Abstract
Isolating a particular strand of DNA from a double stranded DNA duplex is an important step in aptamer generation as well as many other biotechnology applications. Here we describe a microfluidic, flow-through, dialysis device for isolating single-stranded DNA (ssDNA) from double-stranded DNA (dsDNA). The device consists of two channels fabricated in polydimethylsiloxane (PDMS) separated by a track etched polycarbonate membrane (800 nm pore size). To isolate ssDNA, dual-biotin labelled dsDNA was immobilized onto streptavidin-coated polystyrene beads. Alkaline treatment was used to denature dsDNA, releasing the non-biotinylated ssDNA. In the flow-through dialysis device the liberated ssDNA was able to cross the membrane and was collected in an outlet channel. The complementary sequence bound to the bead was unable to cross the membrane and was directed to a waste channel. The effect of NaOH concentration and flow rate on purity and yield were compared. >95% ssDNA purity was achieved at 25mM NaOH. However, lower flow rates were necessary to achieve ssDNA yields approaching the 50% theoretical maximum of the concurrent-flow device. Under optimized conditions the microfluidic isolation achieved even higher purity ssDNA than analogous manual procedures.
INTRODUCTION
ssDNA has broad applications in therapeutics1–5, diagnostics6–10, biosensing11–17 and sequencing technologies18–21. Unlike dsDNA, the unpaired bases of ssDNA allow it to participate in complex three dimensional folding giving rise to unique structural conformations including hairpins8, 22, pseudoknots23, 24 and quadruplexes25, 26. Formation of conformations such as these are key to the success of technologies such as Systematic Evolution of Ligands by Exponential enrichment (SELEX)27–29 and single-stranded conformation polymorphism (SSCP)30, 31. ssDNA templates are also critical components of hybridization-based detection schemes to detect single-nucleotide polymorphisms (SNP)32–34, pyrosequencing technology19, 35, 36, solid phase DNA sequencing37, 38, and sensing in DNA chips and microarrays17, 39–41.
Aptamers are 40–120 nucleotide sequences of ssDNA or RNA that have been selected to fold into conformations with affinity for specific targets27–29, 42, 43, 44. Aptamers have been selected for a variety of targets ranging from small inorganic molecules to whole cells23, 45–51. Applications of aptamers are broad ranging from purification52 and detection53 to diagnostic54 and therapeutic agents55. The SELEX process used to isolate aptamers begins with a ssDNA or RNA library of sequences containing a 20–60 bp random region flanked by forward and reverse primers. Binding sequences are enriched using an affinity separation and then amplified using PCR. A key step of the process is recovering the forward strand ssDNA from the dsDNA generated by PCR allowing the aptamers to refold into their binding conformations prior to further rounds of enrichment. There are several methods for recovering ssDNA from dsDNA including denaturing urea-polyacrylamide gel (PAGE)56–59, asymmetrical PCR60–64, Lambda exonuclease digestion65–68, and separation with streptavidin-coated beads 20, 40, 51, 69–72. In denaturing PAGE, a functional group is conjugated to one of the PCR primers. The resulting forward and reverse ssDNA have different sizes allowing them to be separated by denaturing gel electrophoresis57. While effective, denaturing PAGE is a labour intensive and expensive approach73. Asymmetrical PCR preferentially amplifies one strand over the other by adding different concentrations of the two PCR primers60. While this provides a simple and economic solution, the enriched ssDNA pool also contains dsDNA, necessitating further purification using gel electrophoresis. The recovery process is slow, generates low yields of ssDNA and is not amenable to integration into a microfluidic format74, 75. Lambda exonuclease digestion selectively digests the 5’-phosphorylated strands of dsDNA. To recover ssDNA the complementary sequence is preferentially digested by introducing a 5’-phosphate group during PCR amplification65, 66. Phenol-chloroform extraction followed by ethanol precipitation is necessary to remove the lambda exonuclease enzyme at the conclusion of the digestion. dsDNA may also accumulate in the reaction mixture due to incomplete digestion of the phosphorylated double-stranded PCR product76.
The most common method for isolating ssDNA in aptamer selections is immobilization of biotinylated dsDNA onto streptavidin-coated beads followed by alkaline denaturation51. Many aptamers have been isolated using this method20, 40, 51, 69. In the current paper, we have incorporated this approach into a flow through, microfluidic dialysis device, providing a simple procedure for isolating ssDNA from dsDNA.
A Lab on a Chip or Micro Total Analysis System (µTAS), is a miniaturized analysis system integrated onto a single chip, often conducting multiple functions such as sample preparation, separation, detection, etc. Microfluidic devices offer a number of important advantages including short analysis time, high sensitivity, low consumption of sample and reagents, and low cost77, 78. Several groups have reported microfluidic devices that are useful for aptamer selections. Park and Ahn developed a microfluidic device for binding nucleic acids to multiple proteins immobilized in sol-gel arrays followed by aptamer elution and recovery79. The Soh group demonstrated an M-SELEX system which manipulated small numbers of beads to isolate high-affinity aptamers rapidly80, 81. Jing and Bowser isolated aptamers after a single round of selection in a µFFE device82. Huang et al. have used a magnetic bead-based microfluidic system which integrated a ssDNA incubation module with an on-chip nucleic acid amplification module83. Ferguson et al. developed a microfluidic electrochemical DNA sensor which integrated symmetric PCR, ssDNA generation and sequence-specific electrochemical detection onto a single chip 84. Lambda exonuclease digestion was used to generate ssDNA. While impressive, each of these microfluidic SELEX systems only addressed isolated steps of aptamer selection, not the entire SELEX process. Identifying strategies for incorporating ssDNA isolation into a more complex device is a key step in developing a fully integrated microfluidic SELEX system. For example, integration of µFFE selection82 and continuous PCR85 with the online ssDNA purification described here suggests that a simple, continuous flow strategy for microscale aptamer generation may be feasible.
EXPERIMENTAL
Reagents and Chemicals
PDMS was made using a Sylgard 184 silicone elastomer kit (Dow Corning, Midland, MI). Piranha solution (3:1 H2SO4:H2O2, Ashland Chemical, Dublin, OH) was used to clean silicon wafers. Diluted HCl was used to reactivate the PDMS surface. SU8-2050 (Microchem Corp., Newton, MA) was used to fabricate the master. All DNA samples were purchased from Integrated DNA technologies (Coralville, IA). Tris EDTA (TE) buffers consisted of 10mM Tris(hydroxymethyl)aminomethane(Tris), 1mM Ethylenediamenetetraacetic acid disodium salt (EDTA), 150mM NaCl (Spectrum Chemical, New Brunswick, NJ) and 0.002% Triton X-100, adjusted to pH 7.5 using 1M HCl. TE buffer was degassed before use. 12.5mM, 18.75mM, 25mM, 50mM, 100mM and 300mM NaOH were prepared to denature dsDNA. 1.26µm streptavidin-coated polystyrene beads (Spherotech, Lake Forest, IL) were used to capture biotinylated dsDNA. Wash buffer comprised of 5mM Tris, 0.5mM EDTA, 1mM NaCl and 0.05% Tween 20, adjusted to pH 7.5 using 1M HCl. Binding buffer consisted of 10mM Tris, 1mM EDTA and 2M NaCl, adjusted to pH=7.5 using 1M HCl. TBE buffer (Tris 89mM, Boric acid 89mM, EDTA 2mM) with 10% glycerol (Invitrogen, Grand Island, NY, purity≥99.5%) was used as CE running buffer. 8% Linear polyacrylamide (LPA) (MW=600,000–1,000,000, Polysciences, Inc, Warrington, PA) in TBE buffer with 10% glycerol was prepared and filtered through a 0.45µm membrane filter. Poly(ethylene oxide) (PEO) was dissolved in deionized water to 0.2% by mass with 0.1M HCl in a water bath at 95°C with stirring. All buffer solutions were prepared using nuclease free water (Integrated DNA technologies, Coralville, IA) and filtered through a 0.2µm membrane before use. All other reagents and chemicals were from Sigma-Aldrich (St. Louis, MO).
Fabrication
Fabrication of the microfluidic dialysis device was performed following a previously described procedure86. A two-step soft lithography process was used.87 An SU8 master was fabricated and multiple PDMS replicas were then cast from this master. The chip geometry was designed using CAD software which was then used to print a transparency film mask. Standard photolithography procedures were used to make the SU8 master. The master was pre-treated with fluorinated silanes (UCT, Bristol, PA). Degassed PDMS prepolymer (10:1 ratio) was then poured onto the master and baked at 65°C for 3 hours. The cured PDMS was removed from the master. A second PDMS layer was made using the same process. The resulting channels were 250 µm wide and 110 µm deep. The total channel length was 17 cm with an overlapped channel length of 15.3 cm. Inlet and outlet holes were punched into one PDMS layer using a piece of 21 gauge stainless steel tubing (McMaster-Carr, Chicago, IL). Both PDMS layers were sonicated in dilute HCl for 10 minutes to reactivate prior to bonding88. A 12mm×20mm×6µm polycarbonate membrane with 800 nm pore diameter (Sterlitech Corp., Kent, WA) was placed on one layer of PDMS over the overlapped area of the channel. The PDMS layers were O2 plasma treated at 600 mTorr and 75 W for 10 s. The two layers were then quickly aligned and bonded irreversibly. The entire assembly was placed in an oven at 65 °C for 2 hours to complete the bonding process. Two glass substrates were used to support the device and eight screws on the edges of the substrates were finger tightened to seal the membrane and the PDMS slabs. 0.5" long pieces of stainless steel tubing with the same inner and outside diameter as a 23 gage needle (New England Small Tube Corp., Litchfield, NH) were inserted into in the inlet and outlet holes of the device allowing connections to be made with Tygon tubing (0.02 ID, 0.06 OD, Small parts, Inc, Miramar, FL). Buffer was pumped into the device using a syringe pump (Harvard Apparatus, Holliston, Massachusetts).
Instrumentation and Data Collection
A Synergy2 multi-mode microplate reader (BioTek, Winooski, VT) mounted with a Tungsten Halogen lamp was used for fluorescence intensity measurements (λex=485nm, λem=528nm). Fluorescence data were recorded and processed using Gen5 Reader Control and Data Analysis software (BioTek, Winooski, VT). Capillary electrophoresis (CE) was performed on a P/ACE MDQ system (Beckman Coulter Inc., Fullerton, CA) equipped with a laser induced fluorescence (LIF) detection module (λex=488nm, λem=520nm). 32 Karat Software (Beckman Coulter Inc., Fullerton, CA) was used for collecting and recording data.
Sample preparation
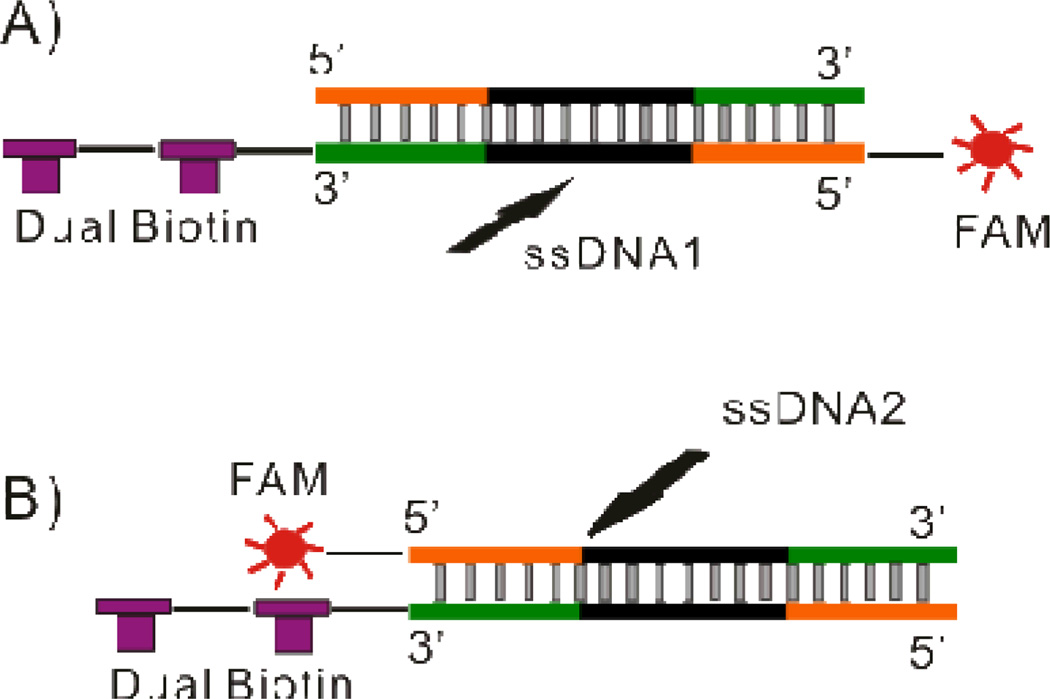
Two complementary 80nt ssDNA sequences (5’-biotin-biotin- TT CAC GGT AGC ACG CAT AGG CGA TCA TAA AGC CTC CAT AGC ATT GTA CGT CGA AAC CTG CCA TCT GAC CTC TGT GCT GCT-FAM-3’ and 5'- AGC AGC ACA GAG GTC AGA TGG CAG GTT TCG ACG TAC AAT GCT ATG GAG GCT TTA TGA TCG CCT ATG CGT GCT ACC GTG AA -3') were synthesized by Integrated DNA technologies (Coralville, IA) and annealed to generate dsDNA1 manually in the following steps. Both ssDNA sequences were dissolved in Nuclease Free Duplex Buffer (Integrated DNA technologies, Coralville, IA) at concentrations of 100µM. Equal volumes of each ssDNA solution were mixed together and heated to 94°C for 2 min followed by gradual cooling to room temperature. The resulting dsDNA1 product was diluted in TE buffer to 10µM and stored at −20°C for future use. A second pair of 80nt ssDNA sequences (5’biotin-biotin- TT CAC GGT AGC ACG CAT AGG CGA TCA TAA AGC CTC CAT AGC ATT GTA CGT CGA AAC CTG CCA TCT GAC CTC TGT GCT GCT-3’ and 5’FAM- AGC AGC ACA GAG GTC AGA TGG CAG GTT TCG ACG TAC AAT GCT ATG GAG GCT TTA TGA TCG CCT ATG CGT GCT ACC GTG AA −3’) were synthesized by Integrated DNA technologies (Coralville, IA) and annealed to generate dsDNA2 using the same procedure. The only difference between dsDNA1 and dsDNA2 was the location of the FAM label allowing the path of each sequence to be tracked through the device independently depending on the specific experiment (see Figure 1).
Figure 1.
Schematic demonstrating the labeling configuration of (A) dsDNA1 and (B) dsDNA2. In dsDNA1 dual biotin and FAM were labeled on the same strand allowing the complementary strand to be followed using fluorescence. In dsDNA2 dual biotin and FAM were labeled on opposite strands allowing the forward sequence to be followed.
Binding dsDNA to beads
A 150 µL suspension of streptavidin-coated polystyrene beads was pelleted by centrifugation (15,000 rpm for 5 min) to remove the storage buffer. The beads were resuspended in washing buffer followed by pelleting (15,000 rpm for 5 min) three times. After the final pelleting beads were resuspended in 300 µL binding buffer. 200 pmol dsDNA in 300 µL nuclease-free water was mixed with the bead suspension and incubated at room temperature for 15 min. The supernatant was collected after centrifugation and the fluorescence intensity was measured to determine how much free dsDNA existed in the supernatant. By comparing how much dsDNA was added with how much dsDNA remained in solution after incubation, the amount of dsDNA immobilized onto the beads could be determined. Washing buffer was used to remove any unbound dsDNA with two centrifugation, resuspension cycles. Finally the beads were resuspended in TE buffer with a dsDNA concentration of 100 nM.
ssDNA generation by alkaline denaturation
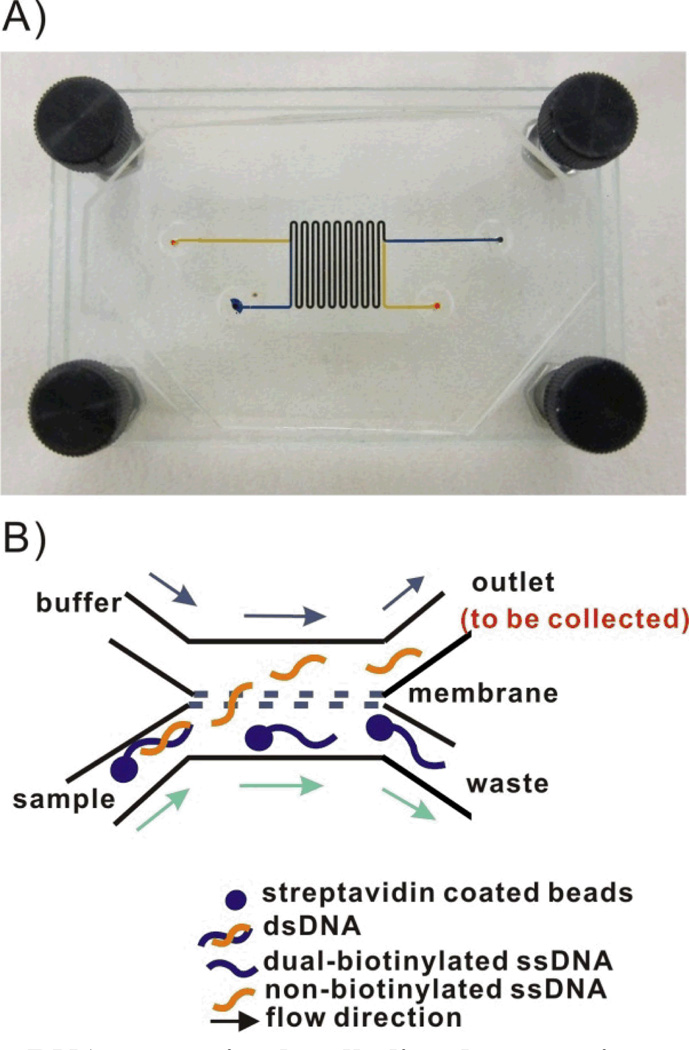
The dsDNA-bead suspension was mixed with a 1:1 volume of freshly prepared NaOH solution. This mixture was continuously pumped into the sample channel of the microfluidic dialysis device (see Figure 2). A 1:1 mixture of TE buffer and NaOH was pumped into the buffer channel of the device at the same flow rate. The buffer composition on each side the membrane was kept the same to simplify the fluorescence analysis. Since the buffers were identical the ratio of DNA in each channel was simply the ratio of observed fluorescence intensities. The distribution of each of the two ssDNA in each channel was determined at six different NaOH concentrations (12.5mM, 18.75mM, 25mM, 50mM, 100mM and 300mM). Concurrent flow was adopted in the device (see Figure 2) to eliminate pressure differences across the membrane which caused beads to be trapped in the pores. The flow rate in each channel was set to be 0.2 ml/hr which corresponded to 2.2 mm/s and a 70 sec. equilibration time.
Figure 2.
A) Image of the microfluidic dialysis device. Two 250 µm wide × 110 µm deep PDMS channels were separated by a polycarbonate membrane (800 nm pore size). The total channel length was 17 cm with a 15.3 cm long overlapping region. Channels were filled with food coloring to facilitate visualization of the sample and buffer channels. B) Schematic of the anticipated flow paths of the denatured ssDNA in the microfluidic dialysis device. The forward sequence is freed from the bead during denaturation and is small enough to diffuse across the membrane. The biotinylated complementary sequence remains immobilized on the bead and is carried to the waste channel.
An analogous manual method for isolating ssDNA was performed in parallel as a point of reference. Aliquots of the dsDNA-bead suspension were transferred into seven centrifuge tubes and six tubes were spiked with an equal volume of NaOH (12.5 mM, 18.75 mM, 25 mM, 50 mM, 100 mM and 300 mM). The control experiment was performed by mixing the dsDNA-beads suspension with an equal volume of TE buffer. After incubating for 10 min at room temperature, the solution was centrifuged at 15,000 rpm for 5 min and the supernatant was collected. CssDNA2/CssDNA1 was determined using fluorescence. All experiments were conducted three times.
Determining the effect of flow rate
Equal volumes of dsDNA-bead suspension and 25mM NaOH solution were mixed and the resulting solution was pumped into the sample channel of the device. Equal volumes of TE buffer and 25mM NaOH were mixed and the resulting solution was pumped into the buffer channel of the device. CssDNA2/CssDNA1 in the outlet channel was calculated at five different flow rates (0.5 ml/hr, 0.3 ml/hr, 0.2 ml/hr, 0.1 ml/hr and 0.05 ml/hr). Samples were collected from the outlet of the buffer channel after 15–20 min from the moment of adding NaOH into dsDNA-bead suspension for consistency.
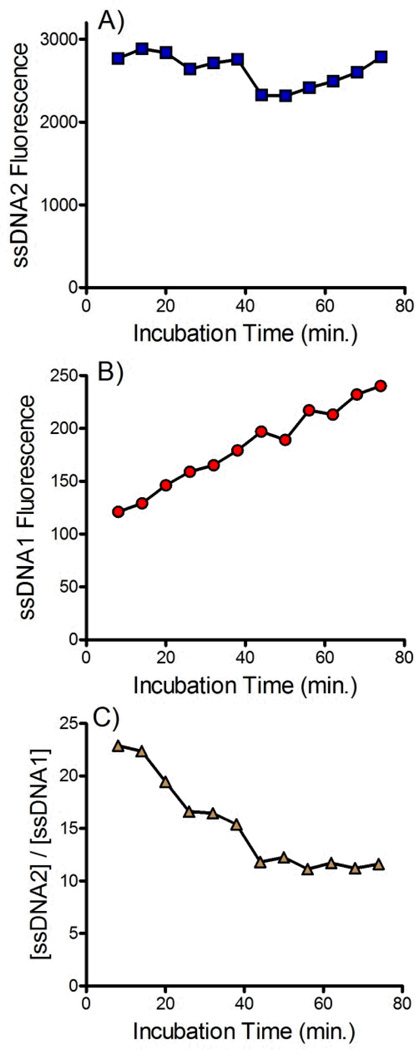
Determining the effect of incubation time
Equal volumes of the dsDNA-bead suspension and 25 mM NaOH solution were mixed and the resulting solution was pumped into the sample channel of the device as the sample. Equal volumes of TE buffer and 25 mM NaOH were mixed and the resulting solution was pumped into the buffer channel of the device. Aliquots were collected from the outlet of the buffer channel every 6 min starting at 8 min after NaOH was added to the dsDNA-bead suspension. Separated experiments were conducted for dsDNA1 and dsDNA2 allowing the distribution of both the forward and complementary sequences to be determined.
Capillary electrophoresis (CE) characterization
A 40 cm-long, 75 µm i.d., 360 µm o.d. uncoated fused-silica separation capillary (Polymicro Technologies, Phoenix, AZ) was used. Electroosmotic flow (EOF) was suppressed by coating the capillary surface with PEO according to Iki and Yeung’s method89. The capillary was rinsed fir 10 minutes each of 0.1 M NaOH, H2O, 1 M HCl, 0.2% PEO/0.1 M HCl and TBE buffer containing 10% glycerol. Barron’s method for separating ssDNA and dsDNA was adopted to confirm the purity of the ssDNA isolation90, 91. 8% w/v LPA (MW600000-1,000,000) in TBE buffer containing 10% glycerol was used as the sieving matrix for separating 80 nt ssDNA and 80 bp dsDNA. Glycerol has previously been reported to improve resolution of ssDNA and dsDNA92, 93. Linear polyacrylamide (LPA) was filtered through a 0.45 µm membrane filter and degassed by centrifuging at 12000 rpm for 1 min. All the other solutions were filtered through a 0.2 µm membrane filter. LPA was loaded into the 40 cm capillary in the MDQ capillary electrophoresis system at 20 psi for 20 min. Samples were injected using 2 kV for 15 s. The separation was performed under the following conditions: 10 kV voltage (reverse polarity) thermostatted to 15°C with laser induced fluorescence (LIF) detection at 488/520 nm. TBE buffer containing 10% glycerol was used as the separation buffer. Before performing CE separations, the LPA-loaded capillary was conditioned under 10 kV voltage (reverse polarity) overnight in order to maintain a consistent gel structure for improved run-to-run reproducibilty.
RESULTS AND DISCUSSION
Design Considerations
An important consideration in a continuously flowing device such as the one described here is the potential for beads to sediment over time. In particular, if beads settle to the bottom of the syringe it becomes difficult to introduce a consistent concentration of beads into the device, especially over longer time periods. The settling velocity is related to the balance of viscous drag and gravitational force. According to Stoke’s law, which is derived by solving the general Navier-Stokes equation at the Stokes flow limit for small Reynolds numbers94–97:
where Vm is the maximum settling velocity (cm/sec), g is the acceleration of gravity (980.7 cm/sec2), η0 is the viscosity of the external phase in poises(g/cm·sec), ρsis the density of solid sphere (g/cm3), ρm is the density of the medium(g/cm3) and d is the mean diameter of the suspended particles (cm). The rate of sedimentation is determined by the density and the diameter of the particles. Therefore, choosing the appropriate size and material of the beads is crucial to make long term operation of the device feasible. According to the manufactures’ data, streptavidin-coated polystyrene beads had a lower density than comparable silica or magnetic beads. Polystyrene (PS) beads are available in a series of diameters ranging from low nm to high µm. For the current application beads should have a diameter larger than the membrane pore diameter while maintaining a low density to keep the beads in suspension for long periods. To maximize ssDNA recovery across the membrane a relatively large pore size was chosen (800 nm). Taking this into consideration, 1.26 µm streptavidin coated polystyrene beads were chosen to immobilize dsDNA.
Alkaline denaturation was chosen to isolate ssDNA due to its simplicity. This method makes integration of heating elements into the device unnecessary. Similarly, reestablishing re-naturing conditions is as simple as neutralizing the solution with no further purification necessary. A potential drawback to the alkaline denaturation approach is the addition of NaOH could also denature streptavidin, disrupting its interaction with biotin and releasing dsDNA from the beads. To address this a dual-biotin label was used to increase the stability of the streptavidin-biotin interaction. Similar approaches have proven successful in stabilizing streptavidin-biotin interactions through multiple PCR cycles98. Ideally a single dsDNA would be used with a fluorescent label attached to each strand allowing the path of each sequence to be tracked through the device simultaneously. In practice synthetic limitations, the potential for signal cross talk and impact on quantum yield made using two dsDNA’s, each with a single label to track either the forward or complementary strand, a much more reliable approach (see Figure 1).
NaOH concentration
Alkaline treatment is known to be a reliable and convenient way to denature double-stranded DNA in a mechanism related to the deprotonation of guanine and cytosine99–102. However, if the denaturation conditions are too harsh, the streptavidin-biotin interaction may be disrupted103–105, allowing dsDNA to cross the membrane into the outlet channel. Therefore, careful consideration must be given to optimize the denaturation procedure to ensure effective recovery of the desired ssDNA strand.
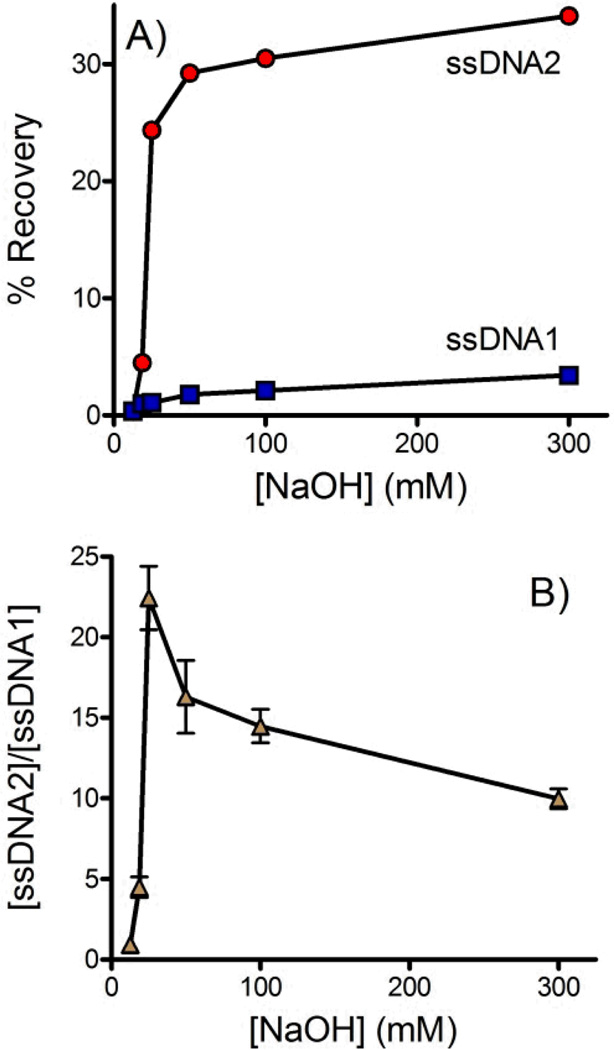
Six different concentrations of NaOH were assessed. In each case solution from the outlet of the buffer channel was collected and its fluorescence measured to determine the concentration of each strand of DNA that crossed the membrane (see Figure 3). In ssDNA2 the forward sequence was fluorescently labeled. Figure 3A shows that the forward sequence was efficiently denatured and crossed the membrane at NaOH concentrations of 25 mM or higher. In dsDNA1 the fluorescent label was on the biotinylated complementary strand. Overall the amount of complementary DNA crossing the membrane was low, indicating that the biotin-streptavidin interaction remained effective even in the presence of NaOH. A trend of increasing complementary DNA crossing the membrane with increasing NaOH was observed. Figure 3B plots the ratio of forward and complementary sequences that crossed the membrane. A clear optimum is observed at 25mM NaOH. dsDNA is not effectively denatured at lower NaOH concentrations. At higher NaOH concentrations the forward sequence is already completely denatured but the biotin-streptavidin interaction is increasingly disrupted. At 25 mM NaOH the concentration of forward sequence ssDNA was 22-fold higher than the observed complementary sequence resulting in a purity of >95%.
Figure 3.
Effect of NaOH concentration on ssDNA isolation. Solution was collected from the outlet of the buffer channel and fluorescence was measured to quantify the DNA that crossed the membrane. A) The forward sequence is labeled in dsDNA2 (red circles) and the biotinylated complementary sequence is labeled in dsDNA1 (blue squares). B) The ratio of the forward and complementary ssDNA sequences ([ssDNA2]/[ssDNA1]) that crossed the membrane into the buffer channel. >95% purity was achieved at 25 mM NaOH. All experiments were performed in triplicate. In some cases error bars were smaller than the symbols. The flow rate was 0.2 mL/hr and buffer contained 10 mM Tris, 1 mM EDTA, 150 mM NaCl and 0.002% Triton X-100.
Flow rate
Flow rate through the device impacts the equilibration time and therefore recovery of ssDNA across the membrane. The overlapping region of the channels is 15.3 cm long. Equilibration time therefore ranges from 30.6 s to 4.2 min for flow rates of 0.5 mL/hr to 0.06 mL/hr (5 mm/s to 0.61 mm/s).
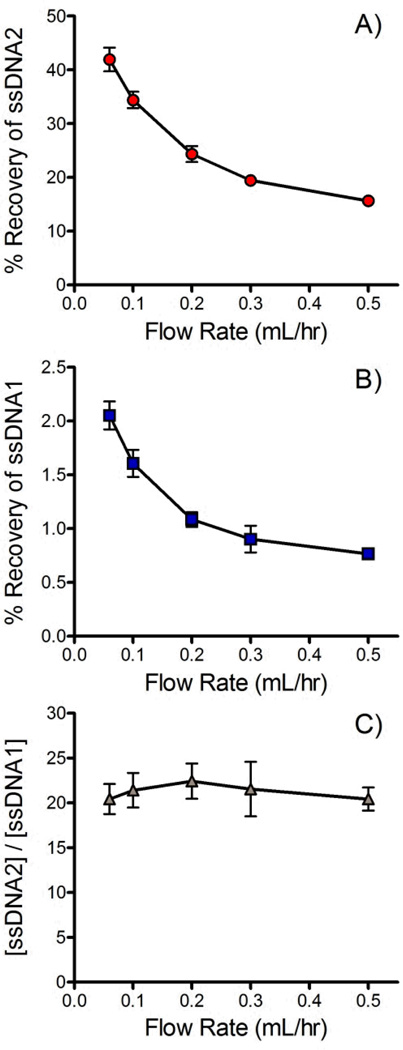
Figure 4A and B show the impact of flow rate on the % recovery of the forward and complementary DNA sequences across the membrane. As expected the recovery of both sequences trends lower as flow rate increases and equilibration time decreases. A concurrent flow configuration was chosen for this application. Concurrent dialysis is limited to a maximum % recovery of 50% since complete equilibration is achieved at this point. Counter current devices are able to achieve nearly 100% recovery.86 Unfortunately, counter current devices generate a pressure differential across much of the membrane. In the current application it was found that this pressure differential was enough to trap passing beads into the pores, effectively blocking ssDNA transport within a short period of time. Bead adsorption onto the membrane was minimal when a concurrent flow configuration was adopted. At 0.06 ml/hr the % recovery of the forward sequence reached 42±3%, 84% of the theoretical maximum of a concurrent flow device. At a more practical 0.2 ml/hr, the % recovery was 24±0.2%, 48% of the theoretical maximum.
Figure 4.
Effect of flow rate on % recovery of A) the forward sequence (ssDNA2) and B) the biotinylated complementary sequence (ssDNA1). C) Ratio of forward and complementary sequences ([ssDNA2]/[ssDNA1]) that crossed the membrane. All data were recorded in triplicate. 25 mM NaOH was used for denaturation and the buffer contained 10 mM Tris, 1 mM EDTA, 150 mM NaCl and 0.002% Triton X-100.
It should be noted that similar trends in recovery were observed for the forward and complementary sequences (see Figure 4A & B). Figure 4C plots the ratio of the forward and complementary sequences that crossed the membrane at different flow rates. Although the recovery of each sequence decreased with increasing flow rate the purity of the recovered ssDNA was largely unaffected since the recovery of each sequence was affected equally.
Incubation time
The incubation time between the addition of NaOH and delivery into the device is an important variable. This time must be long enough to achieve complete denaturation of the dsDNA but not be so long as to promote disruption of the biotin-streptavidin interaction. The effect of incubation time on the amount of the forward sequence ssDNA2 and the complementary sequence ssDNA1 is shown in Figure 5. No trend in the recovery of the forward sequence was observed (see Figure 5A) suggesting that complete denaturation of the dsDNA occurred within several minutes. Conversely, the amount of complementary DNA crossing the membrane increased linearly with time, suggesting a constant, continual denaturation of streptavidin was occurring with prolonged exposure to NaOH. Figure 5C shows the ratio of forward and complementary sequences that cross the membrane. Over longer periods the purity of the recovered ssDNA decreases due to the slow release of the complementary sequence. Combined this data suggests that beads should be delivered to the device immediately after addition of NaOH. Future devices would benefit from incorporation of an online mixing tee to ensure fast, reproducible, delivery of beads into the device after addition of NaOH.
Figure 5.
Effect of incubation time on amount of A) forward sequences (ssDNA2) and B) biotinylated complementary sequences (ssDNA1) that cross the membrane into the buffer channel. C) Effect of incubation time on the ratio of forward and complementary sequences ([ssDNA2]/[ssDNA1]) that crossed the membrane. 25 mM NaOH was used for denaturation and the buffer contained 10 mM Tris, 1 mM EDTA, 150 mM NaCl and 0.002% Triton X-100. The flow rate was 0.2 mL/hr.
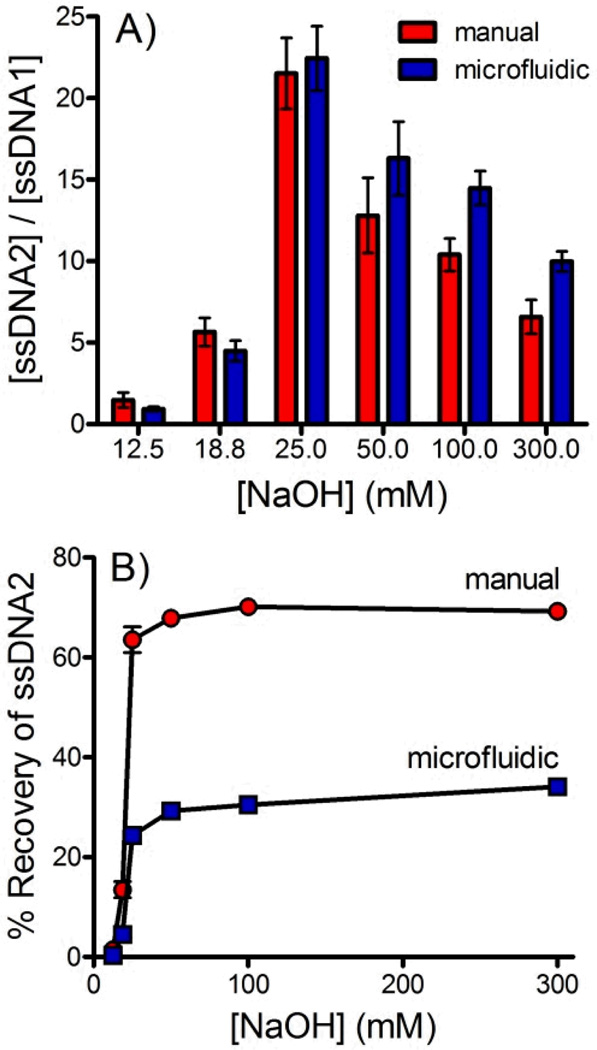
Comparison with Manual Methods
The purity of the ssDNA recovered using the flow through, microfluidic dialysis device was compared with ssDNA recovered using an analogous manual method. For the manual method dsDNA-bead suspension was incubated with NaOH at room temperature for 10 minutes. The beads were pelleted using centrifugation and the amount of labeled DNA in the supernatant was assessed using fluorescence.
Figure 6A compares the ratio of forward and complementary sequence DNA obtained by each method across a range of NaOH concentrations. Overall, the microfluidic device performed similarly to the manual procedure. The microfluidic device does consistently outperform the manual procedure at NaOH concentrations ≥50 mM. The additional steps of the manual procedure make it difficult to isolate the ssDNA quickly, likely giving rise to increased disruption of the biotin-streptavidin interaction, releasing the complementary sequence. Integration into a single, flow-through microfluidic device would allow these steps to be performed more efficiently, potentially improving purity. Figure 6B compares the % recovery of the microfluidic and manual approaches. The manual approach significantly outperforms in this regard. This is not surprising considering that the concurrent approach used in the microfluidic device described here is limited to a maximum recovery of 50%. Further development of counter current devices will be necessary to match the % recovery of the manual procedure.
Figure 6.
A) A comparison of the ratio of forward and complementary sequences ([ssDNA2]/[ssDNA1]) isolated using the manual (red) and continuous flow microfluidic (blue) techniques. B) A comparison of the % recovery of ssDNA2 sequences isolated using the manual (red) and continuous flow microfluidic (blue) techniques.
CE Characterization
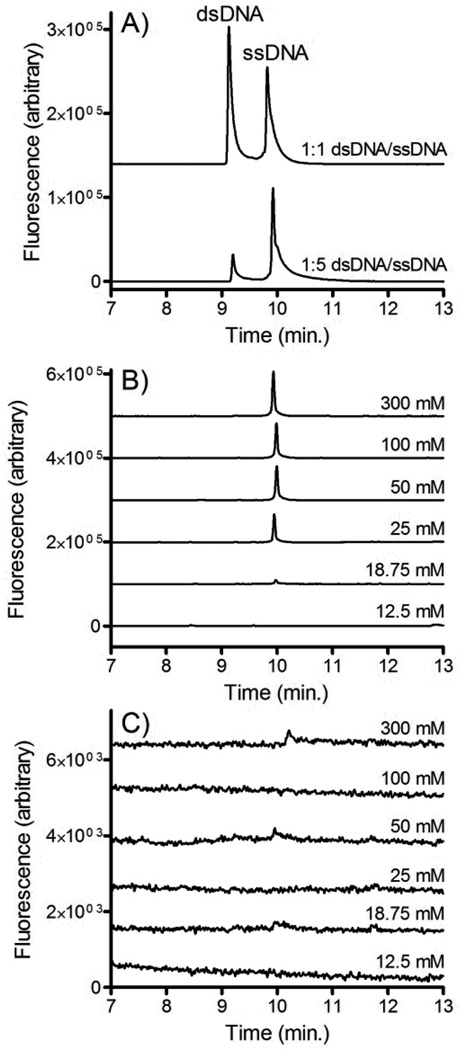
CE was used to confirm that the isolated DNA was indeed single stranded. This confirmation is important since if NaOH significantly denatures streptavidin on the timescale of the isolation, the dsDNA released from the beads would be small enough to cross the membrane. Figure 7A shows electropherograms of fluorescently labeled single stranded and double stranded DNA. Clear resolution is observed between the two peaks suggesting that CE is an appropriate assay for confirming the relative abundance of ssDNA and dsDNA.
Figure 7.
A) CE-LIF analysis of ssDNA and dsDNA. In the top trace 100 nM dsDNA2 was mixed with an equal volume of 100 nM ssDNA2. In the bottom trace 20 nM dsDNA2 was mixed with an equal volume of 100nM ssDNA2. B) dsDNA2-bead and C) dsDNA1-bead suspensions were incubated with varying concentrations of NaOH and pumped into the microfluidic dialysis device at 0.2 ml/hr. Solution from the buffer outlet channel was collected, neutralized and analyzed using CE-LIF.
Solutions of dsDNA were incubated with varying concentrations of NaOH and pumped into the microfluidic dialysis device. Aliquots were collected from the outlet of the buffer channel and analyzed using CE. Figure 7B shows the electropherograms that were observed when dsDNA2 (i.e. the forward sequence is fluorescently labeled) is pumped into the device. Similar to the fluorescence experiments, at low NaOH concentrations no DNA crosses the membrane into the buffer channel, confirming that the forward sequence remained bound to the bead through its complementary sequence. At NaOH concentrations greater than 25 mM a clear ssDNA peak is observed. This confirms that the forward sequence is freed from the beads and is able to cross the membrane. dsDNA was not observed suggesting that denaturation of the streptavidin was minimal.
Figure 7C show the electropherograms that were observed when dsDNA1 (i.e. the biotinylated complementary strand is fluorescently labeled) is introduced into the device. In contrast to dsDNA2, very little DNA crosses the membrance at any NaOH concentration. This confirms that the complementary sequence largely remains bound to the bead, even at NaOH concentrations where significant concentrations of the forward sequence is collected.
CONCLUSIONS
We have demonstrated the application of a microfluidic dialysis device for the isolation of a specific ssDNA strand from dsDNA. This is an important step of the SELEX process for isolating aptamers as well as many other biotechnology procedures. NaOH denaturation was optimized to maximize recovery of the forward sequence while minimizing release of the complementary strand. The purity of the correct forward sequence ssDNA collected reached >95% under optimized conditions, higher than that obtained using an analogous manual procedure. Yield was closely tied to flow rate through the device with 24% recovery achieved at 0.2 mL/hr. This relatively low throughput does restrict the potential of the device somewhat. Further development of membrane materials with higher pore densities that are robust enough to survive the fabrication and denaturation procedures would allow reasonable recoveries to be achieved at higher flow rates.
It should be noted that the major innovation reported here is not in regards to the performance of the device itself. In many ways the observed performance is similar or even somewhat worse that what can be performed using currently available manual approaches. This is not surprising for a technique such as this that is amenable to manual intervention. The advantage of the microfluidic dialysis approach described here is the potential for further integration. The flow through design provides simple operation without requiring any special timing or valving. In particular the microfluidic dialysis design is especially well suited for integration with other flow through devices such as µFFE aptamer selection82 and flow through PCR85. Combining these microfluidic modules suggests a unique approach towards designing a fully integrated microfluidic SELEX device.
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health (GM063533). Fabrication was performed using facilities at the University of Minnesota Nanofabrication Center, a NNIN funded site.
REFERENCES
- 1.Lee JF, Stovall GM, Ellington AD. Current Opinion in Chemical Biology. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Wang P, Yang Y, Hong H, Zhang Y, Cai W, Fang D. Current Medicinal Chemistry. 2011;18:4169–4174. doi: 10.2174/092986711797189673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nimjee SM, Rusconi CP, Sullenger BA. Annual Review of Medicine. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 4.Lee J-H, Canny MD, De Erkenez A, Krilleke D, Ng Y-S, Shima DT, Pardi A, Jucker F. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 6.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Biosensors and Bioelectronics. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Collett JR, Cho EJ, Lee JF, Levy M, Hood AJ, Wan C, Ellington AD. Analytical Biochemistry. 2005;338:113–123. doi: 10.1016/j.ab.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 8.Dominik K, Yin-Shan EN, TShima DT. Biochemical Society Transactions. 2009;37:1201–1206. doi: 10.1042/BST0371201. [DOI] [PubMed] [Google Scholar]
- 9.Hesselberth J, Robertson MP, Jhaveri S, Ellington AD. Reviews in Molecular Biotechnology. 2000;74:15–25. doi: 10.1016/s1389-0352(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 10.Brody EN, Willis MC, Smith JD, Jayasena S, Zichi D, Gold L. Molecular Diagnosis. 1999;4:381–388. doi: 10.1016/s1084-8592(99)80014-9. [DOI] [PubMed] [Google Scholar]
- 11.Kong R-M, Zhang X-B, Chen Z, Tan W. Small. 2011;7:2428–2436. doi: 10.1002/smll.201100250. [DOI] [PubMed] [Google Scholar]
- 12.Odenthal KJ, Gooding JJ. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- 13.Jhaveri S, Rajendran M, Ellington AD. Nat Biotech. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Yang R, Yang L, Tan W. ACS Nano. 2009;3:2451–2460. doi: 10.1021/nn9006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiddiky MJA, Torriero AAJ, Zeng Z, Spiccia L, Bond AM. Journal of the American Chemical Society. 2010;132:10053–10063. doi: 10.1021/ja1021365. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y, Wang X, Xiao Y, Feng L, Zhao C, Ren J, Qu X. Journal of the American Chemical Society. 2009;131:13813–13818. doi: 10.1021/ja9051763. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W-J, Halpern AR, Seefeld TH, Corn RM. Analytical Chemistry. 2012;84:440–445. doi: 10.1021/ac202863k. [DOI] [PubMed] [Google Scholar]
- 18.Katz E, Willner I. Angewandte Chemie International Edition. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 19.Groth M, Huse K, Reichwald K, Taudien S, Hampe J, Rosenstiel P, Birkenmeier G, Schreiber S, Platzer M. Analytical Biochemistry. 2006;356:194–201. doi: 10.1016/j.ab.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 20.Hultman T, Stahl S, Hornes E, Uhlen M. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stahl S, Hultman T, Olsson A, Moks T, Uhlen M. Nucleic Acids Res. 1988;16:3025–3038. doi: 10.1093/nar/16.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou S-H, Chin K-H, Wang AH-J. Nucleic Acids Research. 2003;31:2461–2474. doi: 10.1093/nar/gkg367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuerk C, MacDougal S, Gold L. Proceedings of the National Academy of Sciences. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider DJ, Feigon J, Hostomsky Z, Gold L. Biochemistry. 1995;34:9599–9610. doi: 10.1021/bi00029a037. [DOI] [PubMed] [Google Scholar]
- 25.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Proceedings of the National Academy of Sciences. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing N, Rando RF, Pommier Y, Hogan ME. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 27.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 29.Ellington AD, Szostak JW. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 30.Aguade M, Meyers W, Long AD, Langley CH. Proceedings of the National Academy of Sciences. 1994;91:4658–4662. doi: 10.1073/pnas.91.11.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Proceedings of the National Academy of Sciences. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto A, Tanaka K, Fukuta T, Saito I. Journal of the American Chemical Society. 2003;125:9296–9297. doi: 10.1021/ja035408l. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Yuan M, Lou X, Mao H, Zheng D, Zou R, Zou N, Tang X, Zhao J. Biosensors and Bioelectronics. 2011;26:4294–4300. doi: 10.1016/j.bios.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Ji M, Hou P, Li S, He N, Lu Z. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2004;548:97–105. [Google Scholar]
- 35.Pourmand N, Elahi E, Davis RW, Ronaghi M. Nucleic Acids Research. 2002;30:e31. doi: 10.1093/nar/30.7.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen R, Ishak HD, Estrada D, Dowd SE, Hong E, Mueller UG. Proceedings of the National Academy of Sciences. 2009;106:17805–17810. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hultman T, Stahl S, Hogan E, Uhlen M. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy M, Hammond H, Caskey C. Methods in Molecular Biology. 1996;65:163–176. doi: 10.1385/0-89603-344-9:163. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Onoshima D, Aki M, Okamoto Y, Kaji N, Tokeshi M, Baba Y. Analytical Chemistry. 2011;83:3528–3532. doi: 10.1021/ac200236r. [DOI] [PubMed] [Google Scholar]
- 40.Donhauser SC, Niessner R, Seidel M. Analytical Chemistry. 2011;83:3153–3160. doi: 10.1021/ac2002214. [DOI] [PubMed] [Google Scholar]
- 41.Famulok M, Mayer G. Accounts of Chemical Research. 2011;44:1349–1358. doi: 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- 42.Syed M, Pervaiz S. Oligonucleotides. 2010;20:215–224. doi: 10.1089/oli.2010.0234. [DOI] [PubMed] [Google Scholar]
- 43.Mendonsa SD, Bowser MT. Journal of the American Chemical Society. 2005;127:9382–9383. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 44.Mosing RK, Mendonsa SD, Bowser MT. Analytical Chemistry. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 45.Famulok M. Current Opinion in Structural Biology. 1999;9:324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 46.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proceedings of the National Academy of Sciences. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berens C, Thain A, Schroeder R. Bioorganic & Medicinal Chemistry. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 48.Famulok M. Journal of the American Chemical Society. 1994;116:1698–1706. [Google Scholar]
- 49.Sassanfar M, Szostak JW. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 50.Burgstaller P, Famulok M. Angewandte Chemie International Edition in English. 1994;33:1084–1087. [Google Scholar]
- 51.Mayer G, Ahmed M-SL, Dolf A, Endl E, Knolle PA, Famulok M. Nat. Protocols. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 52.Deng Q, German I, Buchanan D, Kennedy RT. Analytical Chemistry. 2001;73:5415–5421. doi: 10.1021/ac0105437. [DOI] [PubMed] [Google Scholar]
- 53.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Journal of the American Chemical Society. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 54.Tombelli S, Minunni M, Mascini M. Biomolecular Engineering. 2007;24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Fichou Y, Ferec C. Trends in Biotechnology. 2006;24:563–570. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Walder R, Hayes J, Walder J. Nucleic Acids Res. 1993;21:4339–4343. doi: 10.1093/nar/21.18.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams K, Bartel D. Nucleic Acids Res. 1995;23:4220–4221. doi: 10.1093/nar/23.20.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordstrom LJ, Clark CA, Andersen B, Champlin SM, Schwinefus JJ. Biochemistry. 2006;45:9604–9614. doi: 10.1021/bi052469i. [DOI] [PubMed] [Google Scholar]
- 59.Stoltenburg R, Reinemann C, Strehlitz B. Analytical and Bioanalytical Chemistry. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 60.Gyllensten U, Elrlich H. Proc natl Acad Sci USA. 1988;85:7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boiziau C, Dausse E, Yurchenko L, Toulme J-J. Journal of Biological Chemistry. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 62.Fukusaki E-i, Kato T, Maeda H, Kawazoe N, Ito Y, Okazawa A, Kajiyama S-i, Kobayashi A. Bioorganic & Medicinal Chemistry Letters. 2000;10:423–425. doi: 10.1016/s0960-894x(00)00013-5. [DOI] [PubMed] [Google Scholar]
- 63.Low SY, Hill JE, Peccia J. Biochemical and Biophysical Research Communications. 2009;378:701–705. doi: 10.1016/j.bbrc.2008.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Javaherian S, Musheev MU, Kanoatov M, Berezovski MV, Krylov SN. Nucleic Acids Research. 2009;37:e62. doi: 10.1093/nar/gkp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reske T, Mix M, Bahl H, Flechsig G-U. Talanta. 2007;74:393–397. doi: 10.1016/j.talanta.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Citartan M, Tang T-H, Tan S-C, Gopinath S. World Journal of Microbiology and Biotechnology. 2011;27:1167–1173. [Google Scholar]
- 67.Higuchi RG, Ochman H. Nucleic Acids Res. 1989;17:5865. doi: 10.1093/nar/17.14.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dastjerdi K, Tabar GH, Dehghani H, Haghparast A. Biotechnology and Applied Biochemistry. 2011;58:226–230. doi: 10.1002/bab.36. [DOI] [PubMed] [Google Scholar]
- 69.Wilson R. Nucleic Acids Therapeutics. 2011;21:437–440. doi: 10.1089/nat.2011.0322. [DOI] [PubMed] [Google Scholar]
- 70.Espelund M, Stacy R, Jakobsen K. Nucleic Acids Res. 1990;18:6157–6158. doi: 10.1093/nar/18.20.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dwivedi H, Smiley R, Jaykus L-A. Applied Microbiology and Biotechnology. 2010;87:2323–2334. doi: 10.1007/s00253-010-2728-7. [DOI] [PubMed] [Google Scholar]
- 72.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proceedings of the National Academy of Sciences. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim YS, Hyun CJ, Kim IA, Gu MB. Bioorganic & Medicinal Chemistry. 2010;18:3467–3473. doi: 10.1016/j.bmc.2010.03.074. [DOI] [PubMed] [Google Scholar]
- 74.Liu D. Handbook of nucleic acid purification. Boca Raton: CRC Press; 2009. ch. 23. [Google Scholar]
- 75.Heller M, Guttman A. In: Integrated microfabricated biodevices. Ramanujam R, Sacks W, Kang J, editors. New York: Marcel Dekker, Inc.; 2002. pp. 71–84. [Google Scholar]
- 76.Marimuthu C, Tang T-H, Tominaga J, Tan S-C, Gopinath SCB. Analyst. 2012;137:1307–1315. doi: 10.1039/c2an15905h. [DOI] [PubMed] [Google Scholar]
- 77.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 78.Arora A, Simone G, Salieb-Beugelaar GB, Kim JT, Manz A. Analytical Chemistry. 2010;82:4830–4847. doi: 10.1021/ac100969k. [DOI] [PubMed] [Google Scholar]
- 79.Park S-m, Ahn J-Y, Jo M, Lee D-k, Lis JT, Craighead HG, Kim S. Lab on a Chip. 2009;9:1206–1212. doi: 10.1039/b814993c. [DOI] [PubMed] [Google Scholar]
- 80.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT. Proceedings of the National Academy of Sciences. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qian J, Lou X, Zhang Y, Xiao Y, Soh HT. Analytical Chemistry. 2009;81:5490–5495. doi: 10.1021/ac900759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jing M, Bowser MT. Lab on a Chip. 2011;11:3703–3709. doi: 10.1039/c1lc20461k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang C-J, Lin H-I, Shiesh S-C, Lee G-B. Biosensors and Bioelectronics. 2012 doi: 10.1016/j.bios.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 84.Ferguson BS, Buchsbaum SF, Swensen JS, Hsieh K, Lou X, Soh HT. Analytical Chemistry. 2009;81:6503–6508. doi: 10.1021/ac900923e. [DOI] [PubMed] [Google Scholar]
- 85.Hashimoto M, Chen PC, Mitchell MW, Nikitopoulos DE, Soper SA, Murphy MC. Lab on a Chip. 2004;4:638–645. doi: 10.1039/b406860b. [DOI] [PubMed] [Google Scholar]
- 86.Sheng YX, Bowser MT. Analyst. 2012;137:1144–1151. doi: 10.1039/c2an15966j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDonald J, Duffy D, Anderson J, Chiu D, Wu H, Schueller O, Whitesides G. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Jo B, Van Lerberghe L, Motsegood K, Beebe D. Journal of microelectromechanical systems. 2000;9:76–81. [Google Scholar]
- 89.Iki N, Yeung ES. Journal of Chromatography A. 1996;731:273–282. [Google Scholar]
- 90.Hestekin CN, Lin JS, Senderowicz L, Jakupciak JP, O'Connell C, Rademaker A, Barron AE. Electrophoresis. 2011;32:2921–2929. doi: 10.1002/elps.201100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kourkine IV, Hestekin CN, Buchholz BA, Barron AE. Analytical Chemistry. 2002;74:2565–2572. doi: 10.1021/ac020025b. [DOI] [PubMed] [Google Scholar]
- 92.Albarghouthi MN, Buchholz BA, Huiberts PJ, Stein TM, Barron AE. Electrophoresis. 2002;23:1429–1440. doi: 10.1002/1522-2683(200205)23:10<1429::AID-ELPS1429>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 93.O'Connell CD, Atha DH, Oldenburg MC, Tian J, Siebert M, Handrow R, Grooms K, Heisler L, de Arruda M. Electrophoresis. 1999;20:1211–1223. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1211::AID-ELPS1211>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 94.Steinour HH. Industrial & Engineering Chemistry. 1944;36:618–624. [Google Scholar]
- 95.Lamb H. Hydrodynamics. 6th edition ed. Cambridge University Press; 1994. [Google Scholar]
- 96.Buscall R, White LR. Journal of the Chemical Society, Faraday Transactions 1: Physical Chemistry in Condensed Phases. 1987;83:873–891. [Google Scholar]
- 97.Richardson JF, Zaki WN. Chemical Engineering Science. 1954;3:65–73. [Google Scholar]
- 98.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Proceedings of the National Academy of Sciences. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ageno M, Dore E, Frontali C. Biophysical journal. 1969;9:1281–1311. doi: 10.1016/S0006-3495(69)86452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bimboim HC, Doly J. Nucleic Acids Research. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luck G, Zimmer C, Snatzke G, Söndgerath G. European Journal of Biochemistry. 1970;17:514–522. doi: 10.1111/j.1432-1033.1970.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 102.Barber R. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 1971;238:60–66. doi: 10.1016/0005-2787(71)90009-8. [DOI] [PubMed] [Google Scholar]
- 103.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlén M. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 104.Weber P, Ohlendorf D, Wendoloski J, Salemme F. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 105.Bulmus V, Ding Z, Long CJ, Stayton PS, Hoffman AS. Bioconjugate Chemistry. 1999;11:78–83. doi: 10.1021/bc9901043. [DOI] [PubMed] [Google Scholar]