Abstract
Folding and unfolding rates for the ultrafast folding villin subdomain were determined from a photon-by-photon analysis of fluorescence trajectories in single molecule FRET experiments. One of the obstacles to measuring fast kinetics in single molecule fluorescence experiments is blinking of the fluorophores on a timescale that is not well separated from the process of interest. By incorporating acceptor blinking into a two-state kinetics model, we show that it is possible to extract accurate rate coefficients on the microsecond time scale for folding and unfolding using the maximum likelihood method of I.V. Gopich and A. Szabo. This method yields the most likely parameters of a given model that can reproduce the observed photon trajectories. The extracted parameters agree with both the decay rate of the donor-acceptor cross correlation function and the results of ensemble equilibrium and kinetic experiments using nanosecond laser temperature jump.
1. Introduction
The power of single molecule fluorescence spectroscopy is the ability to investigate distributions in molecular behavior for intrinsically heterogeneous systems. One such system is protein folding, in which theory predicts many different sequences of structural changes in the pathways that connect the folded and unfolded states [1]. The -helical, 35-residue villin subdomain (Fig. 1) is currently the most extensively studied protein by experiment, theory, and simulations (see bibliography in Supplementary Material). The reasons are that it has equilibrium properties of a much larger single-domain protein [2,3], is among the fastest folding proteins [4,5], and exhibits unusual kinetics such as a denaturant independent relaxation rate [6] and an apparent increase in the internal friction with temperature in a Kramers description of the barrier crossing [7]. Our ultimate goal for single molecule experiments on this protein is to observe the distribution of transition paths – a uniquely single molecule property. Such measurements would provide a very demanding test of the accuracy of the mechanisms found in molecular dynamics simulations, but represent a major challenge since transition paths have not been observed for any molecular system in the condensed phase. New and sensitive tests of simulation are important because, if accurate, everything one would ever want to know about the folding mechanism of a particular protein is contained in a sufficiently long atomistic trajectory [8–10] or Markov state modeling on many short trajectories [11].
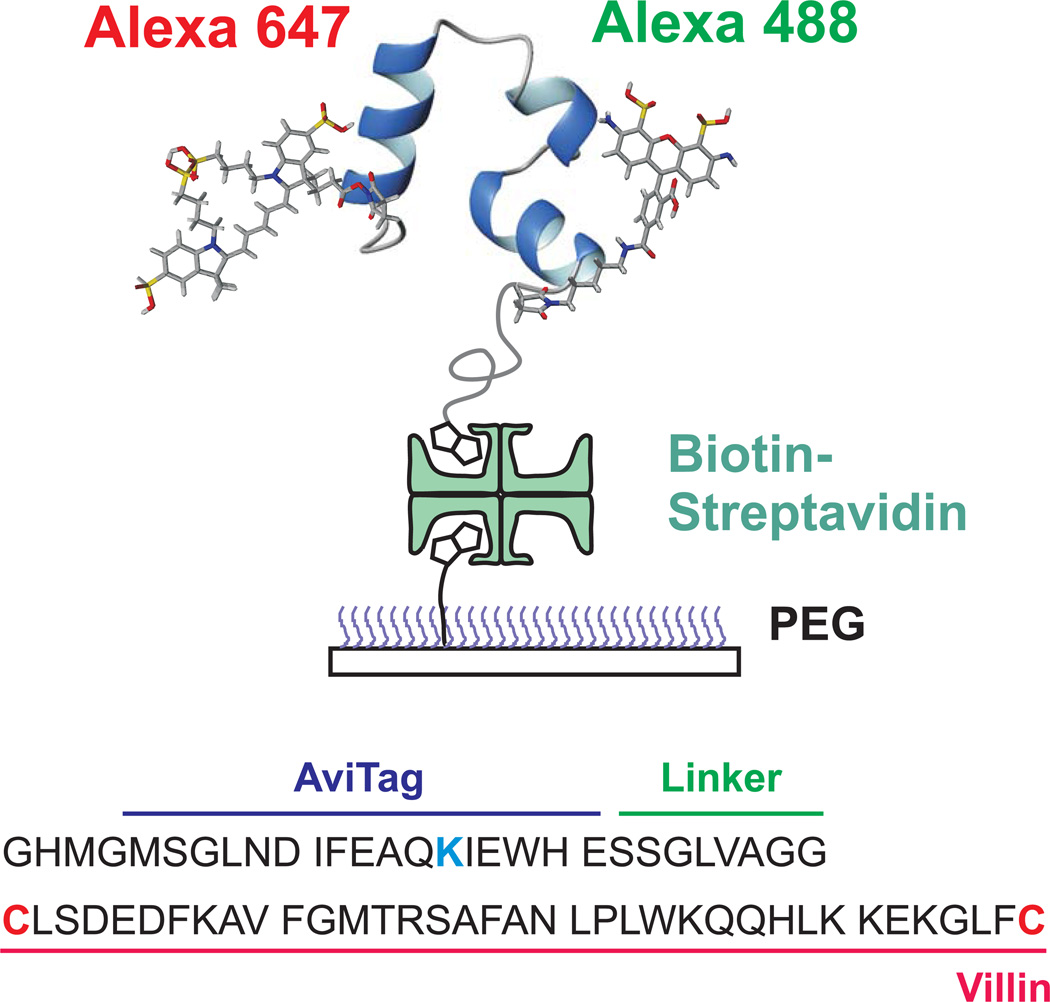
Fig. 1.
Schematic of immobilized folded villin subdomain showing donor (green-emitting) and acceptor (red-emitting) fluorophores. Protein molecules were attached to a polyethyleneglycol-coated glass surface via a biotin-strepavidin-biotin linkage. Dyes were attached to cysteine residues (red) at the N- and C-termini of villin and a biotin molecule to the lysine residue (blue) within the AviTag sequence.
Studies of the villin subdomain are particularly challenging because of its very rapid kinetics, with folding times of the wild-type on the order of 5–50 µs [3,6,12]. Until quite recently the time resolution in single molecule FRET experiments [13] has been limited by the bin time of the measurement, which is usually 1 – 10 ms at the moderate illumination intensities that have been employed to avoid photochemical problems such as bleaching and blinking of the dyes [14]. For residence times much longer than these bin times, two distinct peaks will appear in a histogram of the FRET efficiencies and the rate coefficients can be simply determined from the FRET efficiency trajectories as the reciprocal of the average residence times (also called waiting or dwell times). When the average residence time is much shorter than the bin time, however, transitions occur within every bin, and rate coefficients cannot be simply extracted from the residence time distributions in the FRET efficiency trajectories. To extract kinetics from the data under these circumstances, Gopich and Szabo have developed maximum likelihood methods for analyzing photon trajectories without binning [15]. We previously used this method on the helical protein 3D, and showed that the method produced accurate folding times of ~1 ms, as judged by the excellent agreement of the sum of the rate coefficients obtained from the maximum likelihood analysis for this two-state system and the decay of the donor-acceptor cross-correlation function [16]. For the faster-folding villin subdomain, blinking occurs on a similar time scale to folding and unfolding, making it problematic to distinguish dye blinking from protein dynamics. We show how to extract the kinetics of folding and unfolding from the data that is complicated by blinking, and confirm the accuracy of the extracted rate coefficients by comparison with both the donor-acceptor cross-correlation functions and the results of ensemble equilibrium and kinetic experiments.
2. Methods
Materials
The structure of the 35-residue long villin headpiece subdomain (PDB 1YRF), together with the amino acid sequence of the construct used in the single molecule experiments, is shown in Fig. 1 [17]. For the laser temperature jump measurements, the chemically-synthesized 35-residue sequence at a purity >98% was purchased from California Peptide (Napa, CA). For single molecule studies, the complete sequence in Fig. 1 (avitag + linker + cysteine + villin subdomain sequence + cysteine) was expressed in E. coli. Details of the expression, purification and dye labeling are given in the Supplementary Material.
Nanosecond laser temperature-jump measurements
Ensemble kinetic measurements were carried out on solutions containing 300 M of the 35-residue sequence (Fig. 1) or N-acetyl tryptophanamide (NATA) using a nanosecond-laser-temperature-jump instrument previously described [5]. All solutions were buffered to pH = 4.9 with 20 mM sodium acetate and flowed through the illuminated region to eliminate effects of tryptophan photodamage. Temperature jumps of ~7–10°C to a final temperature of 20°C were generated by Raman shifting pulses of a Nd:YAG fundamental at 1064 nm to 1560 nm using D2 gas. To ensure a consistent temperature jump in the presence of changing solvent conditions, the temperature jump was calibrated using NATA. A frequency-doubled Kr laser with an output at 284 nm was used to excite Trp fluorescence. In each experiment, four to eight traces of 512 laser shots were collected. Rate constants and amplitudes were calculated by a least-squares exponential fit of the data at times >3 µs and baseline from a NATA trace.
Single molecule spectroscopy
Single molecule FRET experiments were performed using a confocal microscope system (MicroTime200, Picoquant). The CW mode of a dual mode (CW/pulsed) 485 nm diode laser (LDH-D-C-485, PicoQuant) was used to excite donor dyes (Alexa Fluor 488) through an oil-immersion objective (PlanApo, NA 1.4, × 100, Olympus). Donor and acceptor (Alexa Fluor 647) fluorescence was collected by the same objective, split into two channels, and focused through optical filters (ET525/50m for the donor and E600LP for the acceptor, Chroma Technology) onto photon-counting avalanche photodiodes (SPCM-AQR-15, PerkinElmer Optoelectronics). Additional details for the optical setup and single molecule experiments can be found elsewhere [18,19].
In the free diffusion experiment, where the fluorescence bursts of single freely diffusing molecules entering into the focal volume were measured, protein concentration was 40 pM in 50 mM HEPES buffer (pH 7.6). A surfactant, 0.01% Tween-20 (Thermo Scientific) was used to prevent sticking of proteins on a glass surface. The illumination power of the laser was 2.9 µW.
In the immobilization experiment, protein molecules were immobilized on a biotin-embedded, polyethyleneglycol (PEG)-coated glass coverslip (Bio_01, Microsurfaces Inc.) via a biotin (surface)-streptavidin-biotin (protein) linkage. To reduce dye bleaching and blinking, 2 mM L-ascorbic acid (A92902, Sigma) and 2 mM methyl viologen (856117, Sigma) were added [20] to the 50 mM HEPES buffer (pH 7.6) solution.
Fluorescence trajectories were collected using an automated data collection scheme. An area of 10 × 10 µm2 was raster scanned at low intensity and the location of molecules was determined. To ensure the single molecule detection, molecules immobilized too close to one another were identified by an image larger than a threshold size and were excluded. The piezo-controlled stage was then moved to locate each molecule and the trajectory was collected at high illumination intensity. The laser was turned off during movement of the stage to prevent photobleaching. After the collection of trajectories for all identified molecules was completed, the procedure was repeated for the next 10 × 10 µm2 area. Before raster scanning each area, the focus along z-axis (perpendicular to the surface) was set at the position with minimum variance of the reflected image from the surface recorded by a CCD camera. The illumination intensity for the raster scan (0.4 kW/cm2, the average power entering the microscope was 1.3 µW) and for the trajectory collection (4 kW/cm2) were adjusted by inserting a neutral density filter (OD1) in the laser path using a home-built mechanical shutter.
3. Results
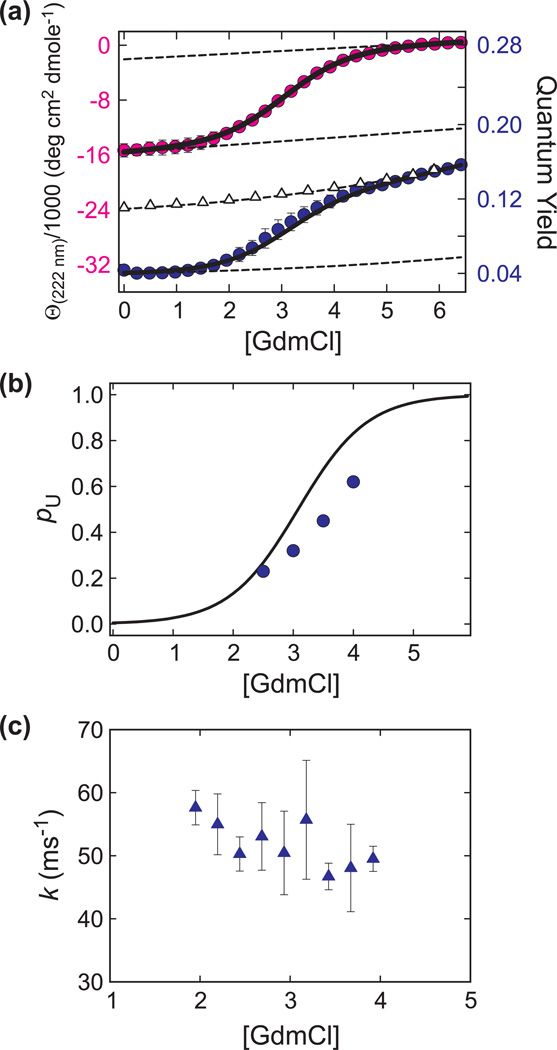
Figure 2 shows the ensemble equilibrium and kinetic data at 20°C on the 35-residue sequence that has not been labeled with the donor and acceptor fluorophores. The population versus GdmCl concentration in Fig 2b is obtained from a two-state fit to the circular dichroism and fluorescence unfolding curves. The relaxation rate, measured from the time course of the tryptophan quantum yield in nanosecond laser temperature jump experiments, is nearly independent of GdmCl concentration at ~50 ms−1 in the range 2–4 M GdmCl (Fig. 2c). Since we do not anticipate a major effect from either the attachment of the fluorophores or the additional residues of the complete construct in Fig. 1, the residence times in single molecule experiments are expected to be sub-100 µs.
Fig. 2.
(a) Guanidinium chloride (GdmCl) induced unfolding curve measured by circular dichroism (CD) (pink circles) and tryptophan fluorescence quantum yield (Φ, blue circles). The measurements were carried out at 20°C in 20 mM sodium acetate buffer, pH 4.9. The plotted values are the average of three independent measurements and the error bars correspond to the standard deviation. The dotted lines are the baselines and the triangles correspond to the quantum yield of a reference peptide (AcWKQQH) (Φref). The data were fit simultaneously to a two-state model (continuous black lines), Θ222 nm = [(a + bC) + (c + dC)K]/(1 + K) and = (e Φref + f ΦrefK)/(1 + K), where Φref = 0.0794 + 0.0027C + 0.0002C2 + 0.00003C3 and K = exp[m(C – Cm)/RT]. C and Cm are the concentration of GdmCl and the mid point concentration, respectively. The fitting yields Cm = 3.07 M and m = 1.01 kcal mol−1 M−1. The CD and fluorescence data collection and fitting procedures are the same as those used in ref. [6]. Note that the CD data have smaller uncertainties and thus are fit slightly better than the fluorescence data. However, in all cases the calculated value is less than one standard deviation away from the measured value, yielding a normalized 2 value of less than one. (b) Plot of the population of unfolded molecules (pu) vs. the concentration of GdmCl. The black line is calculated using the thermodynamic parameters from the unfolding of unlabeled villin (Fig. 2a). The blue circles are obtained from the maximum likelihood analysis of the single molecule photon trajectories (see Table 1). (c) Measured relaxation rates following laser temperature-jump to 20 °C. The plotted values are the average of at least three independent measurements and the error bars correspond to the standard deviation.
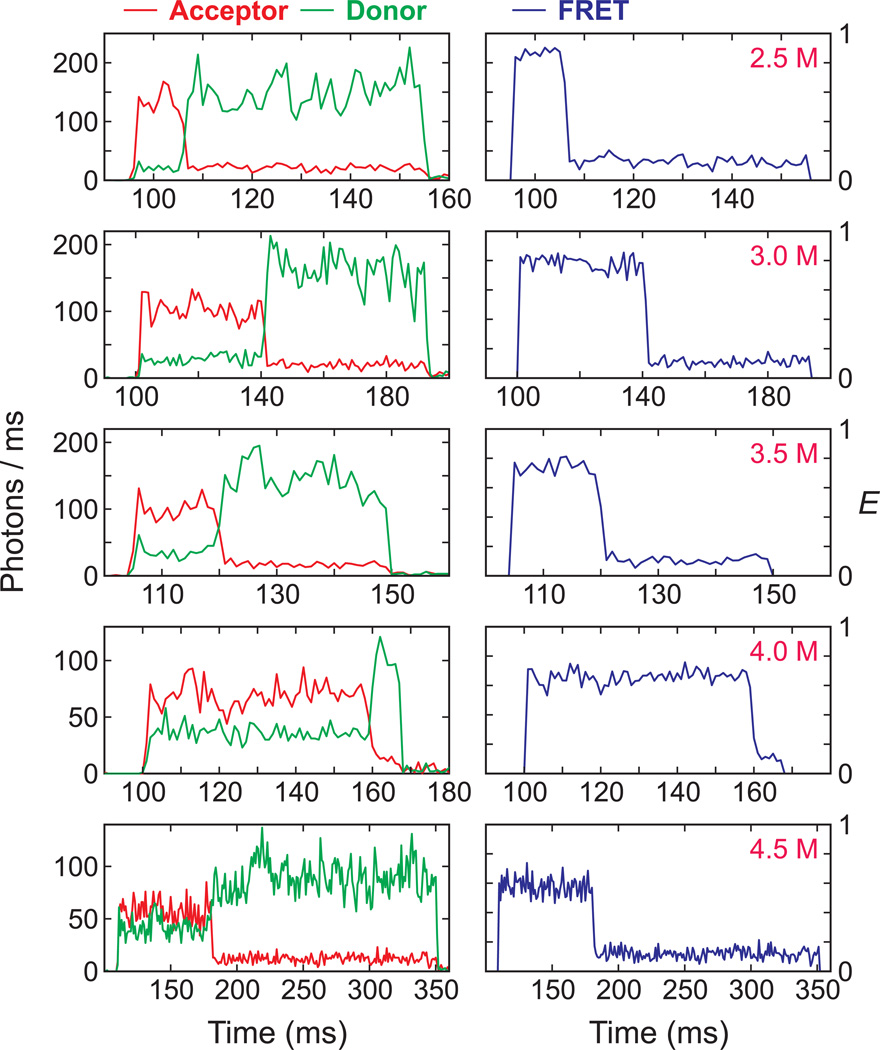
To measure single molecule equilibrium folding/unfolding trajectories using FRET spectroscopy, the recombinant villin construct was labeled with two fluorescent dyes, Alexa Fluor 488 as donor and Alexa Fluor 647 as acceptor, and immobilized on a PEG-coated glass surface by a biotin-streptavidin linkage (Fig. 1). Since the N- and C-termini of the protein where the two dyes were attached are close to each other when the protein is folded, the FRET efficiency is expected to be higher in the folded state than in the unfolded state. Fig. 3 shows representative donor fluorescence, acceptor fluorescence, and FRET efficiency trajectories at different guanidinium hydrochloride (GdmCl) concentrations. Because the average residence times in the folded and unfolded states (inverse of the unfolding and folding rate coefficients, respectively) are much shorter than the bin time of 1 ms, the trajectories show constant fluorescence intensity for the donor and acceptor, and correspondingly constant FRET efficiency, with small fluctuations due to shot noise without any indication of folding and unfolding transitions. However, the gradual shift of the FRET efficiency to lower values with increasing GdmCl concentration indicates an increasing unfolded population with increasing GdmCl concentration, as expected from the ensemble results (Fig. 2a). This progressive increase of the unfolded population is more easily seen in the distribution of the FRET efficiencies in Fig. 4a calculated from the total number of donor and acceptor photons in an entire individual trajectory.
Fig. 3.
Representative binned fluorescence trajectories of donor (green) and acceptor (red) and FRET efficiency trajectories at various GdmCl concentrations. Photons were collected in 1 ms bins at the illumination intensity of 4 kW/cm2. In each trajectory the acceptor bleaches before the donor. The apparent FRET efficiency near zero when the acceptor bleaches indicates that the donor does not have the light-induced red-shifted spectrum [19].
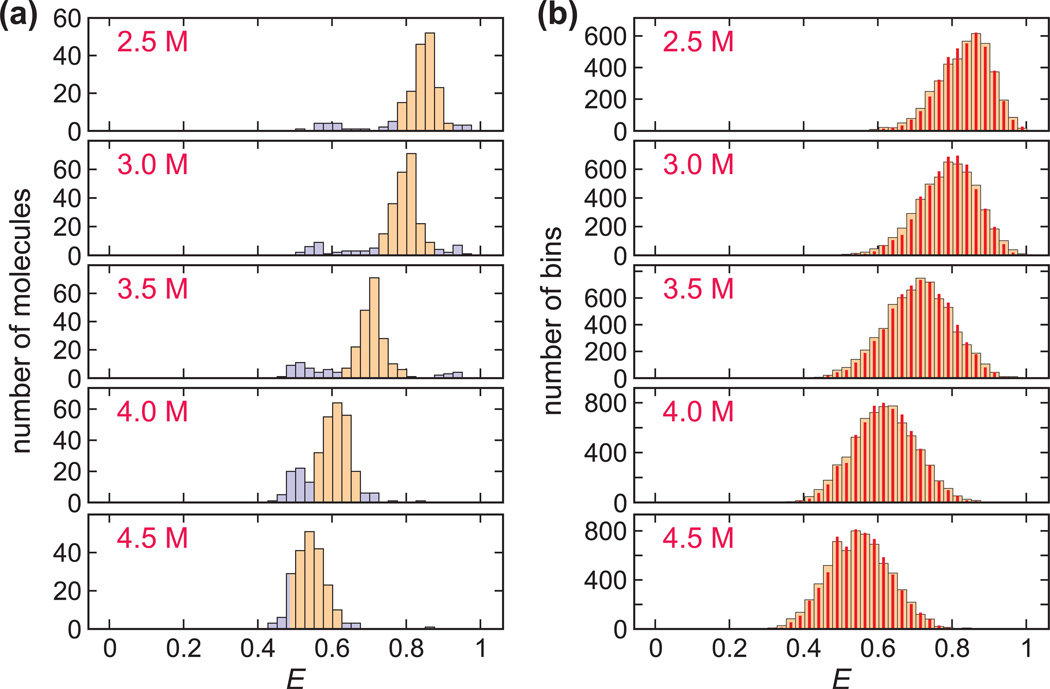
Fig. 4.
FRET efficiency histograms at various GdmCl concentrations. (a) The FRET efficiency histograms were obtained from the FRET efficiencies calculated for the first segment in each trajectory until either a bleach or a blink. Although a very narrow unimodal distribution is expected at each concentration due to the fast folding-unfolding compared to 0.5 ms bin time, an additional distribution appears at E ~ 0.5 colored in light blue. The trajectories with FRET efficiencies within the specific range (colored in orange) at each GdmCl concentration were used in plot (b) and analyzed further. These ranges are: 0.775 < E < 0.925 (2.5 M); 0.725 < E < 0.875 (3 M) ; 0.625 < E < 0.8 (3.5 M) ; 0.55 < E < 0.675 (4 M) ; 0.485 < E < 0.625 (4.5 M). (b) The FRET efficiency histograms (wide bars) were constructed from the FRET efficiencies of individual 0.5 ms bins of segments in the main distribution (orange) in (a) with the mean photon count rate > 75 ms−1. The histograms constructed from re-colored photon trajectories (red narrow bars) using the parameters obtained from the maximum likelihood method (4-state kinetic model) coincide extremely well with the experimental histograms.
Although a narrow distribution is expected due to fast folding and unfolding (Fig. 2c), we observed an additional distribution at lower FRET efficiency (colored in light blue) in addition to the main peak (colored in orange), which increases with increasing GdmCl concentration. Suspecting that this additional peak arises from molecules containing cis-isomers, which are expected to have a lifetime in the unfolded state >100 ms, the approximate length of the longest trajectories before bleaching, we carried out free diffusion experiments. As shown in Fig. S1, there is a long tail on the unfolded side (lower FRET efficiency) in the free diffusion FRET efficiency histograms especially at 2.5 and 3 M GdmCl, indicating that the additional peak is not caused by protein-surface interactions, and consistent with the explanation of cis isomers.
We therefore analyzed only those trajectories in the immobilization experiments, with a FRET efficiency within the range of the main distribution. As shown in Fig. 4a, the main peak overlaps the lower FRET efficiency peak at 4.5 M GdmCl, so the parameters at this concentration are expected to be the least accurate. In addition, we only analyzed the trajectories in which the acceptor photobleached earlier than the donor to clearly detect and exclude trajectories affected by the light-induced red shift of the donor Alexa 488 emission spectrum [19] using the increased donor leak into the acceptor channel. Fig. 4b shows the FRET efficiency distributions calculated from each of 0.5 ms bins of the ~200 selected trajectories at each GdmCl concentration. Due to multiple folding and unfolding transitions within the 0.5 ms bin time, only one peak is present, which is a phenomenon similar to fast chemical exchange in NMR [15,16,21].
To extract parameters for kinetics that occur much faster than the bin time, we used the Gopich-Szabo maximum likelihood method and analyzed the photon trajectories without time binning. This method yields the most likely parameters of an assumed model that can reproduce the observed photon trajectories. The likelihood function for the jth photon trajectory is [15]
| (1) |
where Nj is the number of photons in a trajectory, ci is the color of the ith photon (donor or acceptor), and i is a time interval between the ith and (i-1)th photons (Fig. 5a). The photon color matrix F depends on the color of a photon as F(acceptor) = E and F(donor) = I – E, where E is a diagonal matrix with elements that are FRET efficiencies of the individual states. I is the identity matrix, peq is a vector consisting of the equilibrium population of each state, and 1T is a row vector with elements of 1. The calculation of likelihood values were performed using the diagonalization of the matrix exponential in Eq. (1) as described in ref. [15]. Practically, the log likelihood function was calculated and the total log likelihood function of all trajectories was calculated by the summation of individual log likelihood functions as .
Fig. 5.
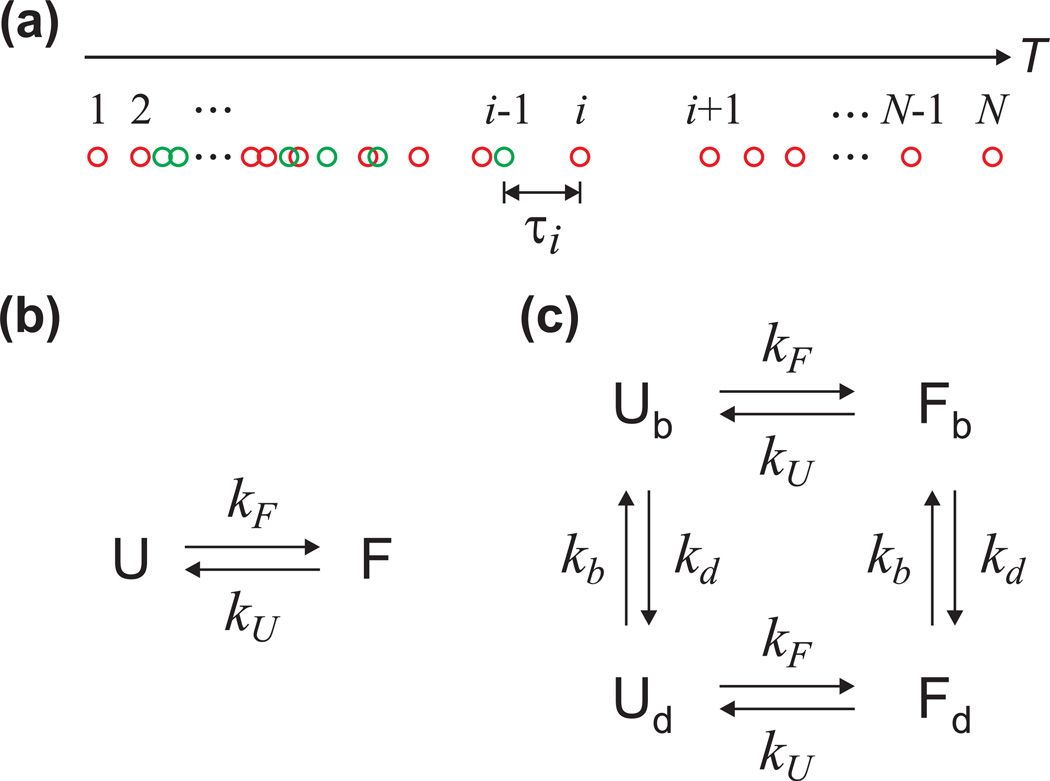
(a) The definition of photon indices and time interval used in the calculation of the likelihood function in Eq. (1). Red circles are acceptor photons and green donor photons. (b) Two-state kinetics model. (c) Four-state kinetics model including acceptor blinking. Subscripts b and d stand for the bright and dark states, respectively.
For a two-state system, as is the case for the villin subdomain [5], there are four independent parameters - the apparent FRET efficiencies and the rate coefficients of the folded and unfolded states (EF, EU, kF, kU). In analyzing the data, instead of varying the two rate coefficients, it is more convenient to vary the sum of the rate coefficients (k = kF + kU) and the fractional population of folded molecules (pF = kF/[kF + kU]). For the two-state kinetic model (Fig. 5b), the rate matrix is given by
| (2) |
The extracted parameters are summarized in Table 1. There is a decrease in the folded fraction (pF) with the increasing GdmCl concentration, as expected from the ensemble measurements (Fig. 2b) and from the decrease in the average FRET efficiency in Figs. 3 and 4. The FRET efficiencies of the folded and unfolded states are high and low as predicted. The decrease in EU with increasing GdmCl concentration results from the continuous expansion of the unfolded polypeptide chain [18,22–27]. Unless there is partial unfolding of the structures on the folded side of the free energy barrier, EF is expected to vary only slightly due to the change in the refractive index of the GdmCl solution [22]. Consequently, the decrease in EF, especially at high GdmCl concentration (>= 4 M), probably results from the inclusion of the unfolded molecules in the analysis. We also fit the data by fixing EF to that of the lowest GdmCl concentration, which results only slight changes in the parameters (Table S1).
Table 1.
Comparison of the parameters from experimental data obtained from the maximum likelihood method and the donor-acceptor cross correlation function.a
| GdmCl (M) | |||||||
|---|---|---|---|---|---|---|---|
| 2.5 | 3 | 3.5 | 4 | 4.5 | |||
| Maximum Likelihood Method |
2-state | EF | 0.91 ( 0.002) |
0.90 ( 0.002) |
0.88 ( 0.002) |
0.84 ( 0.004) |
0.77 ( 0.005) |
| EU | 0.57 ( 0.008) |
0.56 ( 0.005) |
0.51 ( 0.003) |
0.47 ( 0.003) |
0.43 ( 0.004) |
||
| k (ms−1) | 50.0 ( 2.3) | 41.3 ( 1.3) | 37.7 ( 0.9) | 45.0 ( 1.2) | 49.1 ( 1.7) | ||
| pF | 0.77 ( 0.009) |
0.68 ( 0.008) |
0.55 ( 0.007) |
0.39 ( 0.008) |
0.35 ( 0.011) |
||
| 2-state with acceptor blinking (4-state kinetic model) |
EF | 0.92 ( 0.003) |
0.92 ( 0.003) |
0.90 ( 0.003) |
0.88 ( 0.005) |
0.81 ( 0.006) |
|
| EU | 0.60 ( 0.009) |
0.58 ( 0.006) |
0.53 ( 0.004) |
0.52 ( 0.004) |
0.48 ( 0.005) |
||
| k (ms−1) | 35.8 ( 1.9) | 34.2 ( 1.2) | 32.2 ( 0.8) | 34.3 ( 1.1) | 36.1 ( 1.5) | ||
| pF | 0.78 ( 0.009) |
0.69 ( 0.009) |
0.56 ( 0.007) |
0.38 ( 0.009) |
0.35 ( 0.012) |
||
| kon (ms−1) | 860 ( 110) | 1170 ( 140) | 1050 ( 110) | 620 ( 70) | 730 ( 80) | ||
| Pb | 0.98 ( 0.002) |
0.97 ( 0.002) |
0.96 ( 0.002) |
0.95 ( 0.003) |
0.94 ( 0.004) |
||
| ln Lb | 95 | 140 | 182 | 233 | 180 | ||
| BICc | −164 | −255 | −338 | −441 | −336 | ||
| Donor- Acceptor Cross Correlation |
Experiment | A | 0.055 ( 0.015) |
0.087 ( 0.008) |
0.093 ( 0.006) |
0.093 ( 0.005) |
0.058 ( 0.006) |
| k (ms−1) | 11.6 ( 9.9) | 30.4 ( 9.6) | 25.1 ( 5.8) | 31.4 ( 5.6) | 24.6 ( 8.9) | ||
| Recolored | A | 0.070 ( 0.006) |
0.074 ( 0.004) |
0.108 ( 0.003) |
0.094 ( 0.003) |
0.058 ( 0.003) |
|
| k (ms−1) | 28.8 ( 6.2) | 24.0 ( 3.8) | 31.3 ( 2.7) | 37.0 ( 3.2) | 31.4 ( 5.1) | ||
Errors are standard deviations obtained from the diagonal elements of the covariance matrix calculated from the likelihood function (maximum likelihood method) or chi-square function (donor-acceptor cross-correlation calculation).
ln L = ln L (4-state) - ln L (2-state).
Baysian inference criterion (BIC) was calculated by BIC = -2ln L + nln Np, where n is the number of parameters and Np is the number of photons. BIC = BIC (4-state) - BIC (2-state).
The relaxation rates are almost independent of GdmCl concentration, as observed for the unlabeled protein in the laser temperature-jump experiments (Fig. 2). To check the accuracy of these relaxation rates, we calculated the donor-acceptor cross-correlation function as CDA(τ) = −A exp(kτ) (Fig.6). The relaxation rates obtained from the single exponential fitting are listed in Table 1. Generally, the relaxation rates obtained from the cross-correlation function are less precise than those from the maximum likelihood method as judged by the much bigger fitting errors, particularly at 2.5 M and 4.5 M GdmCl, where there are the most unequal populations of folded and unfolded molecules. In addition, the values from the cross-correlation analysis are smaller than those obtained from the maximum likelihood method. Furthermore, the amplitude of the donor-acceptor cross correlation function is much smaller than the theoretically expected values calculated from the average FRET efficiency values as
| (3) |
Fig. 6.
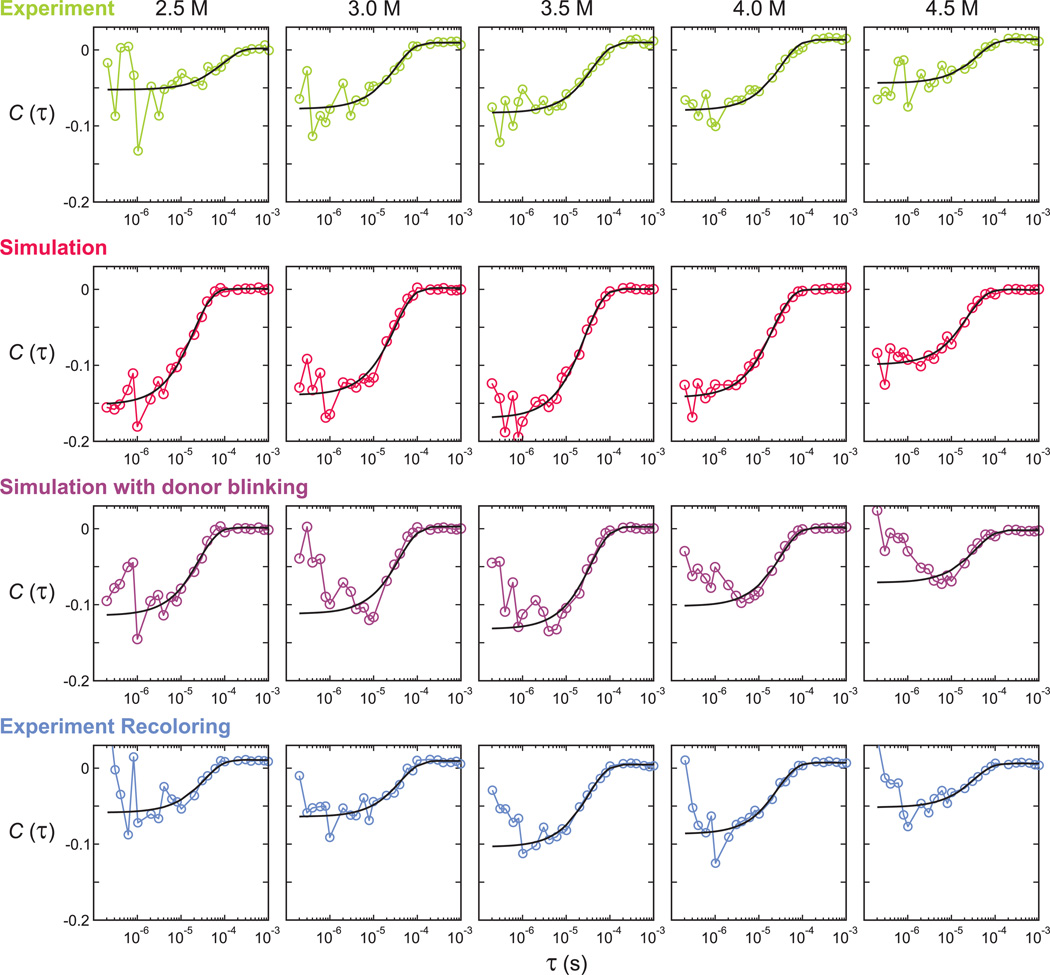
Donor-acceptor cross-correlation functions calculated from the experimental data (first row), simulated photon trajectories using 2-state kinetic model maximum likelihood parameters without (second row) and with (third row) donor blinking, and re-colored experimental trajectories using the 4-state kinetic model that includes acceptor blinking (bottom row). Black solid lines are exponential fitting curves and the fitting parameters are listed in Table 1 and 2. The decay time ranges in the fitting were either 400 ns 1 ms (first and second row) or 2 s 1 ms (third and bottom row).
Here, NA and ND are the number of donor and acceptor photons, <E> = pFEF + (1 − pF) EU, and <E2> = pFEF2 + (1 − pF)EU2, where EF and EU are the apparent FRET efficiencies of the folded and unfolded states, and pF is the folded fraction. The photon count rates in the folded and in the unfolded state are assumed to be the same. The correlation amplitude CDA(0) is the asymptotic value due to folding and unfolding of proteins without including the anti-bunching effect of the fluorescence dyes. The amplitudes calculated using the parameters obtained from the maximum likelihood method (Table 2) are almost twice as large as the amplitudes from the exponential fit to the cross-correlation data (Fig. 6, first row). We also simulated folding/unfolding photon trajectories with parameters obtained from the two-state maximum likelihood method and with experimental photon count rates (~ 125 ms−1 on average), and the calculated donor-acceptor cross-correlation as plotted in Fig. 6 (second row). In this simulated data, the correlation amplitudes are similar to the calculated values using Eq. (3) and the relaxation rates (Table 2) are very similar to the input parameters of the simulation. Therefore, the discrepancies in the time scale and the amplitude of the cross-correlation between the experiment and the simulation indicate that processes other than folding and unfolding are occurring.
Table 2.
Comparison of the parameters of simulated data obtained from the maximum likelihood method and the donor-acceptor cross correlation function.a
| GdmCl (M) | |||||||
|---|---|---|---|---|---|---|---|
| 2.5 | 3 | 3.5 | 4 | 4.5 | |||
| Maximum Likelihood Method |
2-state simulation |
EF | 0.92 ( 0.002) |
0.90 ( 0.002) |
0.88 ( 0.002) |
0.85 ( 0.003) |
0.77 ( 0.005) |
| EU | 0.57 ( 0.007) |
0.56 ( 0.005) |
0.50 ( 0.003) |
0.47 ( 0.003) |
0.43 ( 0.003) |
||
| k (ms−1) | 51.9 ( 1.8) | 40.0 ( 1.1) | 37.8 ( 0.8) | 47.0 ( 1.1) | 47.7 ( 1.5) | ||
| pF | 0.76 ( 0.009) |
0.68 ( 0.008) |
0.55 ( 0.007) |
0.39 ( 0.007) |
0.35 ( 0.010) |
||
| 2-state simulation with donor blinking |
EF | 0.92 ( 0.002) |
0.90 ( 0.002) |
0.88 ( 0.002) |
0.85 ( 0.004) |
0.77 ( 0.006) |
|
| EU | 0.57 ( 0.008) |
0.55 ( 0.005) |
0.50 ( 0.004) |
0.47 ( 0.003) |
0.43 ( 0.004) |
||
| k (ms−1) | 53.0 ( 2.0) | 39.1 ( 1.2) | 37.4 ( 0.9) | 47.9 ( 1.2) | 48.0 ( 1.6) | ||
| pF | 0.77 ( 0.010) |
0.68 ( 0.009) |
0.55 ( 0.007) |
0.38 ( 0.008) |
0.36 ( 0.012) |
||
| Donor- Acceptor Cross Correlation |
Calculated from Eq. 3b |
A | 0.15 | 0.15 | 0.17 | 0.14 | 0.11 |
| 2-state Simulation |
A | 0.153 ( 0.006) |
0.141 ( 0.008) |
0.170 ( 0.006) |
0.142 ( 0.004) |
0.099 ( 0.004) |
|
| k (ms−1) | 50.7 ( 7.1) | 32.1 ( 5.6) | 37.0 ( 4.5) | 46.8 ( 4.5) | 43.0 ( 6.5) | ||
| 2-state simulation with donor blinking |
A | 0.116 ( 0.005) |
0.115 ( 0.010) |
0.135 ( 0.007) |
0.104 ( 0.005) |
0.069 ( 0.005) |
|
| k (ms−1) | 38.0 ( 4.4) | 25.7 ( 5.3) | 27.1 ( 3.7) | 31.3 ( 3.6) | 27.9 ( 5.1) | ||
Errors are standard deviations obtained from the diagonal elements of the covariance matrix calculated from the likelihood function (maximum likelihood method) or chi-square function (donor-acceptor cross-correlation calculation).
Using the FRET efficiencies in the first two rows of Table 1, i.e. the maximum likelihood analysis without blinking.
The most likely additional process that would affect the determination of rates is blinking of donor and acceptor dyes. Although we added ascorbic acid and methyl viologen [20], these chemicals could not completely suppress blinking on the sub-10 µs time scale.
When blinking of the acceptor occurs, the anti-correlation of the donor and acceptor intensity from acceptor blinking will increase the relaxation rate and the correlation amplitude and will, consequently, also affect the maximum likelihood analysis. We therefore modified the kinetic model in the likelihood function to include blinking of the acceptor. In this four-state kinetic model (Fig. 5c), each of the folded and unfolded states exists in both “bright” (fluorescing) and “dark” (non-fluorescing) states. The folding and unfolding rate coefficients in the bright state are assumed to be the same as those in the dark state. We also assumed, for simplicity, that the residence time in both bright and dark states is exponentially distributed. For the four-state kinetic model (Fig. 5c), the rate matrix is given by
| (4) |
Here, kb is the rate coefficient for the transition from the dark state to the bright state of the acceptor, which is independent of the illumination intensity. On the other hand, as the probability of the transition from the bright state to the dark state will increase linearly with the time spent in the excited state, the rate coefficient of a transition to the dark state of the acceptor is proportional to the photon count rate as kd = k0 (n/n0), where n is the average photon count rate of a photon trajectory and k0 is the rate coefficient at the reference photon count rate (n = n0). Consequently, kd is different for each photon trajectory. Since the photon count rate of the acceptor is different in the folded state and in the unfolded state, kd should also be different for the folded and the unfolded states. However, these values were set to be equal to avoid breaking the detailed balance condition and to simplify the analysis. The parameters obtained from the four-state model are listed in Table 1. As expected, the relaxation rates extracted were smaller than those of the two-state model because acceptor blinking transitions do not appear as folding transitions, but there is only minor increase in the FRET efficiencies and virtually no changes in the folded fraction.
However, the results show that only 2 – 5% of the time is spent in the dark state. Therefore, the reliability of these blinking parameters is questionable. As listed in Table 1, Bayesian inference criterion (BIC) values for the 4-state kinetic model are smaller than those of the two-state model, which indicates that the 4-state model is more consistent with the data than the 2-state model. We further checked the reliability of the blinking parameters by re-extracting parameters from the photon trajectories simulated using the extracted 4-state model parameters. Instead of generating new photon trajectories, we used a method of recoloring the experimental photon trajectories [15,16]. In this procedure, the color of the experimental trajectories were removed and randomly assigned according to a given kinetics model (see an example recolored trajectory in Fig. S2). The re-extracted parameters from three different recolored data sets are summarized in Table S2 of the Supplementary Material. We tested both 2-state and 4-state likelihood methods as done for the experimental data. In the 4-state method, both folding and blinking parameters were similar to the originally extracted parameters in Table 1. In the 2-state method, the increased apparent folding/unfolding rate coefficients were reproduced as expected. Although the models and extracted parameters are self-consistent, and the re-extracted parameters agree well with the original parameters, the blinking kinetics seem too fast. The average time spent in the dark state is only 1 – 2 µs, several times shorter than the average photon interval of 8 µs (average photon count rate is 125 ms−1). Therefore, the stability of this method in extracting the folding and blinking kinetics may not guarantee the accuracy of the blinking model (i.e. exponential distribution of dark residence times for the acceptor) and the accuracy of the blinking parameters, but it appears to accurately correct the folding parameters.
When the donor blinking happens, no photon is emitted, which results in an intensity fluctuation. Since the sign of the correlation from this fluctuation is positive, it will cancel the early part (< 10 µs) of the anti-correlation due to folding and unfolding. The net result is a longer apparent decay time. To test the influence of donor blinking, we simulated time trajectories of donor blinking, which consists of alternating bright and dark states. The length of the bright and dark states was exponentially distributed with assumed average lifetimes of 30 µs and 3.3 µs, respectively. The lifetime in the dark state and the fraction of molecules in the dark state (10%) were chosen arbitrarily from the assumption that blinking kinetics is faster than the folding kinetics and do not significantly alter the donor-acceptor cross correlation. The blinking trajectory was overlapped with the simulated photon trajectories with a two-state model above and the photons belong to the dark state were deleted. The cross-correlation function in Fig. 6 (third row) and the fitting parameters in Table 2 indeed show the reduced correlation amplitude and the longer relaxation time. On the other hand, the maximum likelihood parameters extracted from the simulated photon trajectories are almost same for the cases with and without donor blinking (Table 2). It seems that donor blinking does not affect the maximum likelihood analysis because the lost information and the inter-photon gap due to donor blinking do not affect the likelihood function. Therefore, the donor blinking can be another reason for the slower relaxation obtained from the correlation analysis than those from the maximum likelihood method.
Since donor blinking does not affect the maximum likelihood analysis, we did not modify the model to incorporate the donor blinking. Instead, to check the reliability of the extracted parameters, we used the recolored photon trajectories above. The recoloring procedure addresses the problem of acceptor blinking as well as the donor blinking in the most exact way because the experimental photon intervals are preserved. The donor-acceptor cross-correlation function (Fig. 6 bottom row) shows similar amplitudes and relaxation rates with those of the experimental data.
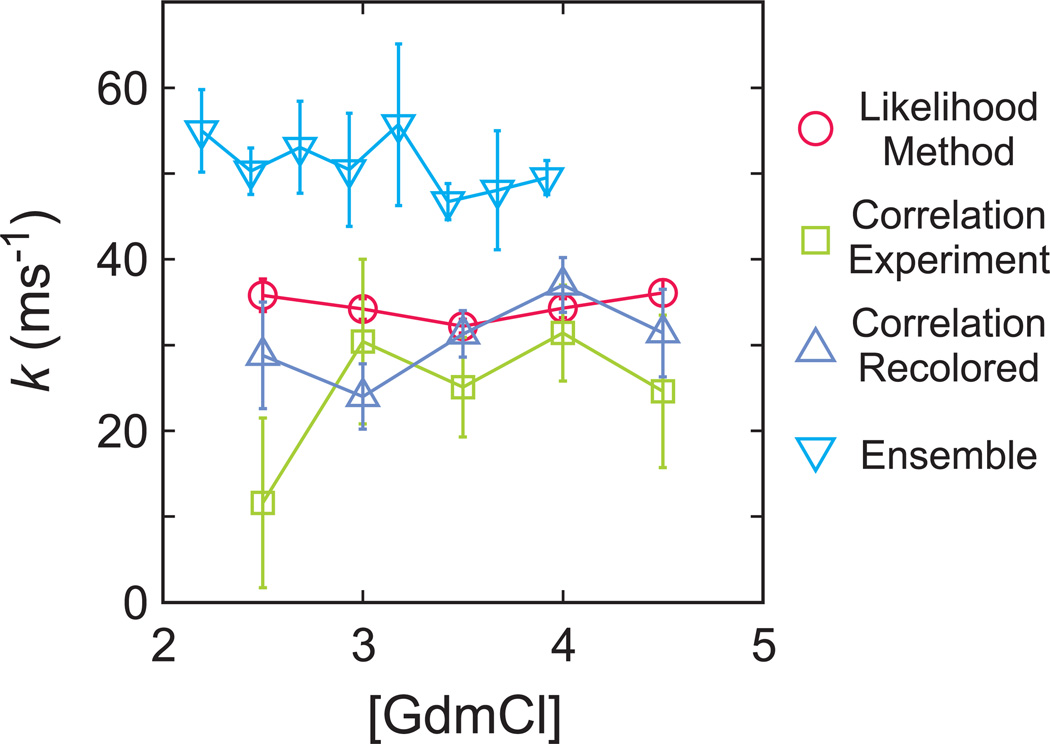
The comparison of the relaxation rates from the 4-state maximum likelihood method and from the donor-acceptor cross correlation of the experimental and recolored data are shown in Fig. 7 and summarized in Table 1. As indicated by the smaller error, as well as the fact that the method is not affected by donor blinking and that acceptor blinking can be incorporated in the model, the parameters obtained from the maximum likelihood method are more reliable than the parameters from the correlation analysis. The FRET efficiency distribution constructed from the recolored trajectories is very similar with the experimental data, providing additional support for the adequacy of the model and accuracy of the model parameters obtained from the maximum likelihood analysis (Fig. 4b).
Fig. 7.
Comparison of relaxation rates obtained from different experimental and analysis methods: maximum likelihood method using 4-state kinetic model (circles), donor-acceptor cross-correlation function (squares), donor-acceptor cross-correlation function for re-colored trajectories using 4-state kinetic model maximum likelihood parameters (triangles), and ensemble temperature-jump method (inverted triangles).
4. Discussion
The main result of this work is that accurate rate coefficients and FRET efficiencies can be obtained from photon trajectories in single molecule FRET experiments for proteins folding on the sub-100 microsecond time scale, even in the presence of dye blinking, by using the maximum likelihood method of Gopich and Szabo [15]. The accuracy of these parameters was confirmed by comparing with the results of a totally independent analysis method – calculation of the donor-acceptor cross-correlation function decay rate and amplitude – and by comparing with the ensemble relaxation rates measured in laser temperature-jump experiments (Fig. 7). The sum of the rate coefficients for this two-state protein corresponds closely to relaxation rates obtained from the decay of the donor-acceptor cross-correlation function, but only after including acceptor blinking in the assumed model for the maximum likelihood analysis (Table 1, Fig. 7). Simulations of photon trajectories with the experimental parameters (Table 2) showed donor blinking can account for most of the difference in the observed amplitudes and those expected from the theoretically-expected values (Eq. 3). Moreover, as an additional check on the maximum likelihood parameters, recoloring of the photon trajectories, using the FRET efficiencies and rate coefficients from the maximum likelihood analysis, did not alter either the amplitude or decay of the donor-acceptor cross-correlation function (Table 1).
A rigorous comparison of the single molecule and ensemble kinetics could not be made because attachment of the dyes does have some, albeit small, effect on the equilibrium and kinetic properties. The fraction unfolded as a function of GdmCl concentration obtained from the maximum likelihood analysis shows that attachment of the dyes slightly stabilizes the protein (Fig. 2b), so differences in relaxation rates are expected. The differences, however, are small, averaging about 30% (Fig. 7). More importantly, like the ensemble measurements, the relaxation rates from the single molecule experiments are independent of the chemical denaturant concentration, in contrast to all other two-state proteins, which have a chevron shape to such a plot. This result is also important for other reasons. For ultrafast folding-proteins it is essential to measure the rate with more than one method, since no single spectroscopic method is capable of monitoring the appearance of the fully folded protein and the disappearance of the unfolded protein in the microsecond time regime. The agreement now among 4 methods - the present single molecule FRET and laser temperature jump, together with the previously reported comparisons using triplet lifetime measurements in photoselection experiments [28] and laser temperature-jump experiments with infrared detection [29] - makes a strong case against the criticism based on limited simulation results that the fluorescence temperature-jump experiments, the most frequently employed method, do not measure overall folding [5,30,31].
As pointed out in the introduction, the long-term goal of single molecule studies on this protein is to obtain information on the transition path time distribution. These distributions have already been obtained by the Shaw group in all atom simulations for both the wild-type [8] and a double mutant of the villin subdomain that folds in 0.7 s [5,8,9,11,32]. An important result of this work is that 4 different force fields, which all predict the correct folding time to within an order of magnitude, yield 3 different pathway distributions as measured by the order in which the 3 helices form [9]. In the past, the rate of folding has frequently been used as a criterion for the validity of the mechanistic information contained in the simulations, but that appears to be an insufficient condition. This is an area where obtaining information on pathway distributions from single molecule experiments could have a major impact.
The results we have reported here represent the first step toward this goal. The next step would be to establish the average transition path time to determine how much the viscosity of the solvent would have to be increased to resolve individual transition paths. We have already accomplished this for an all- protein, the 37-residue FBP WW domain, by introducing a virtual intermediate in a kinetics model for the maximum likelihood analysis [33]. Near the midpoint denaturant concentration, the folding time was found to be ~1 ms and the average transition path time to be ~20 µs at ~10 cP, indicating an average transition path time of ~2 µs. Similar experiments are now underway for the villin subdomain.
Supplementary Material
Highlights.
Photon trajectories were measured for an ultrafast folding protein using single molecule FRET.
Folding rates were obtained from a photon-by-photon analysis using a maximum likelihood method.
Incorporating acceptor blinking into the analysis improved the accuracy of the extracted rates.
The rates agree with the results from both the correlation analysis and ensemble laser T-jump.
Acknowledgement
This paper is dedicated to Robin M. Hochstrasser, who has produced a constant stream of pioneering works that have defined the direction of research in molecular spectroscopy and its applications in biophysics for the past 60 years. We thank Irina Gopich and Attila Szabo for numerous helpful discussions and Annie Aniana for excellent technical assistance. This work was supported by the Intramural Research Program of the NIDDK, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding - a synthesis. Proteins-Struct. Funct. Gen. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.McKnight CJ, Doering DS, Matsudaira PT, Kim PS. A thermostable 35-residue subdomain within villin headpiece. J. Mol. Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 3.Kubelka J, Henry ER, Cellmer T, Hofrichter J, Eaton WA. Chemical, physical, and theoretical kinetics of an ultrafast folding protein. Proc. Natl. Acad. Sci. USA. 2008;105:18655–18662. doi: 10.1073/pnas.0808600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubelka J, Hofrichter J, Eaton WA. The protein folding 'speed limit'. Curr. Opin. Struct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Cellmer T, Buscaglia M, Henry ER, Hofrichter J, Eaton WA. Making connections between ultrafast protein folding kinetics and molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2011;108:6103–6108. doi: 10.1073/pnas.1019552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cellmer T, Henry ER, Kubelka J, Hofrichter J, Eaton WA. Relaxation rate for an ultrafast folding protein is independent of chemical denaturant concentration. J. Am. Chem. Soc. 2007;129:14564–14565. doi: 10.1021/ja0761939. [DOI] [PubMed] [Google Scholar]
- 7.Cellmer T, Henry ER, Hofrichter J, Eaton WA. Measuring internal friction of an ultrafast-folding protein. Proc. Natl. Acad. Sci. USA. 2008;105:18320–18325. doi: 10.1073/pnas.0806154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piana S, Lindorff-Larsen K, Dror RO, Shaw DE. Folding kinetics and thermodynamics from atomistic simulation. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1201811109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 11.Bowman GR, Pande VS. Protein folded states are kinetic hubs. Proc. Natl. Acad. Sci. USA. 2010;107:10890–10895. doi: 10.1073/pnas.1003962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubelka J, Eaton WA, Hofrichter J. Experimental tests of villin subdomain folding simulations. J. Mol. Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 13.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acd. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopich IV, Szabo A. Decoding the pattern of photon colors in single-molecule FRET. J. Phys. Chem. B. 2009;113:10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung HS, Gopich IV, McHale K, Cellmer T, Louis JM, Eaton WA. Extracting rate coefficients from single-molecule photon trajectories and FRET efficiency histograms for a fast-folding protein. J. Phys. Chem. A. 2011;115:3642–3656. doi: 10.1021/jp1009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu TK, Kubelka J, Herbst-Irmer R, Eaton WA, Hofrichter J, Davies DR. High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein. Proc. Natl. Acad. Sci. USA. 2005;102:7517–7522. doi: 10.1073/pnas.0502495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogelsang J, Kasper R, Steinhauer C, Person B, Heilemann M, Sauer M, Tinnefeld P. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew.Chem. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 21.Gopich I, Szabo A. Theory of photon statistics in single-molecule Förster resonance energy transfer. J. Chem. Phys. 2005;122:014707. doi: 10.1063/1.1812746. [DOI] [PubMed] [Google Scholar]
- 22.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 23.Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 24.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nettels D, Hoffmann A, Schuler B. Unfolded protein and peptide dynamics investigated with single-molecule FRET and correlation spectroscopy from picoseconds to seconds. J. Phys. Chem. B. 2008;112:6137–6146. doi: 10.1021/jp076971j. [DOI] [PubMed] [Google Scholar]
- 26.Nettels D, Muller-Spath S, Kuster F, Hofmann H, Haenni D, Ruegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Best RB, Schuler B. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. USA. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buscaglia M, Kubelka J, Eaton WA, Hofrichter J. Determination of ultrafast protein folding rates from loop formation dynamics. J. Mol. Biol. 2005;347:657–664. doi: 10.1016/j.jmb.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 29.Bunagan MR, Gao JM, Kelly JW, Gai F. Probing the folding transition state structure of the villin headpiece subdomain via side chain and backbone mutagenesis. J. Am. Chem. Soc. 2009;131:7470–7476. doi: 10.1021/ja901860f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ensign DL, Kasson PM, Pande VS. Heterogeneity even at the speed limit of folding: Large-scale molecular dynamics study of a fast-folding variant of the villin headpiece. J. Mol. Biol. 2007;374:806–816. doi: 10.1016/j.jmb.2007.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freddolino PL, Schulten K. Common structural transitions in explicit-solvent simulations of villin headpiece folding. Biophys. J. 2009;97:2338–2347. doi: 10.1016/j.bpj.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. Sub-microsecond protein folding. J. Mol. Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 33.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.