6) on the distal end of chromosome (Chr) 15 (Fig. 2 D), with four consecutive SNPs spanning the region from 68–91 megabase pairs (Mbp, Fig. 2 E). LOD scores from the first three SNPs (68, 71, and 78 Mbp) were 8.3, increasing to 12.6 for the distal SNP (91 Mbp). The increased LOD score at 91 Mbp resulted from two mice (#14 and 28) with additional crossovers that were concordant with the observed phenotype. Parental CD22−/−[B6] mice were homozygous B6:B6 within the identified locus (68–91 Mbp), whereas CD22−/−[inbr] mice were homozygous 129:129. All other SNPs on Chr 15 outside of this locus (3.8–53 and 102 Mbp) were homozygous B6:B6 in all mice. Therefore, the primary genetic element responsible for CD22−/−[B6] B cell AICD was located within a 24 Mbp region.
To narrow the 78 to 102 Mbp Chr 15 locus, 23 additional SNPs within this region at 1 Mbp intervals were genotyped in the original 44 N1 mice. Using this method, the AICD phenotype mapped to the 94–98 Mbp region for all 44 CD22−/−[N1] mice (Fig. 2 F), including refined border regions for mice #14 and #28. Mice having apparently discordant phenotypes under low resolution mapping (viable, #2 and 19; nonviable, #44, Fig. 2 E) had concordant genotypes with higher resolution analysis (Fig. 2 F). To further refine and validate this locus as harboring the genetic element controlling AICD, additional SNPs and a total of 103 CD22−/−[N1] mice were analyzed (Fig. 2 G). Genotype and observed phenotypes were 98% (101/103) concordant in the mice, with additional crossovers observed in CD22−/−[N1] mice #21 and 64. Thus, the CD22−/−[N1] survival locus mapped to a refined 5 Mbp region spanning from 93.3 to 98.3 Mbp on Chr 15.
EndoU is overexpressed in B cells that undergo AICD
The 93.3–98.3 Mbp region encodes 17 pseudogenes and 38 known genes, including 10 olfactory receptor genes not expressed in hematopoietic cells. Therefore, the differential expression of transcripts from these genes was compared using BCR-stimulated CD22−/−[inbr] and CD22−/−[B6] B cells that remained similarly viable for 18 h. A single gene was differentially expressed between mouse lines, with 4.4-fold higher EndoU (formerly Pp11r/Tcl-30) levels in CD22−/−[B6] B cells compared with CD22−/−[inbr] B cells (Fig. 3 A). B cells from WT[B6] mice expressed EndoU at lower levels than B cells from WT 129 (WT[129]) mice before and after BCR ligation (Fig. 3 B). EndoU transcript levels in CD22−/−[B6] B cells remained uniquely higher relative to CD22−/−[inbr] mice and all other genotypes after BCR ligation, whereas CD22−/−[F1] B cells expressed intermediate transcript levels. Thus, EndoU overexpression required CD22 deficiency in the context of a B6 genetic background.
Figure 3.
EndoU, a gene within the Chr 15 locus that encodes a novel ssRNA binding protein, is overexpressed in anergic B cells. (A) Relative transcript expression of the 28 genes within the Chr 15 locus in BCR-stimulated spleen B cells from CD22−/−[B6] (closed bars) or CD22−/−[inbr] (open bars) mice. A single gene, EndoU (arrows), was differentially expressed by B cells from CD22−/−[B6] and CD22−/−[inbr] parental mice. (B) RNA was isolated from purified spleen B cells that were untreated (open bars) or were stimulated with F(ab’)2 anti-IgM Abs for 18 h (closed bars), followed by real-time RT-PCR analysis of EndoU gene expression. Values represent mean (±SEM) fold changes in EndoU expression with Cd20 expression as the internal control gene in B cells from three mice of each genotype. All values were normalized relative to WT[129] B cells after BCR stimulation (dashed line). The diploid genotypes of each mouse line at the Chr 15 SNP-defined locus are indicated. (C) Purified spleen B cells or CD4+ T cells were cultured alone or with F(ab’)2 anti-IgM Abs or mitogenic CD3 mAb, respectively, for 18 h. Total cellular proteins were separated by SDS-PAGE, followed by transfer to nitrocellulose membranes and Western blot analysis using polyclonal anti-EndoU Ab. The arrow indicates the position of the major EndoU band, which was dominant in CD22−/−[B6] B cells. A nonspecific band (n.s.) is present in all lanes that is also observed when the secondary detection Ab is used alone as a control to probe CD22−/−[B6] B cell lysates (far right lane). Membranes were reprobed with a polyclonal ERK2 Ab as a control for total protein loading. Bar graphs indicate relative B cell EndoU band densities (±SEM) from three each CD22−/−[B6] and CD22−/−[inbr] mice assessed in independent experiments. **, P < 0.01, Student’s t test. (D) Purified spleen B cells from the indicated mice were cultured alone or with F(ab’)2 anti-IgM Abs for 18 h. Total cellular proteins were separated by SDS-PAGE, followed by transfer to nitrocellulose membranes and Western blot analysis using polyclonal anti-EndoU Ab as described in C. Bar graphs indicate mean EndoU band densities from three each IgTg and IgTgsHEL mice as assessed in two independent experiments producing similar results. **, P < 0.01, Student’s t test. (E) [32P]-labeled ssRNA substrates described for XendoU (Laneve et al., 2008) were incubated with recombinant EndoU protein in the presence or absence of Mn2+ or EDTA as indicated. Reactions were then separated by 15% PAGE and visualized by phosphorimager. The migration positions of EndoU-RNA complexes and of unbound RNA are indicated. Results are representative of those obtained in three experiments producing similar results. (F) c-Myc mRNA (sequence at left) shows an enrichment for poly(U) sequences. Regions of three or more consecutive uridines are highlighted. Bar graph at right shows the binding of in vitro transcribed, [32P]-labeled c-Myc mRNA by recombinant EndoU protein bound to IgG-coated magnetic beads as quantified by scintillation counting. A >100-fold molar excess of nonspecific protein (BSA) and RNA (tRNAs) were used as carriers in these assays. Background c-Myc RNA binding to magnetic beads without EndoU (Carrier) is shown. Bar graphs represent means (±SEM) from three binding assays per group. Results represent one of two experiments producing similar results. (G) NIH-3T3 cells were stably transfected with a plasmid encoding EndoU fused with CFP reporter, or with plasmid expressing a reporter protein alone as a control. Shown are representative histogram overlays of immunofluorescence staining of intracellular c-Myc protein in reporter-positive cells as determined by flow cytometry. The percent decrease in and p-value for the reduction of mean c-Myc MFI values (minus isotype control [Iso ctl] Ab staining) in EndoU-transfected cells (EndoU) relative to control-transfected cells (Vector) from four independent experiments are indicated. The significance level was determined using the Student’s t test.
To ensure that EndoU protein levels correlated with gene overexpression in CD22−/−[B6] B cells, an affinity-purified rabbit polyclonal Ab specific for recombinant EndoU was generated. Western blot analysis of whole cell lysates confirmed increased EndoU expression in both resting and BCR-stimulated CD22−/−[B6] B cells relative to CD22−/−[inbr] B cells (Fig. 3 C). EndoU overexpression in CD22−/−[B6] mice was B cell specific, as EndoU levels were modest in activated spleen T cells from CD22−/−[B6] and CD22−/−[inbr] mice. Remarkably, EndoU was also overexpressed in B cells from IgTgsHEL mice both before and after BCR ligation but was nearly undetectable in B cells from IgTg littermate mice (Fig. 3 D). Thus, B cell EndoU overexpression required both a homozygous B6:B6 EndoU locus in CD22−/−[B6] and IgTgsHEL mice that correlated with their characteristic AICD phenotypes.
Mammalian EndoU is a uridine-directed ssRNA binding protein
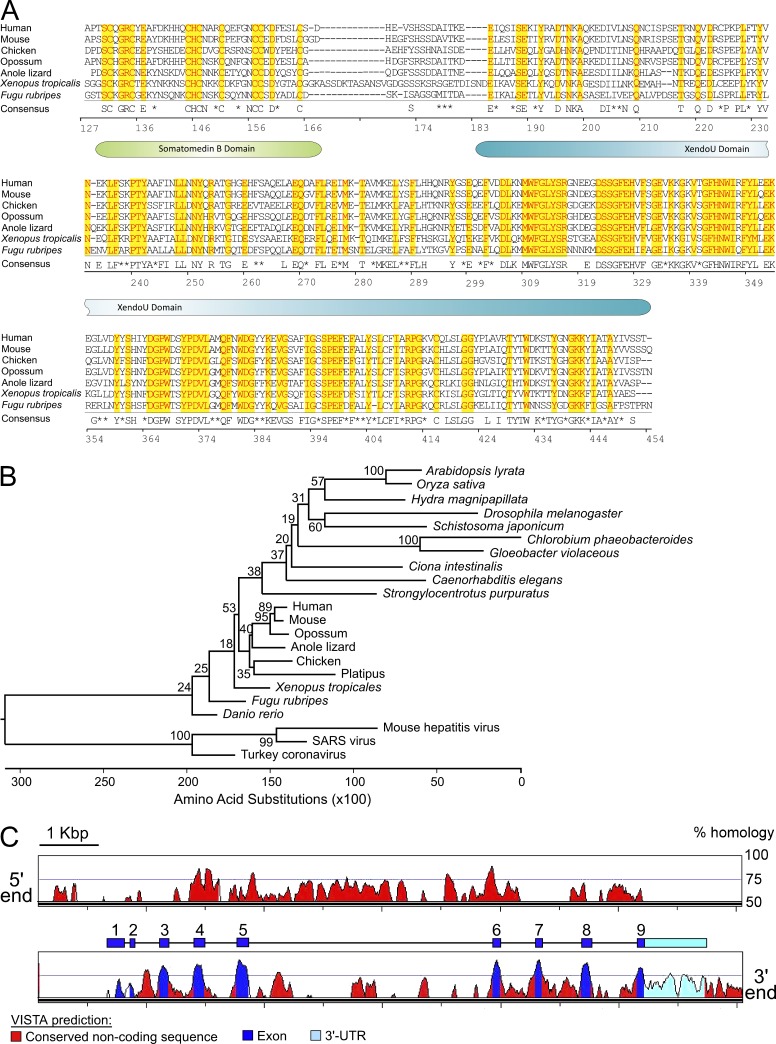
EndoU function in mammals is unknown, although EndoU transcripts are abundant in glucocorticoid-sensitive thymocytes undergoing apoptosis (Baughman et al., 1992). Highly conserved EndoU orthologs are found in many advanced and primitive species (see Fig. 8 A; Renzi et al., 2006), but EndoU paralogs derived from ancestral gene duplications do not exist in mice. The Xenopus ortholog of EndoU, XendoU, has been defined as an endonuclease targeting ssRNA molecules harboring polyuridine (poly(U)) regions (Renzi et al., 2006). Because EndoU substrates have not been defined in other species and EndoU-related molecules that could share redundant activities are not found in mice, the ability of EndoU to bind and/or process XendoU ssRNA substrates was examined. Recombinant EndoU protein specifically bound poly(U) ssRNAs, but not ssRNA species lacking these sites (Fig. 3 E). However, EndoU did not cleave these poly(U) ssRNAs, even in the presence of Mn2+, a condition favorable for XendoU endonuclease activity (Laneve et al., 2003). Mn2+ chelation did not interfere with EndoU binding of ssRNA molecules, as described for XendoU (Gioia et al., 2005). Thus, EndoU binds ssRNAs, likely through its predicted XendoU Superfamily domain (see Fig. 8 A).
Figure 8.
EndoU orthologs are conserved among diverse species. (A) Protein sequences for EndoU and representative vertebrate orthologs were extracted from Ensembl or NCBI databases (human #ENST00000229003; mouse #ENSMUST00000023105; chicken #ENSGALT00000010269; opossum #ENSMODT00000012388; anole lizard #ENSACAT00000014936; Xenopus tropicalis #XM_002935652.1; Fugu rubripes #ENSTRUT00000035070). Yellow highlighted residues with red font are conserved among all species. The consensus sequence at the bottom shows residues conserved in six out of seven species. Asterisks indicate conserved residues within similar functional groups. Numbers indicate the residue positions within mouse EndoU. The positions of the somatomedin B and XendoU domains are shown for reference. (B) Phylogenetic gene tree comparison of EndoU and its orthologs in eukaryotes, prokaryotes, and members of the coronaviruses. The neighbor-joining method was used to construct a phylogenetic tree using outgroup sequences for mouse endoribonucleases Ern1 and Dicer1. The outgroups were removed from the diagram for clarity. Bootstrap values from 1,000 replications are noted near branches. (C) Gene sequence homology between mouse and human EndoU. The region of mouse Chr 15 containing the entire EndoU locus was analyzed against the human genome using the VISTA Browser whole genome comparative analysis tool (http://genome.lbl.gov/vista/index.shtml), with a calculation window and minimum conservation width of 100 bp. Homologous regions of >50% identity are indicated by colored peaks, with predictions of exons, the 3′-UTR, and conserved noncoding regions as indicated. For reference, the cartoon graphic above the bottom panel shows the actual positions of EndoU exons and the 3′-UTR to scale.
Because c-Myc expression is reduced in CD22−/−[B6] and IgTgsHEL B cells after BCR ligation and c-Myc transcripts contain numerous stretches of poly(U) residues within the 3′ untranslated region (3′-UTR, Fig. 3 F), c-Myc mRNA interactions with EndoU were examined. Like the XendoU ssRNA substrate, c-Myc mRNA was also bound by recombinant EndoU (Fig. 3 F), but mRNA cleavage was not detectable (not depicted). Thus, c-Myc mRNA is a potential EndoU substrate, which may explain reduced c-Myc expression when B cells overexpress EndoU. Further supporting this notion, ectopic EndoU expression in mouse NIH-3T3 fibroblasts significantly reduced c-Myc protein levels (Fig. 3 G; 32% decrease, P < 0.05).
EndoU deficiency reverses CD22−/−[B6] B cell AICD
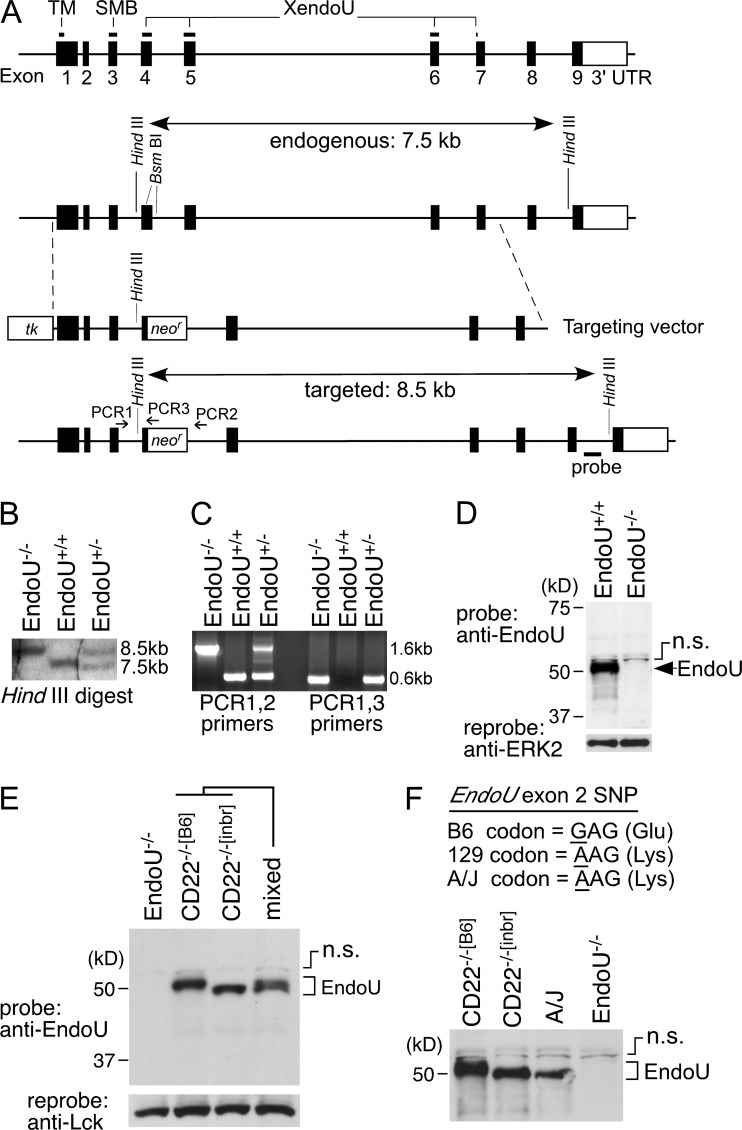
Because EndoU expression is low in WT and CD22−/−[inbr] B cells after BCR ligation (Fig. 3, B and C), the effect of reduced EndoU expression on CD22−/−[B6] B cell AICD was assessed. EndoU-deficient mice were generated by appropriately targeting the EndoU gene on B6 Chr 15 (Fig. 4, A–C). PCR-based SNP genotyping of genomic DNA from targeted mice confirmed that the targeted EndoU locus was B6 in origin. The chimeric mice were then bred onto a B6 genetic background to generate EndoU−/−[B6] mice that were homozygous B6:B6 throughout the 93.3–98.3 Mbp Chr 15 region (unpublished data). The absence of EndoU protein in homozygous gene-targeted mice was confirmed by Western blot analysis, which identified an appropriately sized protein in WT mice (Fig. 4 D). EndoU from CD22−/−[B6] B cells and thymocytes migrated more slowly on SDS-PAGE gels (∼50 kD band) than EndoU from CD22−/−[inbr] thymocytes (∼48 kD band, Figs. 3 C and 4 E), due to a nonsynonymous SNP present within EndoU exon 2 which switches a single Glu (B6 mice) to Lys (129 mice). This was verified using A/J mice that share the EndoU SNP sequence with 129 mice, with EndoU from both CD22−/−[inbr] and A/J mice migrating identically (Fig. 4 F). Splenocytes from CD22−/−[B6] and CD22−/−[inbr] mice expressed a single EndoU cDNA species as confirmed using 5′ RACE procedures (unpublished data). Thus, EndoU−/−[B6] mice do not express EndoU protein.
Figure 4.
Generation of EndoU-deficient (EndoU−/−) mice. (A) EndoU gene targeting strategy. The EndoU gene contains nine exons. Positions encoding a predicted transmembrane (TM) domain, the somatomedin B-like (SMB) domain, and the XendoU superfamily domain are indicated. A targeting vector was generated that contained a neomycin-resistance gene (neor) introduced into two unique BsmBI restriction sites, which introduced an in-frame termination codon. A thymidine kinase gene (tk) was inserted at the 5′ end of the EndoU gene sequence. (B) Homologous recombination of the targeting vector into the EndoU gene of heterozygous and homozygous offspring expanded the endogenous 7.5 kb HindIII digested genomic DNA fragment into an 8.5 kb fragment as assessed by Southern blot. (C) Appropriate targeting of the EndoU gene was further confirmed by PCR analysis (primer locations shown in A), whereby a common forward primer (PCR1) was used in combination with reverse primers either adjacent to (PCR2) or within (PCR3) the neor gene. Results show amplification of genomic DNA from WT, heterozygous, and homozygous offspring. (D) Western blot analysis of thymocyte lysates confirmed the absence of EndoU protein in EndoU−/− mice. The presence of a nonspecific band (n.s.) as described in Fig. 3 is indicated. (E) The major EndoU protein band in CD22−/−[B6] thymocytes (lane 2) migrates slower than the EndoU protein from CD22−/−[inbr] thymocytes (lane 3). A 1:1 mixture of lysates from CD22−/−[B6] and CD22−/−[inbr] thymocytes generates a double band (lane 4). The presence of a nonspecific band (n.s.) also described above is indicated. (F) The difference in EndoU protein migration results from a nonsynonymous SNP in the second exon of the EndoU gene. EndoU protein of CD22−/−[inbr] and A/J mice are identical in sequence, but different from that of CD22−/−[B6] mice as indicated by nucleotide sequence shown and protein migration by SDS-PAGE. The presence of a nonspecific band (n.s.) as also described above is indicated.
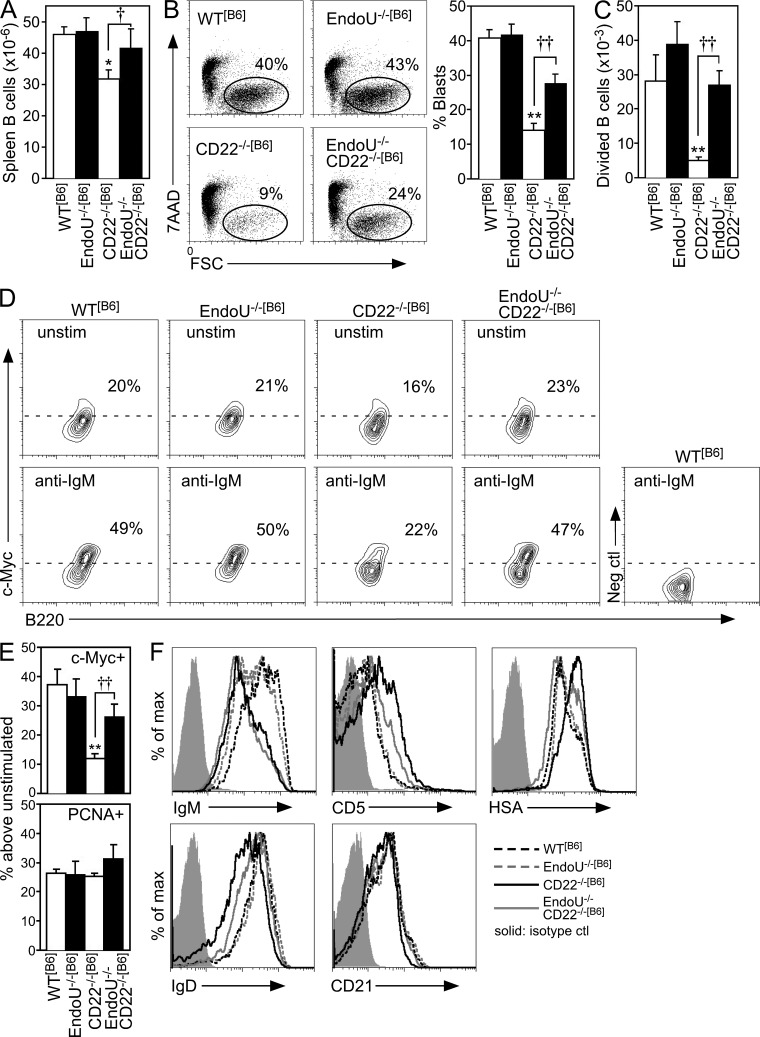
EndoU−/−[B6] and CD22−/−[B6] mice were crossed to generate double-deficient (EndoU−/−CD22−/−[B6]) progeny. A modest but significant reduction of total spleen B cell numbers is observed in CD22−/−[B6] mice, which normalized in EndoU−/−CD22−/−[B6] mice (Fig. 5 A). AICD was also reversed in B cells from EndoU−/−CD22−/−[B6] mice with normal BCR-induced blast development (Fig. 5 B) and proliferation (Fig. 5 C). c-Myc expression was also normalized in BCR-stimulated EndoU−/−CD22−/−[B6] B cells relative to CD22−/−[B6] B cells (Fig. 5, D and E). EndoU may preferentially regulate c-Myc expression in CD22−/−[B6] B cells after BCR ligation because proliferating cell nuclear Ag (PCNA) was expressed at normal levels (Fig. 5 E), as reported for other molecules critical for cell cycle progression (Poe et al., 2004). The rescue of EndoU−/−CD22−/−[B6] B cells from AICD appeared to be B cell intrinsic because lymphoid tissue and thymic development and cellularity were normal in EndoU−/− mice (unpublished data).
Figure 5.
EndoU deficiency eliminates the anergic phenotype of CD22−/−[B6] B cells. (A) Bar graphs show mean (±SEM) total spleen B cell numbers (≥10 mice of each genotype) in WT[B6], EndoU−/−[B6], CD22−/−[B6], and EndoU−/−CD22−/−[B6] mice (*, P < 0.05 compared with WT[B6] mice, Student’s t test; †, P = 0.06, Student’s t test). All mice were homozygous (B6:B6) within the Chr 15 locus. (B) AICD was assessed (7AAD vs. FSC) for spleen B cells from WT[B6], EndoU−/−[B6], CD22−/−[B6], and EndoU−/−CD22−/−[B6] mice after 48 h stimulation with F(ab’)2 anti-IgM Abs. Bar graph indicates mean (±SEM) frequencies of B cell blasts (7AAD−FSChigh) from ≥6 mice of each genotype assessed in independent experiments. Mean values significantly different from WT B cells are indicated by asterisks (**, P < 0.01, Student’s t test). Significant differences between means for CD22−/−[B6] and EndoU−/−CD22−/−[B6] B cells are indicated by crosses (††, P < 0.01, Student’s t test). (C) Purified spleen B cells from mice of the indicated genotypes were labeled with CFSE and cultured with F(ab’)2 anti-IgM Abs. After 72 h, CFSE dilution was assessed by flow cytometry analysis. Values represent mean (±SEM) numbers of divided cells out of 100,000 live cells analyzed for each genotype in three independent experiments. Mean values significantly different from those of WT B cells are indicated by asterisks (**, P < 0.01, Student’s t test). Significant differences between means for CD22−/−[B6] and EndoU−/−CD22−/−[B6] B cells are indicated by crosses (††, P < 0.01, Student’s t test). (D) Purified spleen B cells from the indicated genotypes were cultured alone or with F(ab’)2 anti-IgM Abs. After 18 h, the cells were analyzed for c-Myc or PCNA expression by intracellular staining and flow cytometry analysis. Gated regions in the contour plots were defined using a negative control Ab for background staining. The percentage of positive cells (above the dashed line) in each histogram is indicated. (E) Bar graphs represent the mean (±SEM) percent increase of c-Myc+ or PCNA+ B cells in BCR-stimulated B cell cultures compared with unstimulated cultures as in D from each genotype in four independent experiments. Mean values significantly different from those of WT B cells are indicated by asterisks (**, P < 0.01, Student’s t test). Significant differences between means for CD22−/−[B6] and EndoU−/−CD22−/−[B6] B cells are indicated by crosses (††, P < 0.01, Student’s t test). (F) Cell surface phenotypes of freshly isolated B cells were assessed as in Fig. 1 C. The isotype control shown is the same for the HSA and IgD groups because a common fluorochrome of the same isotype was used for detection. Results are representative of three mice of each genotype assessed in independent experiments producing similar results.
B cells from both CD22−/−[B6] and EndoU−/−CD22−/−[B6] mice shared a surface IgMlow phenotype (Fig. 5 F) that results from enhanced BCR tonic signals in the absence of CD22 negative regulation (Doody et al., 1995; O’Keefe et al., 1996; Otipoby et al., 1996; Sato et al., 1996; Nitschke et al., 1997; Tedder et al., 1997). Otherwise, the modest reductions in surface IgD and CD21 expression that characterize CD22−/−[B6] B cells (Poe et al., 2004) were normalized in EndoU−/−CD22−/−[B6] B cells (Fig. 5 F). Enhanced heat stable Ag (HSA/CD24) expression is commonly used as a marker for immature/transitional B cells, but as with CD5, enhanced HSA expression is also characteristic of chronically stimulated or anergic B cells (Noorchashm et al., 1999). Regardless, the characteristic CD5highHSAhigh phenotype of CD22−/−[B6] B cells (Poe et al., 2004) was largely normalized in EndoU−/−CD22−/−[B6] B cells (Fig. 5 F), and CD22−/−[B6] mice have a normal ratio of mature to immature/transitional peripheral B cells (Poe et al., 2004). Thereby, elevated cell surface CD5 and HSA densities were phenotypic indicators for B cell AICD after BCR ligation.
EndoU-deficient IgTgsHEL mice produce auto-Abs
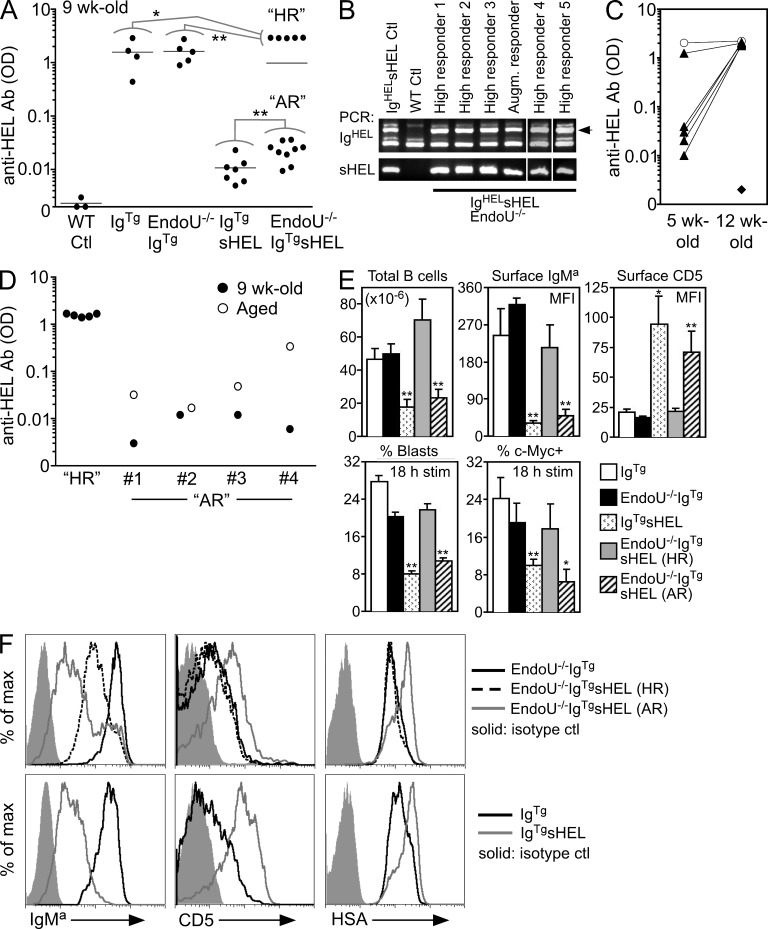
A defining feature of IgTgsHEL mice is the paucity of anti-HEL auto-Ab production. To examine EndoU function in this process, EndoU−/−IgTgsHEL mice were generated. Serum anti-HEL auto-Ab levels were strikingly high in some EndoU−/−IgTgsHEL mice, which were labeled “high responders” (Fig. 6 A). Anti-HEL auto-Ab levels in high-responder EndoU−/−IgTgsHEL mice were even greater than the levels of spontaneous anti-HEL Ab produced in IgTg mice (1.9-fold increase, P = 0.02) and EndoU−/−IgTg mice (1.8-fold increase, P < 0.01) lacking auto-Ag. High-responder EndoU−/−IgTgsHEL mice were present in both sexes, and carried both the IgTg and sHEL transgenes (Fig. 6 B). High serum anti-HEL auto-Ab levels developed in most high-responder mice after 5 wk of age and were maximal by 9–12 wk of age (Fig. 6, A and C). In addition to high-responder EndoU−/−IgTgsHEL mice, their age-matched littermates had augmented anti-HEL auto-Ab levels that were 2.4-fold higher than IgTgsHEL mice (P = 0.01, Fig. 6 A). Anti-HEL auto-Ab levels in augmented-responder mice continued to increase with age (Fig. 6 D), whereas anti-HEL auto-Abs were not enhanced in old IgTgsHEL mice (not depicted). Thus, EndoU overexpression regulates B cell auto-Ag responses in IgTgsHEL mice.
Figure 6.
EndoU deficiency promotes auto-Ab production and normalizes the B cell phenotype in IgTgsHEL mice. (A) Serum anti-HEL auto-Ab levels in the indicated mice at 9 wk of age, with WT littermates assessed as negative controls (WT Ctl). Symbols represent ELISA ODs for individual mice shown on a log10 scale. Horizontal bars represent means for each genotype. For EndoU−/−IgTgsHEL mice, high-responder littermates (HR) were independently compared statistically with IgTg and EndoU−/−IgTg littermates lacking the sHEL transgene. Likewise, augmented-responder (AR) littermates were independently compared with IgTgsHEL mice. Mean values significantly different between groups indicated by lines are represented by asterisks (*, P < 0.05; **, P ≤ 0.01, Student’s t test). (B) PCR genotyping of EndoU−/−IgTgsHEL mice was performed using forward and reverse primers as described in the Materials and methods section, followed by 2.5% agarose gel analysis. The arrow in the top gel indicates the location of the band showing the presence of the IgTg transgene. (C) Serum anti-HEL auto-Ab levels from littermate mice were assessed at 5 wk and again at 12 wk of age. Triangles connected by lines represent individual high-responder EndoU−/−IgTgsHEL mice. A littermate EndoU−/−IgTg mouse assessed at these ages (connected circles) and a WT control mouse assessed at 12 wk (diamond) are shown for comparison. (D) Serum anti-HEL auto-Ab responses were assessed as in A for representative augmented-responder EndoU−/−IgTgsHEL mice at 9 wk of age and again at 12 mo (#1), 8 mo (#2), 6 mo (#3), and 5 mo (#4). 9-wk-old high-responder EndoU−/−IgTgsHEL littermate mice (HR) are shown for comparison. (E) Total spleen B cell number, expression of the IgTg BCR (IgMa) and CD5 on freshly isolated spleen B cells, and c-Myc expression and B cell blast development after BCR stimulation for IgTg, EndoU−/−IgTg, IgTgsHEL, high-responder EndoU−/−IgTgsHEL, and augmented-responder EndoU−/−IgTgsHEL mice. Means (±SEM) represent ≥3 mice assessed for each genotype in all assays. Assessment of B cell c-Myc expression and blast development was as described in Fig. 5. For statistical analysis, IgTgsHEL mice were compared against littermate IgTg mice and high-responder EndoU−/−IgTgsHEL and augmented-responder EndoU−/−IgTgsHEL mice were compared against EndoU−/−IgTg littermate mice (*, P < 0.05; **, P ≤ 0.01, Student’s t test). (F) Spleen cells from the indicated genotypes were stained with fluorescently labeled IgMa, CD5, HSA, and B220 Abs and analyzed by flow cytometry. Histogram overlays represent gating on B220+ cells. Results are representative of three mice of each genotype assessed in independent experiments producing similar results.
EndoU deficiency normalizes the phenotype of IgTgsHEL mice
Compared with IgTgsHEL mice, high-responder EndoU−/−IgTgsHEL mice had a marked increase in B cell blast development and c-Myc expression after BCR ligation, increased spleen B cell numbers and surface IgMa expression, and reduced CD5 and HSA expression (Fig. 6, E and F). In fact, high-responder mice were comparable to IgTg and EndoU−/−IgTg littermates lacking sHEL auto-Ag, except that surface IgMa levels were modestly decreased. In contrast, augmented-responder EndoU−/−IgTgsHEL mice remained phenotypically similar to IgTgsHEL mice. Some B cells in augmented-responder mice exhibited surface IgMa levels comparable to high-responder and IgTg B cells (Fig. 6 F, small peak), but otherwise their B cells had reduced IgMa expression, modest blast development and c-Myc expression after BCR ligation, and high CD5 and HSA expression, with low spleen B cell numbers (Fig. 6, E and F). The frequency of IgMa-high B cells in augmented-responder mice (11% ± 2.1%) was significantly greater than the frequency of IgMa-high B cells present in IgTgsHEL mice (2.2 ± 0.5%, P < 0.01). Thus, the normalized phenotype of high-responder EndoU−/−IgTgsHEL mice and the IgMa-lowCD5highHSAhigh phenotype of augmented-responder EndoU−/−IgTgsHEL B cells suggests differential sensitivities to auto-Ag exposure.
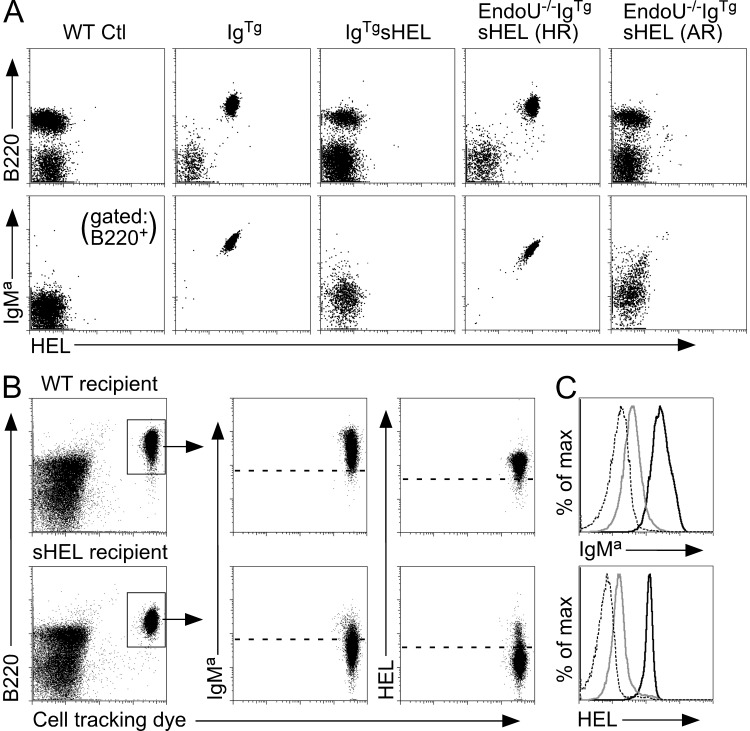
Whether reduced B cell AICD led to the depletion of sHEL auto-Ag in EndoU−/−IgTgsHEL mice with age was examined. Blood B220+ cells isolated from IgTg and high-responder EndoU−/−IgTgsHEL mice expressed relatively high surface IgMa levels and bound fluorescently labeled HEL protein at similar high levels (Fig. 7 A), suggesting that their BCRs were not occupied by sHEL. In contrast, B cells from both IgTgsHEL and augmented-responder EndoU−/−IgTgsHEL mice maintained an IgMa-low phenotype and did not bind labeled HEL protein, indicating BCR occupancy by sHEL. Dye-labeled spleen B cells from high-responder EndoU−/−IgTgsHEL mice were also adoptively transferred into WT or sHEL transgenic mice (Fig. 7 B). There was a dramatic reduction in IgM levels and HEL-binding capacity for high-responder B cells recovered from sHEL recipients in comparison with WT recipients (Fig. 7, B and C). Thus, the absence of AICD in high-responder EndoU−/−IgTgsHEL mice promoted auto-Ab production and endogenous sHEL clearance in vivo.
Figure 7.
sHEL auto-Ag consumption occurs in high-responder EndoU−/−IgTgsHEL mice. (A) Blood cells from the indicated genotypes were stained with fluorescently labeled HEL protein, and with IgMa and B220 Abs. Top panels show dot plots for HEL and B220 staining in the lymphocyte gate, with bottom panels representing the cells in the top panels with additional gating on B220+ cells only. Results are representative of three mice of each genotype producing similar results. (B) Purified spleen B cells from high-responder EndoU−/−IgTgsHEL mice were labeled with cell tracking dye and adoptively transferred into the peritoneal cavities of either WT or sHEL recipient mice. After 36 h, peritoneal cavity cells were harvested from recipient mice and stained with fluorescently labeled HEL protein or IgMa Abs. Results represent one of two independent adoptive transfers producing similar results. (C) Histogram overlays for IgMa expression and HEL-binding levels from the B220+ donor cell populations shown in B. Solid black line, donor cells recovered from the WT recipient mouse; solid gray line, donor cells recovered from the sHEL recipient mouse; dashed line, background staining of B220+ cells in the WT recipient mouse. Results represent one of two independent adoptive transfers producing similar results.
DISCUSSION
EndoU was identified in a gene modifier screen as a unique and potent regulator of B cell AICD. In WT[B6] mice, EndoU transcription and protein expression were suppressed by the balance between tonic BCR signals and their modulation by CD22, permitting normal BCR-mediated responses without blatant AICD after BCR ligation. In contrast, heightened BCR signals in peripheral B cells of CD22−/−[B6] mice drove EndoU overexpression and the CD5highHSAhigh phenotype with predominant AICD after BCR ligation. Genetic deletion of EndoU expression reversed the CD5highHSAhigh phenotype of B cells from CD22−/−[B6] mice and normalized AICD, confirming its important regulatory function in these processes. Bona fide EndoU substrates are likely to include c-Myc and other regulatory transcripts in vivo because c-Myc up-regulation is uniquely impaired in CD22−/−[B6] B cells (Poe et al., 2004), c-Myc expression was normalized in CD22−/−[B6] B cells by EndoU deficiency, EndoU bound c-Myc mRNA in vitro, and ectopic EndoU expression in a fibroblast cell line reduced c-Myc levels. A regulatory role for c-Myc in the mouse WEHI-231 B cell model of AICD has been previously described (Sonenshein, 1997; Donjerković and Scott, 2000). In CD22−/− mice, EndoU overexpression and AICD required B6 homozygosity at the EndoU locus. Therefore, genetic alterations must exist within the EndoU locus between the B6 and 129 genetic backgrounds that positively influences EndoU[B6] allele transcription in B cells, but not T cells. Although the EndoU locus has yet to be fine-mapped or sequenced in 129 mice, these important regulatory elements are likely to be located within the numerous stretches of conserved noncoding DNA present within the EndoU 5′ promoter region and 3′-UTR between B6 mice and humans (Fig. 8 C).
These collective observations support a model in which a CD22, EndoU, and c-Myc expression axis controls B cell fate after BCR ligation in B6 mice. EndoU levels remain low in WT[B6] B cells, allowing robust c-Myc expression and cell cycle progression after BCR engagement. In contrast, chronically high EndoU expression by CD22−/−[B6] B cells prevents c-Myc up-regulation after BCR ligation, resulting in AICD. EndoU was a major regulator of AICD in CD22−/−[B6] and IgTgsHEL mice, although additional unknown mechanisms undoubtedly also contribute to this complex regulatory process.
EndoU overexpression in vivo also occurred in response to BCR signals generated in IgTgsHEL mice, where B cells displayed a CD5highHSAhighIgMlow phenotype and failed to up-regulate c-Myc expression as was observed in CD22−/−[B6] mice. Genetic deletion of EndoU normalized this B cell phenotype in a substantial number of IgTgsHEL mice, which correlated with robust anti-HEL auto-Ab responses at an early age in mice that were defined as high responders. In augmented-responder EndoU−/−IgTgsHEL mice, some B cells expressed normalized phenotypes, suggesting that some B cells escape from an AICD phenotype with age to generate higher anti-HEL auto-Ab levels than IgTgsHEL mice. Given the linkage between EndoU, c-Myc up-regulation and AICD, it is likely that decreased B cell AICD in EndoU−/−IgTgsHEL mice leads to increased numbers of B cells that eventually consume endogenous sHEL auto-Ag, permitting clonal expansion with a resultant increase in anti-HEL auto-Ab production.
The divergence of phenotypes between high-responder and augmented-responder mice has precedent based on previous studies. For example, IgTgsHEL mice deficient in the inositol phosphatase PTEN have reduced BCR occupancy by sHEL and produce anti-HEL auto-Abs at a higher level than IgTgsHEL mice. This results in a subsequent break in tolerance in some mice caused by depletion of available sHEL auto-Ag (Browne et al., 2009). Likewise, IgTgsHEL mice transgenic for CD19 overexpression progressively break tolerance as they age and eventually produce high anti-HEL auto-Ab levels (Inaoki et al., 1997). In fact, remarkably similar to EndoU−/−IgTgsHEL mice, CD19TgIgTgsHEL mice develop as both high- and augmented-responder mice by 2 mo of age. Thus, modest increases in B cell numbers or enhancement in auto-Ab production resulting from altered B cell signaling can lead to auto-Ag depletion. Once this initial reduction in sHEL levels occurs, the resulting lack of B cell anergy enables peripheral B cell numbers to be maintained at a level that exceeds sHEL production, resulting in sustained and high-level anti-HEL auto-Ab production. Thereby, the EndoU regulatory pathway is likely to be an important component of B cell tolerance regulation, which is consistent with the known polygenic origins of autoimmunity.
The EndoU gene has an ancient origin, with conserved orthologs found in lower vertebrates (Suzuki et al., 2002; Gioia et al., 2005) and in invertebrates (Fig. 8, A and B). EndoU transcripts expressed in mouse B cells encoded a protein of 454 amino acids, which includes an N-terminal somatomedin B-like domain believed to be involved in protein–protein interactions (Gijsbers et al., 2003), a XendoU Superfamily ssRNA-binding domain, and a predicted transmembrane domain. EndoU specifically interacted with an ssRNA substrate harboring U-rich sequences that is also bound by XendoU, which is implicated in small nucleolar RNA processing in Xenopus (Gioia et al., 2005; Renzi et al., 2006). XendoU apparently generates snoRNAs through the cleavage of pre-mRNAs encoded within introns (Laneve et al., 2003). Human EndoU may exhibit low-level U-directed ssRNA endoribonuclease activity against the same in vitro ssRNA substrates recognized by XendoU (Laneve et al., 2008). However, mouse EndoU did not demonstrate measurable endonuclease activity against XendoU substrates. Mammalian EndoU may thus be functionally distinct from its Xenopus ortholog because human EndoU is excluded from the nucleolus of multiple cell lines (http://www.proteinatlas.org/ENSG00000111405/subcellular).
Viral orthologs of EndoU also exist (Fig. 8 B). Nsp15 (NendoU) is unique to the nidovirus family of ssRNA viruses, which includes the SARS coronavirus (Laneve et al., 2003; Bhardwaj et al., 2004; Ivanov et al., 2004; Gioia et al., 2005). Although Nsp15 is required for viral replication, specific Nsp15 substrates have not been identified and, surprisingly, appear to exclude the viral genome itself (Ivanov et al., 2004). Nsp15 has been postulated to suppress host immune responses (Ricagno et al., 2006) and to facilitate apoptosis in cells expressing the MAVS (mitochondrial anti-viral signaling) adapter protein that induces host anti-viral responses (Lei et al., 2009). Mouse hepatitis virus is also a member of the nidovirus family, is lymphotropic, and induces immunosuppression at least in part through the elimination of developing B cells (Lamontagne et al., 1989; Jolicoeur and Lamontagne, 1994). It therefore remains possible that mouse EndoU and hepatitis virus Nsp15 may target similar ssRNA substrates in B cells despite their evolutionary divergence. If true, each would have the capacity to suppress immune responses by inducing B cell AICD.
The current studies demonstrate for the first time that an RNA-binding protein plays a major role in regulating AICD, c-Myc expression, and the phenotype of peripheral B cells. Furthermore, these studies suggest an EndoU, CD22, c-Myc, and CD19 regulatory loop that directs normal B cell survival and malignant transformation (Fujimoto et al., 1999; Poe et al., 2012). EndoU likely regulates c-Myc and potentially other crucial ssRNA (mRNA or pre-mRNA) transcripts and nonclassical ssRNAs that directly regulate or fine-tune specific subsets of genes required for optimal B cell proliferation and survival. For autoreactive B cells that survive central selection and inhabit peripheral tissues, EndoU overexpression has nonetheless evolved as a checkpoint protein within the complex pathway that keeps potentially pathogenic B cells at bay through deletion via AICD.
MATERIALS AND METHODS
Mice.
All procedures were approved by the Duke University Institutional Animal Care and Use Committee. CD22−/−[B6] and CD22−/−[inbr] mice were as previously described (Poe et al., 2004). EndoU−/−CD22−/−[B6] mice were produced by breeding EndoU−/− mice with CD22−/−[B6] mice, and then crossing heterozygous F1 animals to generate homozygous mice. WT littermates generated from the same F1 intercross served as controls. IgTg and IgTgsHEL mice (B6 background) were as previously described (Goodnow et al., 1989), with the IgTg and sHEL transgenes confirmed using JAX Mice Genotyping Protocols (The Jackson Laboratory).
To disrupt the EndoU gene by homologous recombination, an 8 kb DNA fragment containing exons 1–7 was isolated from a B6 genomic BAC clone (RP24-169P4). Flanking BsmBI restriction sites were used to remove the distal end of exon 4, a portion of intron 4 and the splice donor site, which was replaced with a neomycin-resistance gene (neor) that introduced an in-frame termination codon. A thymidine kinase (tk) gene was inserted at the 5′ end of the EndoU targeting sequence. Appropriate homologous recombination in EF1 (B6:129 hybrid) embryonic stem cells (Duke Transgenic Mouse Facility) was confirmed by Southern blot analysis of HindIII digested genomic DNA using a probe outside of the targeting sequence. The targeted allele generated an expected 8.5 kb band, compared with the WT 7.5 kb fragment. Correct EndoU gene targeting in subsequent offspring was verified by PCR analysis using a common forward primer (PCR1) in combination with reverse primers either 3′ of (PCR2) or within (PCR3) the inserted neor gene.
B cell blast development and proliferation.
Purified spleen B cells (B Cell isolation kit; Miltenyi Biotec) were analyzed by flow cytometry for blast development after 48 h of culture with F(ab′)2 goat anti–mouse IgM Abs as previously described (Poe et al., 2004). Alternatively, B cell proliferation during 72-h cultures was assessed by 3H-thymidine incorporation or CFSE labeling and dilution as previously described (Poe et al., 2004).
Flow cytometry analysis.
Assessment of cell surface Ags was performed by flow cytometry using fluorochrome-conjugated Abs as previously described (Poe et al., 2004). For intracellular c-Myc and PCNA expression, purified spleen B cells were cultured for 18 h in the presence or absence of F(ab′)2 anti–mouse IgM Ab and then fixed and permeabilized using a Cytofix/Cytoperm Plus kit (BD). Intracellular protein staining used either c-Myc Ab (IgG1 clone 9E10; BioLegend), followed by RPE-conjugated goat anti–mouse IgG1 secondary Abs (SouthernBiotech), or PCNA Ab (FITC-conjugated mouse IgG2a clone PC10; eBioscience). Immunofluorescence staining was assessed by flow cytometry, with background staining levels determined using nonreactive isotype-matched, fluorochrome-conjugated control Abs.
SNP genotyping and QTL analysis.
CD22−/−[B6] mice were crossed with CD22−/−[inbr] mice to generate syngeneic F1 progeny (CD22−/−[F1]). CD22−/−[F1] mice were backcrossed to CD22−/−[B6] mice to generate CD22−/−[N1] littermates. Blast development after BCR ligation was assessed for purified spleen B cells from CD22−/−[N1] littermates, along with B cells from single parental CD22−/−[B6] and CD22−/−[inbr] mice as controls. The results of 10 independent experiments were pooled. To reduce variability between mice and experiments, CD22−/−[N1] mice B cell blast frequencies >1 SD above the mean percentage blasts were considered viable, whereas B cell blast frequencies <1 SD below the mean were considered nonviable. Genomic DNA from 22 viable and 22 nonviable CD22−/−[N1] mice was used for genome-wide genotyping of 250 relevant SNPs between B6 and 129 mice (Illumina BeadArray; Duke University Genotyping Facility). QTL mapping used MapManager QTX regression analysis software (version QTXb20; Manly et al., 2001). Informative SNPs between B6 and 129 mice were identified using the Mouse Genome Informatics and dbSNP databases. DNA primer pairs were used to PCR amplify these SNPs and their surrounding sequences, which were sequenced (Duke Cancer Center DNA Analysis Facility) to identify single nucleotide (homozygous B6:B6) or double nucleotide (heterozygous B6:129) peaks at SNP sites.
Gene expression analysis.
Spleen B cells from mice of the indicated genotypes were cultured in triplicate for 18 h in medium alone or with F(ab′)2 anti–mouse IgM Ab, and then total RNA was isolated from pooled cells (RNeasy Plus; QIAGEN). For analysis of c-Myc and other signaling molecules, [32P]-radiolabeled cDNA was generated and hybridized to GEArray NF-κB signaling pathway gene arrays (SABiosciences) according to the manufacturer’s instructions, with hybridization assessed using x-ray film. Gene expression within the Chr 15 locus was analyzed using Mouse 430.2 RNA arrays (Affymetrix; Duke Cancer Inst. DNA Analysis Facility) with GeneSpring GX software (Agilent Technologies) analysis. For real-time PCR analysis, cDNA was synthesized using random primers and analyzed using a LightCycler Instrument (software version 3) and FastStart DNA MasterPLUS SYBR Green I kit (Roche Diagnostics Corp), with EndoU expression quantified using EndoU exon 5 (forward) 5′-CGTCAACGAGAAGCTGTTCTCCAAG-3′ and EndoU exon 6 (reverse) 5′-CCACATGTTCTTCAAATCGTCCAC-3′ primers. Cd20 expression was the internal control. Relative EndoU expression was quantified using the REST program (version 2).
Western blot analysis.
Polyclonal antiserum reactive with EndoU was generated using purified recombinant EndoU protein (347 amino acids, 107–454) fused at the C terminus with a 6-His tag and produced in the BL21 strain of E. coli. EndoU protein was affinity-purified on His-agarose, yielding a single band of correct size on SDS-PAGE gels. Rabbits were immunized and boosted with EndoU protein in adjuvant (Maine Biotechnology Services, Inc.). Anti-EndoU Ab was affinity-purified from immune sera using recombinant EndoU protein covalently bound to agarose beads. Purified spleen B cells or CD4+ T cells (CD4+ T Cell isolation kit; Miltenyi Biotec) were cultured with F(ab′)2 anti-IgM Abs for B cells or CD3e mAb for T cells (16.7 µg/ml, clone 500A2; BD) for 18 h. 106 cell equivalents of detergent lysates were separated by SDS-PAGE, followed by transfer to nitrocellulose membranes, with EndoU detected using anti-EndoU Ab, HRP-conjugated donkey anti–rabbit IgG (H+L) secondary Ab (Jackson ImmunoResearch Laboratories), and a Pico Enhanced Chemiluminescence kit (Pierce). The blots were stripped and reprobed with a polyclonal ERK2 Ab (C-14; Santa Cruz Biotechnology, Inc.) to ensure equivalent sample loading. Thymocytes (1.5 × 106 cell equivalents/lane) were also analyzed, with the blots reprobed using either polyclonal ERK2 Ab or polyclonal Lck Ab (2102, Santa Cruz Biotechnology, Inc.).
EndoU binding to RNA substrates and transfection of NIH-3T3 cells.
To assess EndoU binding activity, [32P]-labeled ssRNA substrates for XendoU (Laneve et al., 2008) were generated in vitro using dsDNA template oligonucleotides with the SP6 promoter to drive transcription. Full-length [32P]-labeled c-Myc mRNA was transcribed in vitro. Recombinant EndoU protein with a fused IgG binding domain tag was adsorbed onto MagnaBind goat IgG beads (Thermo Fisher Scientific), and incubated with substrate RNAs. Molar excess (100-fold) BSA and yeast tRNA were used as carriers. For XendoU substrates, the reactions were performed for 45 min in the presence or absence of 5 mM Mn2+ or 20 mM EDTA before analysis by 15% PAGE, with phosphorimager visualization. For c-Myc RNA, binding was assessed by scintillation counting.
To induce EndoU expression in mouse NIH-3T3 fibroblasts, cells were transfected (FuGENE HD; Promega) with a plasmid containing full length EndoU fused at the N terminus with CFP using the pECFP-N1 expression vector backbone (Takara Bio Inc.). Control-transfected reporter-positive NIH-3T3 cells were generated independently using the CFP (pECFP-N1) vector alone, or by co-transfection using the vector alone expressing GPF (pEGFP-C1), with similar results. Cells with stable plasmid incorporation were selected for appropriate reporter expression in the presence of 1 mg/ml Geneticin (Life Technologies).
ELISAs.
Serum anti-HEL Ab levels in mice expressing the IgTg transgene were measured using serum diluted 1:100 in Tris-buffered saline containing 1% BSA as previously described (Inaoki et al., 1997).
Fluorescent labeling of HEL protein for flow cytometry analysis.
Fluorescently labeled HEL protein (Sigma-Aldrich) was made using Pacific blue succinimidyl ester (Invitrogen) according to the manufacturer’s instructions, and dialyzed 4× against a large volume of room temperature PBS.
Adoptive transfer experiments.
Purified spleen B cells (15 × 106) from high-responder EndoU−/−IgTgsHEL mice were labeled with eFluor 670 cell tracking dye (eBioscience) and injected intraperitoneally in sterile PBS into WT or sHEL recipient mice. After 36 h, peritoneal cavity and spleen leukocytes were harvested and analyzed by flow cytometry for the presence of the adoptively transferred B cells along with assessment of their IgMa surface expression levels and HEL-binding capacity.
Acknowledgments
We thank Drs. Bruce Beutler (Scripps Research Institute) and Jack Keene (Duke University) for advice, and Latoya McElreth and Isaac Sanford for technical assistance.
These studies were supported by National Institutes of Health grants AI057157, AI56363, and The Lymphoma Research Foundation.
The authors have no conflicting financial interests.
Author contributions: J.C. Poe, E.I. Kountikov, A. Natarajan, D.A. Marchuk, and T.F. Tedder conceived of and performed the experiments; J.C. Poe, E.I. Kountikov, D.A. Marchuk, J.M. Lykken, and T.F. Tedder analyzed the data; and all authors contributed to writing the manuscript.
Footnotes
Abbreviations used:
- 3′-UTR
- 3′ untranslated region
- 7AAD
- 7-amino-actinomycin d
- Ab
- antibody
- Ag
- antigen
- AICD
- activation-induced cell death
- Chr
- chromosome
- FSC
- forward scatter
- HEL
- hen egg lysozyme
- HSA
- heat stable Ag
- LOD
- logarithm of odds
- Mbp
- megabase pair
- PCNA
- proliferating cell nuclear Ag
- poly(U)
- polyuridine
- QTL
- quantitative trait locus
- sHEL
- soluble HEL
- SNP
- single nucleotide polymorphism
- ssRNA
- single-stranded RNA
References
- Baughman G., Lesley J., Trotter J., Hyman R., Bourgeois S. 1992. Tcl-30, a new T cell-specific gene expressed in immature glucocorticoid-sensitive thymocytes. J. Immunol. 149:1488–1496 [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218–12224 10.1128/JVI.78.22.12218-12224.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne C.D., Del Nagro C.J., Cato M.H., Dengler H.S., Rickert R.C. 2009. Suppression of phosphatidylinositol 3,4,5-trisphosphate production is a key determinant of B cell anergy. Immunity. 31:749–760 10.1016/j.immuni.2009.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A.L., Kung J.T., Paul W.E. 1982. Regulation of growth and proliferation in B cell subpopulations. Immunol. Rev. 64:161–182 10.1111/j.1600-065X.1982.tb00423.x [DOI] [PubMed] [Google Scholar]
- Donjerković D., Scott D.W. 2000. Activation-induced cell death in B lymphocytes. Cell Res. 10:179–192 10.1038/sj.cr.7290047 [DOI] [PubMed] [Google Scholar]
- Doody G.M., Justement L.B., Delibrias C.C., Matthews R.J., Lin J., Thomas M.L., Fearon D.T. 1995. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 269:242–244 10.1126/science.7618087 [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Bradney A.P., Poe J.C., Steeber D.A., Tedder T.F. 1999. Modulation of B lymphocyte antigen receptor signal transduction by a CD19/CD22 regulatory loop. Immunity. 11:191–200 10.1016/S1074-7613(00)80094-1 [DOI] [PubMed] [Google Scholar]
- Gay D., Saunders T., Camper S., Weigert M. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008 10.1084/jem.177.4.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R., Ceulemans H., Bollen M. 2003. Functional characterization of the non-catalytic ectodomains of the nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem. J. 371:321–330 10.1042/BJ20021943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., Caffarelli E. 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 280:18996–19002 10.1074/jbc.M501160200 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Crosbie J., Adelstein S., Lavoie T.B., Smith-Gill S.J., Brink R.A., Pritchard-Briscoe H., Wotherspoon J.S., Loblay R.H., Raphael K., et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682 10.1038/334676a0 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Crosbie J., Jorgensen H., Brink R.A., Basten A. 1989. Induction of self-tolerance in mature peripheral B lymphocytes. Nature. 342:385–391 10.1038/342385a0 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Cyster J.G., Hartley S.B., Bell S.E., Cooke M.P., Healy J.I., Akkaraju S., Rathmell J.C., Pogue S.L., Shokat K.P. 1995. Self-tolerance checkpoints in B lymphocyte development. Adv. Immunol. 59:279–368 [DOI] [PubMed] [Google Scholar]
- Hippen K.L., Tze L.E., Behrens T.W. 2000. CD5 maintains tolerance in anergic B cells. J. Exp. Med. 191:883–890 10.1084/jem.191.5.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoki M., Sato S., Weintraub B.C., Goodnow C.C., Tedder T.F. 1997. CD19-regulated signaling thresholds control peripheral tolerance and autoantibody production in B lymphocytes. J. Exp. Med. 186:1923–1931 10.1084/jem.186.11.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA. 101:12694–12699 10.1073/pnas.0403127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolicoeur P., Lamontagne L. 1994. Impaired T and B cell subpopulations involved in a chronic disease induced by mouse hepatitis virus type 3. J. Immunol. 153:1318–7 [PubMed] [Google Scholar]
- Lamontagne L., Descoteaux J.P., Jolicoeur P. 1989. Mouse hepatitis virus 3 replication in T and B lymphocytes correlate with viral pathogenicity. J. Immunol. 142:4458–4465 [PubMed] [Google Scholar]
- Laneve P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., Caffarelli E. 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 278:13026–13032 10.1074/jbc.M211937200 [DOI] [PubMed] [Google Scholar]
- Laneve P., Gioia U., Ragno R., Altieri F., Di Franco C., Santini T., Arceci M., Bozzoni I., Caffarelli E. 2008. The tumor marker human placental protein 11 is an endoribonuclease. J. Biol. Chem. 283:34712–34719 10.1074/jbc.M805759200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.P. 2009. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE. 4:e5466 10.1371/journal.pone.0005466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly K.F., Cudmore R.H., Jr, Meer J.M. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome. 12:930–932 10.1007/s00335-001-1016-3 [DOI] [PubMed] [Google Scholar]
- Nemazee D.A., Bürki K. 1989. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 337:562–566 10.1038/337562a0 [DOI] [PubMed] [Google Scholar]
- Nitschke L., Carsetti R., Ocker B., Köhler G., Lamers M.C. 1997. CD22 is a negative regulator of B-cell receptor signalling. Curr. Biol. 7:133–143 10.1016/S0960-9822(06)00057-1 [DOI] [PubMed] [Google Scholar]
- Noorchashm H., Bui A., Li H.L., Eaton A., Mandik-Nayak L., Sokol C., Potts K.M., Puré E., Erikson J. 1999. Characterization of anergic anti-DNA B cells: B cell anergy is a T cell-independent and potentially reversible process. Int. Immunol. 11:765–776 10.1093/intimm/11.5.765 [DOI] [PubMed] [Google Scholar]
- O’Keefe T.L., Williams G.T., Davies S.L., Neuberger M.S. 1996. Hyperresponsive B cells in CD22-deficient mice. Science. 274:798–801 10.1126/science.274.5288.798 [DOI] [PubMed] [Google Scholar]
- O’Keefe T.L., Williams G.T., Batista F.D., Neuberger M.S. 1999. Deficiency in CD22, a B cell–specific inhibitory receptor, is sufficient to predispose to development of high affinity autoantibodies. J. Exp. Med. 189:1307–1313 10.1084/jem.189.8.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otipoby K.L., Andersson K.B., Draves K.E., Klaus S.J., Farr A.G., Kerner J.D., Perlmutter R.M., Law C.-L., Clark E.A. 1996. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 384:634–637 10.1038/384634a0 [DOI] [PubMed] [Google Scholar]
- Poe J.C., Fujimoto M., Jansen P.J., Miller A.S., Tedder T.F. 2000. CD22 forms a quaternary complex with SHIP, Grb2 and Shc. A pathway for regulation of B lymphocyte antigen receptor-induced calcium flux. J. Biol. Chem. 275:17420–17427 [DOI] [PubMed] [Google Scholar]
- Poe J.C., Haas K.M., Uchida J., Lee Y., Fujimoto M., Tedder T.F. 2004. Severely impaired B lymphocyte proliferation, survival, and induction of the c-Myc:Cullin 1 ubiquitin ligase pathway resulting from CD22 deficiency on the C57BL/6 genetic background. J. Immunol. 172:2100–2110 [DOI] [PubMed] [Google Scholar]
- Poe J.C., Smith S.-H., Haas K.M., Yanaba K., Tsubata T., Matsushita T., Tedder T.F. 2011. Amplified B lymphocyte CD40 signaling drives regulatory B10 cell expansion in mice. PLoS ONE. 6:e22464 10.1371/journal.pone.0022464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe J.C., Minard-Colin V., Kountikov E.I., Haas K.M., Tedder T.F. 2012. A c-Myc and surface CD19 signaling amplification loop promotes B cell lymphoma development and progression in mice. J. Immunol. 189:2318–2325 10.4049/jimmunol.1201000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi F., Caffarelli E., Laneve P., Bozzoni I., Brunori M., Vallone B. 2006. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. USA. 103:12365–12370 10.1073/pnas.0602426103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA. 103:11892–11897 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Miller A.S., Inaoki M., Bock C.B., Jansen P.J., Tang M.L.K., Tedder T.F. 1996. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 5:551–562 10.1016/S1074-7613(00)80270-8 [DOI] [PubMed] [Google Scholar]
- Shlomchik M.J. 2008. Sites and stages of autoreactive B cell activation and regulation. Immunity. 28:18–28 10.1016/j.immuni.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Sonenshein G.E. 1997. Down-modulation of c-myc expression induces apoptosis of B lymphocyte models of tolerance via clonal deletion. J. Immunol. 158:1994–1997 [PubMed] [Google Scholar]
- Suzuki T., Srivastava A.S., Kurokawa T. 2002. A homologue of human placental protein, PP11, and mouse T cell-specific protein, Tcl-30, in exocrine pancreas of a teleost (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133:325–329 10.1016/S1096-4959(02)00158-6 [DOI] [PubMed] [Google Scholar]
- Tedder T.F., Tuscano J., Sato S., Kehrl J.H. 1997. CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu. Rev. Immunol. 15:481–504 10.1146/annurev.immunol.15.1.481 [DOI] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020 10.1084/jem.177.4.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]