Abstract
Many current cancer vaccine strategies suffer from the inability to mount a CD8 T cell response that is strong enough to overcome the low immunogenicity of tumors. Viruses naturally possess the sizes, geometries, and physical properties for which the immune system has evolved to recognize, and mimicking those properties with nanoparticles can produce robust platforms for vaccine design. Using the non-viral E2 core of pyruvate dehydrogenase, we have engineered a viral-mimicking vaccine platform capable of encapsulating dendritic cell (DC)-activating CpG molecules in an acid-releasable manner and displaying MHC I-restricted SIINFEKL peptide epitopes. Encapsulated CpG activated bone marrow-derived DCs at a 25- fold lower concentration in vitro when delivered with the E2 nanoparticle than with unbound CpG alone. Combining CpG and SIINFEKL within a single multifunctional particle induced ~ 3-fold greater SIINFEKL display on MHC I by DCs over unbound peptide. Importantly, combining CpG and SIINFEKL to the E2 nanoparticle for simultaneous temporal and spatial delivery to DCs showed increased and prolonged CD8 T cell activation, relative to free peptide or peptide-bound E2. By co-delivering peptide epitopes and CpG activator in a particle of optimal DC-uptake size, we demonstrate the ability of a non-infectious protein nanoparticle to mimic viral properties and facilitate enhanced DC activation and cross-presentation.
Keywords: biomimetic, virus-like particle, nanoparticle vaccine, protein cage, cross-presentation, CpG, dendritic cell
Although recent years have seen advances in cancer therapies, common treatment strategies (e.g., chemotherapy, radiation therapy, and surgical resection) still rely on techniques that lack specificity and risk side effects, including toxicity.1 Recently, a more targeted approach to cancer therapy has included harnessing the body’s immune system for tumor destruction. While cancer vaccination is a promising strategy, critical barriers to becoming a viable treatment include immune tolerance and the inability to provoke a robust enough immune response to overcome the weak immunogenicity of many cancer antigens.1, 2
In contrast, the natural immune system has evolved to become particularly adept at recognizing key geometries and pathogenic features, most notably those of viruses. Viruses, virus-like particles (VLPs), and other protein nanoparticles have proven to be well-suited as vaccine platforms,3 and examples of their development exist in the clinic (e.g., Gardasil) and many others in clinical trials.3, 4 VLPs typically contain a hollow core and multiple interfaces (i.e., internal, external, and inter-subunit) for engineering functional elements,3, 5–7 allowing fine control over physical properties such as particle stability, surface chemistry, and biological interaction.6, 8–11 However, many viral-based vaccine platforms exhibit strong self-adjuvanting properties that may not always be desired, depending on the preferred immunotherapeutic outcome.3, 12 Additionally, VLPs and attenuated viruses may be difficult to produce and purify in large quantities using common protein expression systems, and therefore alternative platforms should be explored.12
Our group has been developing the structural core of the non-viral E2 subunit of pyruvate dehydrogenase as a protein nanoparticle platform for therapeutic application.8–11, 13, 14 E2 is a caged protein structure exhibiting unusually high thermostability and is comprised of 60 identical self-assembling monomers that produce a hollow dodecahedral capsule ~25 nm in diameter.13, 15–17 This size falls within the narrow size range of 20–45 nm, which is reported to be optimal for passive diffusion to regions of high immune activity (i.e. the lymph nodes) for uptake by the body’s most potent antigen presenting cell, the dendritic cell (DC).7, 18, 19 Because E2 is a non-viral particle, it does not possess any infectious ability or native biological function for entrance into mammalian cells. We have engineered an E2 particle that contains recombinantly introduced internal cysteine residues for packaging of bioactive molecules and cellular delivery.11 Other groups have explored E2 as a platform for inducing helper T cell and humoral responses to HIV in vivo.20, 21 These recent studies, along with our demonstrated ability to modulate immune interaction with E2 in vitro and to deliver therapeutic molecules to cells, has prompted us to explore the redesign of our protein nanoparticle as a viral-mimicking DC-based vaccine platform.9–11
DCs have been identified as the key target for cell mediated immunotherapies because of their antigen processing capabilities and orchestration of downstream adaptive immune responses.22–24 Important for cancer, DCs are particularly efficient at capturing and presenting endogenous antigen via MHC I (i.e., cross-presentation), leading to a strong CD8 T cell effector response.22, 23, 25 Viruses are strong inducers of CD8 T cell immunity, and therefore nanoparticles by virtue of their similar size, have been explored for the delivery of antigens to DCs.24, 26, 27 In addition to the packaging of antigen, nanoparticles may also encapsulate DC-activating molecules (e.g., CpG DNA motifs) to mediate the magnitude and type of immune response.28
Reports have shown that a requisite for a strong anti-tumor response includes simultaneous co-delivery of antigen and activator to the same DC subcellular compartment, as would happen with a natural viral infection.29, 30 Many current strategies employ systemic delivery of antigen together with adjuvant, thereby not likely meeting this criteria, as it places a high constraint on both free antigen and activator arriving in the same DC subcellular compartment simultaneously in vivo. Recent attempts to overcome this barrier have included use of nanoparticles for packaged delivery of vaccine components.3, 31, 32 Furthermore, nanoparticles protect the molecular cargo while also shielding the host from toxic or immune-impairing side-effects, which have been linked to systemic delivery of Toll-like receptor (TLR) agonists in humans.33–35 A nanoparticle vaccine to mimic the natural properties of viruses for a cell-mediated immune response should deliver antigen to DCs, facilitate increased levels of antigen cross-presentation, and provide necessary signals to induce immune activation.
Natural viruses display repeating patterns of antigen, and they also package genetic material. Therefore, we hypothesized that by mimicking the simultaneous packaging and transport of a repeated MHC I-restricted peptide epitope and a DNA-based DC activator (CpG) within the non-viral E2 particle, we can induce DC maturation and antigen cross-presentation to a greater extent than with free CpG or free peptide, respectively. No studies to date have demonstrated the modular "reprogramming" of an empty protein nanoscaffold shell of non-infectious origin for eliciting immune response to the most potent antigen-presenting cell. We report, for the first time, the biomimetic design and characterization of a protein nanoparticle that is functionalized with CpG and peptide epitopes, for which release can be triggered by DC uptake (Figure 1A). The DC-activating properties of acid-releasable, encapsulated CpG are measured, and we also evaluate the CD8 T cell-activating properties of DCs that have been loaded with E2 nanoparticles harboring both CpG and antigen.
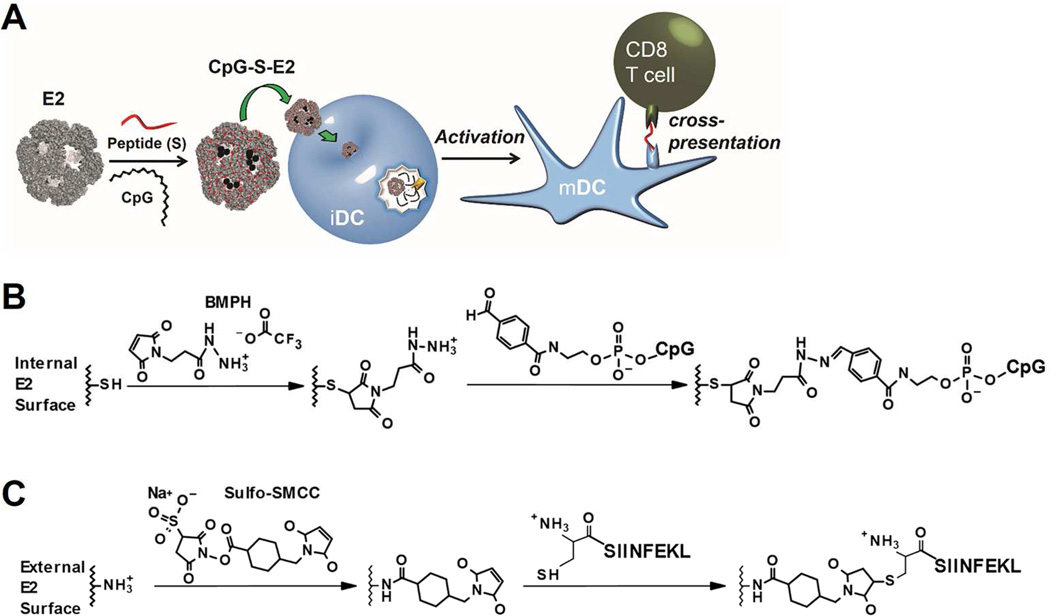
Figure 1.
Schematics of this overall investigation and its chemical conjugation strategies. A) The E2 protein nanoparticle is first covalently combined with CpG activator internally and antigenic peptide (S) externally. The multifunctional particle (CpG-S-E2) is then incubated with immature dendritic cells (iDCs). After entering the acidic endosomal environment, the CpG molecules are released for interaction with Toll-like receptor 9, inducing activation to a mature phenotype (mDC). The co-delivery of CpG and peptide enhance cross-presentation of the associated MHC class I-restricted peptide epitopes to CD8 T cells). B) Conjugation of 5’-aldehyde-terminated CpG to E2’s internal cysteines. C) Conjugation of SIINFEKL peptide to E2’s external lysines.
Results and Discussion
E2 Nanocapsules Can Be Simultaneously Functionalized with CpG Activator and Antigenic Peptide Epitopes
To induce a sufficiently strong CD8 T cell immune response, both antigen and activator molecules should co-localize within the same DC endosomal compartment.30 Therefore, in the design of E2 as a vaccine platform, the combination of antigenic peptides and CpG within the same particle is critical. Our previously characterized E2 containing the functional amino acid mutation D381C (referred to as E2 in this study) was used as the starting scaffold.13 The design of this E2 nanoparticle enabled encapsulation of an endosomal TLR ligand (i.e., CpG for TLR 9) for release in the acidic environment that occurs during DC endocytosis and processing of antigen.36
CpG molecules were successfully and stably encapsulated within core of the E2 nanoparticle. The synthetic CpG molecule was conjugated to the internal E2 cysteine residues (CpG-E2), forming an acid-labile hydrazone bond (Figure 1B). The CpG-E2 lane in the SDS-PAGE gel of Figure 2A revealed two distinct bands, corresponding to an E2 monomer without a CpG conjugate (theoretical molecular weight is 28105 Da for an E2 monomer or 28288 Da for a monomer + cross-linker) and with an attached CpG (theoretical molecular weight of 34879 Da). Dynamic light scattering (DLS) measured a hydrodynamic diameter of 28.0 ± 0.9 nm for CpG-E2 (Supporting Information, Figure S1), which is within the optimal reported DC uptake size range.7, 18, 19
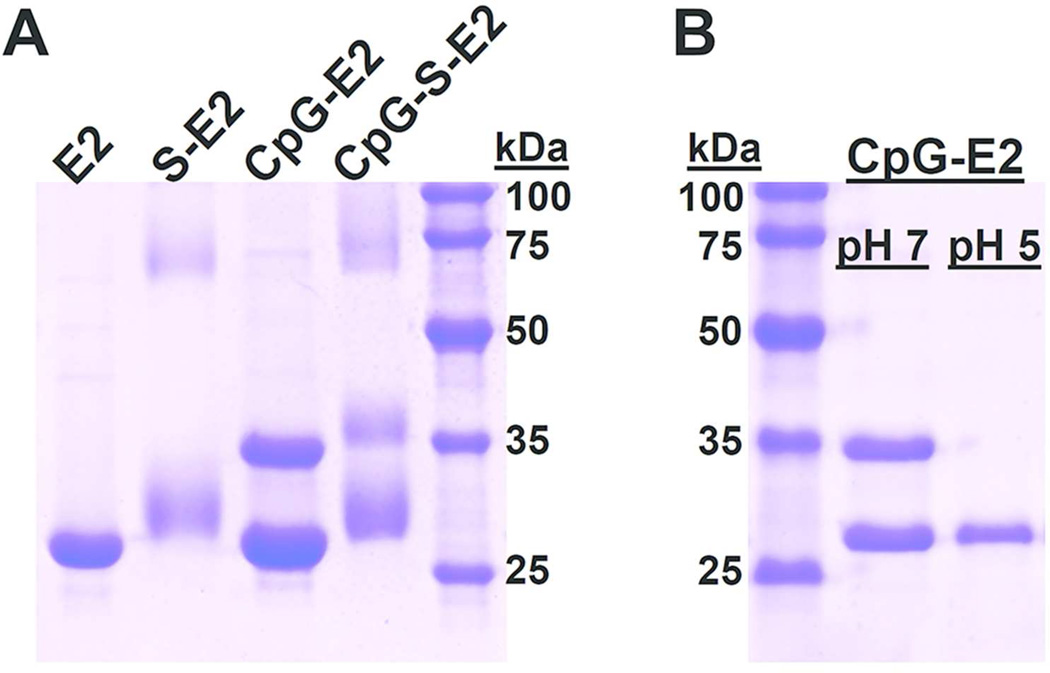
Figure 2.
SDS-PAGE of different E2 protein nanoparticles. A) Functionalization of the E2 nanoparticle (E2) with the SIINFEKL peptide (S-E2), CpG 1826 (CpG-E2), and simultaneous conjugation with CpG and SIINFEKL (CpG-S-E2). The unmodified E2 monomer has a theoretical molecular weight of 28105 Da. The 28–32 kDa band in lane S-E2 supports heterogeneous conjugation of the SIINFEKL peptide to the external E2 lysines, with each peptide adding a mass of 1285 Da to the E2 subunit. The CpG-E2 lane displays two bands, corresponding to an unmodified E2 monomer and a CpG-conjugated monomer (34879 Da). The CpG-S-E2 lane shows bands associated with attachment of both CpG and SIINFEKL peptide, supporting simultaneous conjugation. B) Incubation of CpG-E2 at pH 5 results in near-complete hydrolysis of the CpG activator from the E2 monomer. Incubation at pH 7 retains the CpG-E2 conjugate.
Relatively high amounts of CpG were packaged and contained within the E2 core. The presence of 2 distinct bands in Figure 2A indicates incomplete conjugation to all 60 available internal cysteine residues, likely due to steric limitations within the ~ 12 nm cavity.17 Encapsulation amounts were estimated to be 22 ± 3 CpG molecules per E2 particle, corresponding to ~ 36% of theoretical maximum. The ratio we report here is comparable to those obtained for CpG encapsulation within other VLP systems, which has relied on non-covalent interactions within the viral core, and is even higher than values reported for synthetic nanoparticle systems.34, 37, 38
We observed pH-dependent release of the encapsulated CpG at acidic endosomal conditions. Figure 2B shows that the CpG molecules are fully released from the E2 monomers after 1 hr at pH 5 and 37°C. Our previous studies showed that the E2 particle alone remains intact below pH 5, and therefore the observed CpG release is likely due to hydrazone hydrolysis, rather than protein instability.8, 11 Incubation at 37°C for 1 hr at pH 7.4 did not result in loss of the CpG-conjugates, confirming the stability of the covalent linkage at normal physiologic conditions (Figure 2B). Therefore, we have engineered an E2 particle for pH-responsive encapsulation of CpG activator molecules that should only become available following exposure to an acidic environment (e.g., the endosome). This feature may be important for in vivo application, imparting the potential to protect the host from global immune activation and inflammation, problems of CpG which have been alleviated by nanoparticle delivery.28, 39 While enzymatic degradation is not a major concern with the nuclease-resistant phosphorothioated CpG used in this study, others have shown that porous caged protein complexes can indeed protect the molecular cargo from enzymatic degradation.34, 40
To examine the DC cross-presentation of non-native E2-attached antigen, critical for a CD8 T cell response toward endogenous targets, we conjugated the MHC I-restricted SIINFEKL epitope from the model antigen ovalbumin (OVA) to E2’s external lysines (Figure 1C). The lane in Figure 2A containing the SIINFEKL-conjugated E2 particle (S-E2) shows a broad band in the 28–32 kDa range, consistent with our expected heterogeneous peptide conjugation to the E2 monomer, since crystallographic structure of E2 (PDB code 1B5S) reveals multiple surface lysines as potential conjugation sites.16 The high molecular weight bands of lighter intensities observed for the SIINFEKL-containing constructs (S-E2 and CpG-S-E2) are due to reaction with sulfo-SMCC; these bands are also present in E2 + sulfo-SMCC alone, and suggest a small population of cross-linked E2 subunits. Measurement of peptide conjugation yielded a ratio of 2.9 ± 0.3 peptides per protein monomer, comparable to reported SIINFEKL conjugation with other VLP systems.41 DLS size measurements showed a size of 34.8 ± 4.2 nm (Supporting Information, Figure S1), within the reported optimal size range for vaccine delivery.7, 18, 19
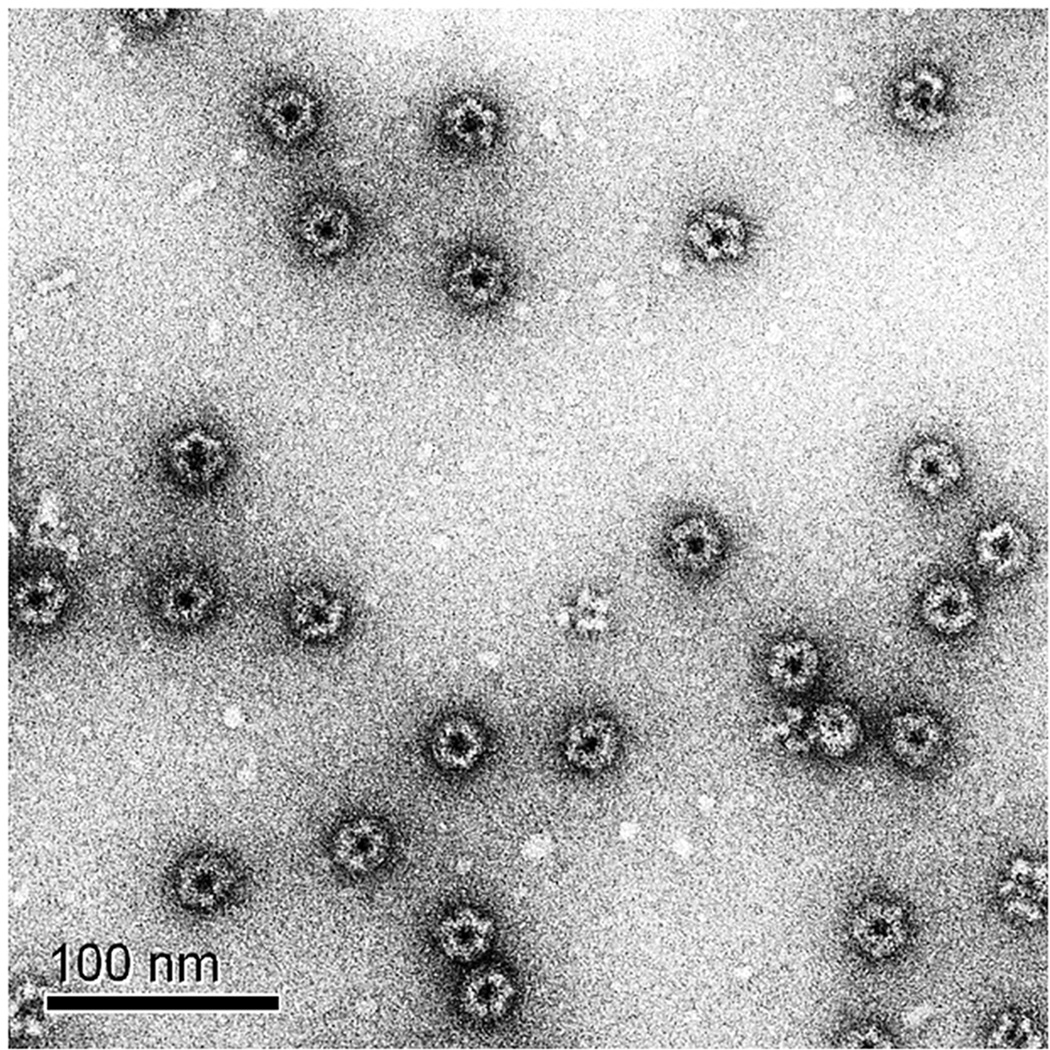
To achieve multiple functionalities, we first encapsulated CpG and subsequently conjugated the SIINFEKL epitope to purified CpG-E2. This multifunctional E2 particle (CpG-S-E2) displayed an average particle diameter of 29.9 ± 1.5 nm (Supporting Information, Figure S1) and SDS-PAGE revealed 2 broad signals, corresponding to E2 monomers (with and without conjugated CpG) with varying peptide conjugation amounts (Figure 2A). Further confirmation of intact particles was obtained with transmission electron microscopy (TEM) (Figure 3), which shows non-aggregated multifunctional particles with a diameter of ~ 30 nm, consistent with DLS data. This demonstrates our ability to combine both antigenic peptides and CpG to a single E2 protein nanocapsule via stable covalent linkages that retain the optimal particle size for DC-based vaccines. Upon mild acidification, CpG is released and available to interact with TLR 9.
Figure 3.
Transmission electron micrograph of 2% uranyl acetate stained CpG-S-E2 confirms monodisperse, intact, non-aggregated nanoparticles with a diameter of ~ 30 nm.
CpG Activation of BMDCs is Enhanced Following E2 Encapsulation
We expected that by combining the CpG activator within a protein nanoparticle of optimal DC uptake size, the concentrations necessary to activate DCs could be decreased, relative to unbound CpG. Delivery of the small CpG oligonucleotides in a 25-nm protein nanoparticle could allow for more efficient shuttling to endosomal compartments where TLR 9 is located. This could also potentially decrease the dose needed in a therapeutic application, while simultaneously shielding interaction of the CpG with systemic components that could degrade the CpG or cause nonspecific immune activation.34
CpG induced greater bone marrow-derived dendritic cell (BMDC) activation following encapsulation within the E2 nanoparticle. BMDCs were incubated with varying amounts of the E2 particle alone, unbound CpG, and the CpG-E2 particle. Flow cytometry was used to measure the fold-change in percent positive cells, relative to immature DCs (iDCs), for the DC activation markers MHC II and CD86. CD11c served as our activation-invariant marker, and LPS served as our positive control for DC activation. The E2 particle alone, at the concentrations tested, did not have any significant effect on the expression levels of CD11c, MHC II or CD86, relative to iDCs (Figure 4, with representative dot plot data set in Supporting Information, Figure S2). Incubation of unbound CpG with BMDCs showed no significant increased expression of activation markers at 0.02 and 0.1 µg/ml and induced only a modest increase in the relative MHC II and CD86 expression levels at 0.5 µg/ml (Figure 4). However, encapsulation of CpG activators within the internal cavity of E2 (CpG-E2) resulted in significant increases in the relative expression levels of MHC II at 0.1 and 0.5 µg/ml CpG and CD86 at 0.02, 0.1, and 0.5 µg/ml CpG, compared to unbound CpG at equivalent concentrations.
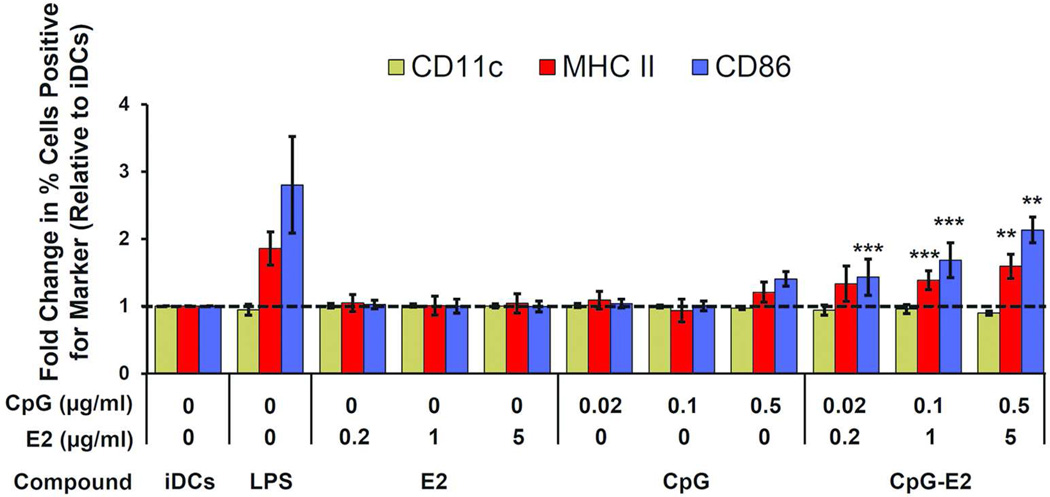
Figure 4.
BMDC activation by CpG is enhanced following encapsulation in the E2 protein nanoparticle (CpG-E2). LPS at 100 ng/ml served as positive control and the activation-invariant marker was CD11c. MFI showed significantly greater DC activation with the CpG-E2 nanoparticle, at 0.1 and 0.5 µg/ml CpG for MHC II and CD86 markers and 0.02 µg/ml for CD86, compared to free CpG at equivalent concentrations. Data is presented as mean ± SD (n ≥ 4 independent experiments). ** p < 0.01, *** p < 0.001, relative to unbound CpG at equivalent concentration for the given marker.
This demonstrates that CpG, at concentrations that do not measurably activate BMDCs in vitro, can do so if combined within the E2 particle, which itself does not cause activation. Significant increases in the activation markers over background can be observed at 25-fold lower concentration of CpG when encapsulated within E2 relative to unbound form, where 0.02 µg/ml E2-encapsulated CpG induces roughly the same amount of increased MHC II and CD86 expression as 0.5 µg/ml unbound CpG. Our observed relative increase in DC-activation is comparable to that reported for nanoparticle studies using alternative activators that also showed significant added therapeutic benefit in vivo.42, 43
One explanation for our observed DC activation increase from CpG in nanoparticle-bound form could be that unbound CpG is below the 20-nm lower reported limit for optimal DC uptake size. This would be consistent with the observations of Wu et al. showing that the aggregation of CpG is necessary for the in vitro activation of DCs.44 Indeed, incubation with fluorescently labeled CpG-E2 (at 0.5 µg/ml CpG) showed greater than 25-fold increase in MFI, relative to unbound fluorescent CpG at an equivalent concentration (Supporting Information, Figure S3). This data supports increased uptake of CpG by BMDC when it is encapsulation within the 25-nm E2 nanoparticle, compared to free CpG. As noted earlier, optimal sizes for vaccine delivery to lymph residing DCs in vivo have been reported to be between 20–45 nm, and the CpG-E2 particle falls within this narrow range.19 Our observations showing specific activation of DCs with a 25-nm CpG-containing E2 protein nanoparticle may be important for vaccine design, since it is mature DCs that display antigen and orchestrate downstream adaptive immune responses.
DCs Can Display E2-bound MHC Class I Epitopes
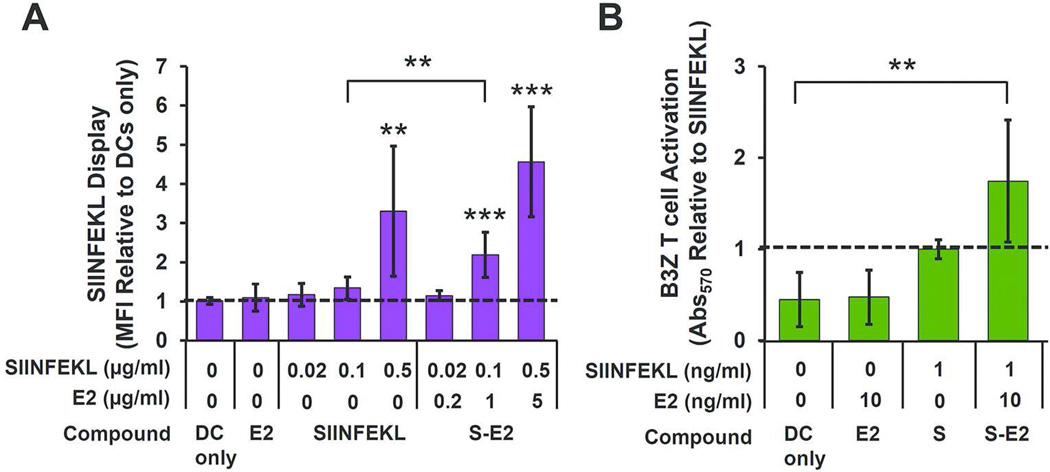
We assessed the ability of DCs to process the OVA257–264 peptide conjugated to E2 and to display this epitope in the context of H-2Kb (MHC I). BMDCs were incubated with the SIINFEKL peptide at varying concentrations in either unbound form or conjugated to E2, followed by antibody staining for MHC I presentation of the epitope. Averaged MFI values are reported as relative to DCs alone (Figure 5A), and representative dot plots are presented in Supporting Information (Figure S4). Unbound SIINFEKL showed a relative MFI increase over background (DC only) of 3.3 ± 1.7 at a concentration of 0.5 µg/ml (p < 0.01). No statistically significant increase in MFI at a concentration of 0.02 or 0.1 µg/ml was observed. At 0.1 and 0.5 µg/ml SIINFEKL delivered as S-E2 to BMDCs, we observed a 2.2 ± 0.6 and 4.6 ± 1.4-fold increase in antibody labeling for SIINFEKL display, respectively, relative to DC only (p < 0.001). No statistically significant display was measured for S-E2 at 0.02 µg/ml SIINFEKL concentration.
Figure 5.
BMDCs process and display E2-bound SIINFEKL (S) epitopes and activate cognate CD8 T cells. A) MFI measurement of BMDCs showed greater SIINFEKL display when incubated with S-E2 compared to free SIINFEKL peptide at 0.1 µg/ml peptide concentration (** p < 0.01). B) The SIINFEKL-bound E2 group (S-E2) in the B3Z assay showed a 1.7 ± 0.7-fold increase in T cell activation, relative to unbound SIINFEKL. The S-E2 group showed significant B3Z activation above background (DC only), whereas unbound SIINFEKL was statistically within background levels. For both panels A & B, data is presented as mean ± SD (n ≥ 4 independent experiments); ** p < 0.01 and *** p < 0.001, relative to DC only.
Our data shows that peptide antigens covalently bound to E2 (S-E2) are able to be processed for display by BMDCs at levels comparable to those of free unbound peptide. In unbound form, peptides can potentially bind surface MHC I markers without needing to be internalized and processed in subcellular compartments. However, in the case of S-E2, the covalent peptide attachment requires intracellular processing. This suggests that the E2 particle is taken up by DC, and the SIINFEKL peptide is cleaved and processed for display on MHC I (i.e., cross-presented). Interestingly, at a SIINFEKL concentration of 0.1 µg/ml there is a significant increase in relative display levels when delivered in E2-bound form (S-E2), relative to unbound form (SIINFEKL, p < 0.01) (Figure 5A). This result shows that conjugating antigen to the E2 protein nanoparticle through stable bonds does not preclude processing and display, and may increase or prolong antigen presentation.
This enhanced presentation effect has been reported before with antigens encapsulated in PLGA nanoparticles and with peptides bound to poly(propylene sulfide) nanoparticles.45, 46 In fact, Hirosue et al. showed that when SIINFEKL peptides were bound to nanoparticles through reducible disulfide bonds rather than more stable linkages, DCs exhibited greater cross-presentation to CD8 T cells.45 In our study, the SIINFEKL peptides are immobilized through succinimidyl thioether bonds, and Baldwin et al. have shown that this type of bond can become labile in physiological reducing environments, such as that of the endosome.47 The kinetics for reduction of disulfide bonds is much faster than that of succinimidyl thioethers, and design of these linkages may be one variable for controlled release of antigen upon cellular uptake.
For a strong cellular mediated immune response against cancer, DCs must display antigen and also engage antigen-specific CD8 T cells. The B3Z hybridoma CD8 T cell, expressing a T cell receptor specific for SIINFEKL in the context of H-2Kb, was incubated overnight with antigen-pulsed BMDC, and SIINFEKL recognition by the CD8 T cell was measured. The results shown in Figure 5B demonstrate that BMDC loaded with the S-E2 nanoparticle have the capability to functionally engage CD8 T cells, a critical event linking innate and adaptive immunity for a cell-mediated effector response. While no statistically significant difference is evident based on averages, the S-E2-pulsed BMDCs exhibited ~ 2-fold greater T cell activation in four out of five separate experiments compared to the free peptide. Therefore, a modest increase in T cell activation may be evident when DCs are incubated with S-E2 rather than unbound peptide, consistent with our observed result of increased antigen display (Figure 5). Taken together, our data shows that the BMDCs can internalize the S-E2 particle, process the covalently bound peptides for display via MHC I, and activate complementary CD8 T cells.
Simultaneously Delivery of Peptide and CpG within E2 Enhances Antigen Display
Since free CpG can enhance cross-presentation in BMDCs,48 and subcellular co-localization of antigen and adjuvant correlates to a more efficacious anti-tumor immune response in vivo,30 we combined the OVA peptide with the CpG activator in a single multifunctional E2 particle (CpG-S-E2). Control studies showed that the conjugation of peptides did not interfere with the DC-activating capacity of the CpG-functionalized nanoparticle (Supporting Information, Figure S5), with no significant difference in relative expression levels of CD11c, MHC II, and CD86 between the CpG-E2 and CpG-S-E2 particles.
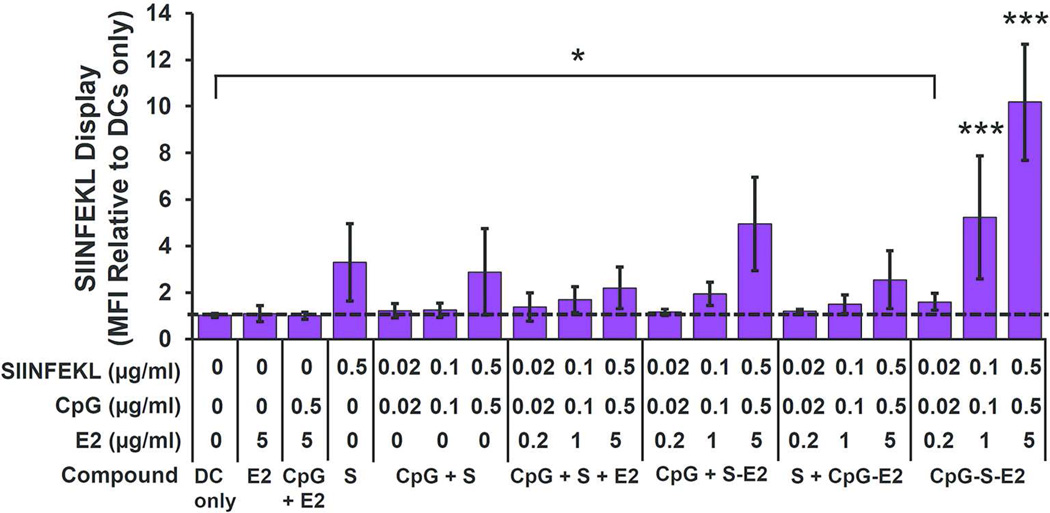
Antigen display experiments showed that the CpG-S-E2 particle induced significantly greater BMDC cross-presentation compared to unbound CpG + S-E2, with ~2.5-fold increase in relative MFI at 0.1 µg/ml and 0.5 µg/ml SIINFEKL. In fact, at SIINFEKL concentrations of 0.1 and 0.5 µg/ml, CpG-S-E2 facilitated significantly greater display than any other SIINFEKL formulation (p < 0.001) (Figure 6; Figure S4 in Supporting Information also shows representative dot plot and histogram overlays). Importantly, while no significant amount of MHC I presentation was detected in BMDCs above background (DC only) at 0.02 µg/ml SIINFEKL for both unbound SIINFEKL peptide and S-E2 (Figure 5A), there is a 1.6 ± 0.4 increase in MFI over background when delivered as CpG-S-E2 (p < 0.05; Figure 6). This shows that combining CpG and peptide within the same E2 particle decreases the amount necessary to induce detectable levels of BMDC antigen display. These results also demonstrate that simultaneous delivery of activator and antigen by use of a protein nanoparticle can significantly enhance the antigen display and cross-presentation capabilities of DCs.
Figure 6.
Simultaneous spatial and temporal delivery of CpG and SIINFEKL (S) with the E2 nanoparticle increases BMDC cross-presentation. MFI showed significantly more SIINFEKL display following simultaneous conjugation of SIINFEKL and CpG to E2 (CpG-S-E2), compared to all other SIINFEKL and CpG delivery formulations at SIINFEKL concentrations of 0.1 and 0.5 µg/ml (*** p < 0.001, for all comparisons at the same SIINFEKL concentration). At a SIINFEKL concentration of 0.02 µg/ml, CpG-S-E2 showed significant MFI above DC-only background (* p < 0.05), whereas MFI of all other SIINFEKL formulations at 0.02 µg/ml SIINFEKL were not statistically different from DC-only background. Data is presented as mean ± SD (n ≥ 4 independent experiments).
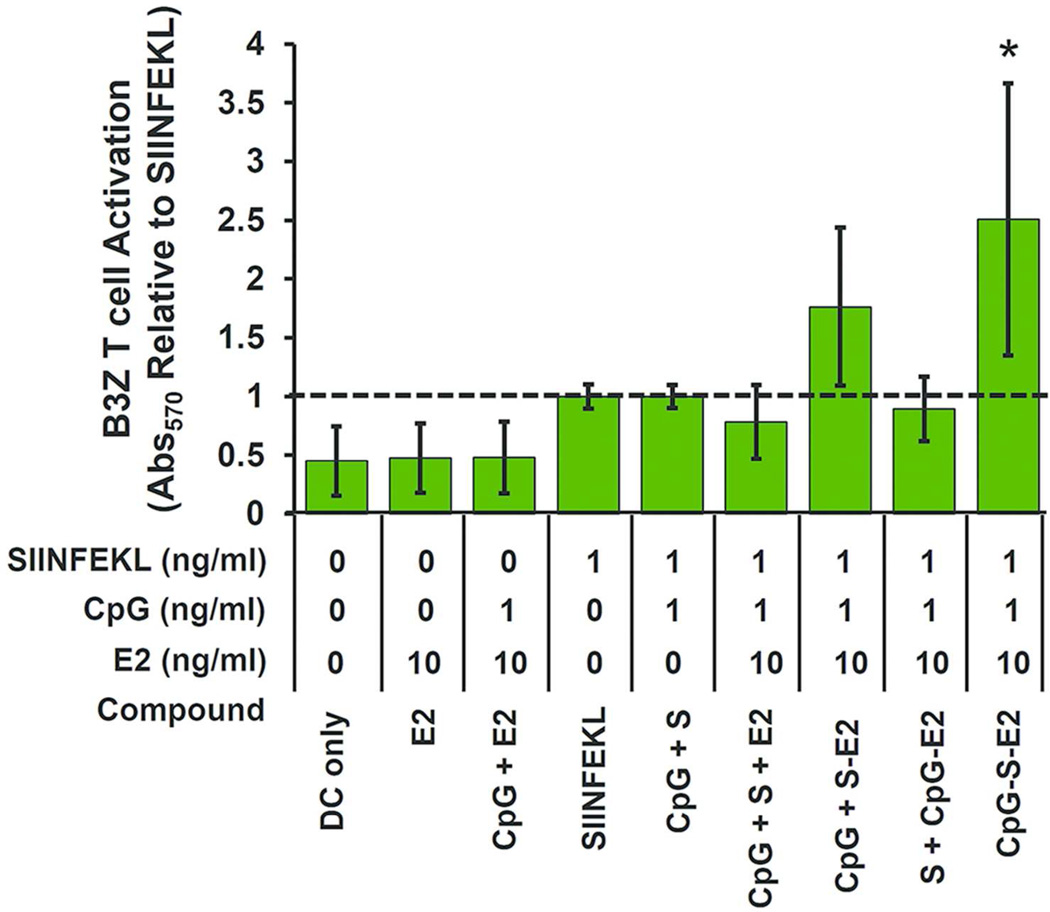
The B3Z assay confirmed these cross-presentation observations, with ~2.5-fold increase in T cell activation at 1 ng/ml peptide concentration delivered as CpG-S-E2, compared to any of the other formulations containing unbound SIINFEKL (p < 0.05, Figure 7). This further confirms our ability to specifically increase antigen display and CD8 T cell activation by simultaneous spatial and temporal dosing of activator and antigen within the E2 nanoparticle. While no statistically significant difference was observed between the CpG-S-E2 or unbound CpG + S-E2 groups within the B3Z assay following 12 hours incubation (Figure 7), T cell activation of the CpG-S-E2 group was higher every time the experiment was performed (n = 5 independent experiments), with an average 1.4-fold increase over CpG + S-E2. Following 48 hour incubation of BMDCs with B3Z T cells, there was indeed a statistically significant increase in T cell activation for the CpG-S-E2 group compared to CpG + S-E2 of ~ 1.5-fold (p < 0.05, Supporting Information, Figure S6). In fact, at 48 hours incubation, CpG-S-E2 was the only SIINFEKL formulation that exhibited statistically greater CD8 T cell activation over DC-only background (p < 0.001). Together, this data shows that simultaneous delivery of CpG and antigen on a nanoparticle (CpG-S-E2) facilitates prolonged antigen display.
Figure 7.
B3Z T cell activation of BMDCs pulsed with different groups. Simultaneous CpG and SIINFEKL conjugation to E2 (CpG-S-E2) showed significantly greater CD8 T cell activation than any other formulation with unbound SIINFEKL (S) peptide after 12 hours incubation. Data is presented as mean ± SD (n ≥ 4 independent experiments for all groups, except n = 3 for groups [S + CpG-E2] and [CpG + S +E2]). Significance was determined by a one-way ANOVA followed by a Bonferroni post-test comparing the set of data for DC-only control and all formulations containing SIINFEKL peptide. * p < 0.05, relative to all formulations containing unbound SIINFEKL.
Peptide epitopes delivered in the E2 nanoparticle enable increased DC-mediated activation of CD8 T cells, and simultaneous delivery of peptide and CpG activator to DCs with E2 further increases and prolongs B3Z activation, compared to unbound peptide and S-E2. While T cell activation differences reported here appear modest, a cancer immunotherapeutic study using free CpG reported relative B3Z activation increases which were similar to our observations using the CpG-S-E2 particle.49 The 2-fold difference reported for the B3Z assay in this previous report translated to animal death or survival, showing that even small relative increases with in vitro CD8 T cell activation can signify noticeable therapeutic immune response differences.
Prior studies with PLGA nanoparticle systems have also shown relative B3Z activation increases comparable to our observations,46 and potent anti-tumor effects following peptide encapsulation.43 These past studies, however, used synthetic nanoparticles that did not covalently associate with the antigen, and thus were subject to loss of the antigen by diffusion over time.43 Therefore, while having similar effects on cross-presentation in vitro, the non-viral E2 nanoparticle has the additional advantage of stable covalent peptide display on the external surface, similar to viruses, ensuring delivery to the BMDCs in a packaged size reported optimal for in vivo DC-based vaccination.7, 19 Our results here show that we can specifically activate BMDCs with the CpG-S-E2 particle, and that associated antigen can be processed for display via MHC I to CD8 T cells at greater levels than with unbound peptides or nanoparticle-conjugated peptides with free CpG.
Conclusions
In this work, we have designed a multifunctional protein nanoparticle platform based on the E2 core scaffold of pyruvate dehydrogenase for immune modulation. This is the first investigation to demonstrate that a non-infectious protein scaffold of non-viral origin can be reengineered to mimic viral properties, giving enhancement of DC response and antigen presentation compared to free DC activator and antigen alone. The nanoparticle structure was designed to allow for simultaneous arrival of both immune-activator and antigen to the DC, which mimics the natural activity of viruses. Upon internalization by DC, release of both DC activator and antigen is specifically triggered by the acidic and reducing environment of the endosome. This controlled release is important because systematic circulation of DC activators may result in inflammation and toxic side effects while weakening the antigen-specific response. CpG DNA motifs were covalently bound internally within the E2 capsule through acid-labile hydrazone linkages that hydrolyze at acidic endosomal conditions. Delivery of CpG to BMDCs in vitro was able to induce cell activation at concentrations lower than required in unbound form, effectively decreasing the dose needed for DC maturation. Peptide epitopes that were bound to the external surface of the E2 nanoparticle were able to be processed by BMDCs in vitro for MHC I display, likely through cross-presentation, and the DCs were able to further functionally engage antigen specific CD8 T cells.
The combination of both CpG and peptide epitopes on a single multifunctional E2 particle increased MHC I display and CD8 T cell activation, relative to unbound forms of the individual components. This shows the ability to enhance cross-presentation of nanoparticle-associated antigens by co-delivering an endosomally restricted TLR 9 ligand. With the use of a model antigen, we demonstrate the potential of engineering a non-viral protein nanoparticle system to induce a CD8 T cell mediated immune response, which is necessary for anti-cancer responses and mimics the activity of viruses, without being infectious. Decreasing the amount of adjuvant molecules needed for DC-activation while increasing antigen cross-presentation may help reduce unwanted side effects or altered immune responses (e.g., tolerance) evident with the systemic administration of these individual components. This provides the groundwork for optimizing the design of an E2 nanoparticle for targeting therapeutically relevant antigenic peptides to the immune system for anti-cancer responses. More broadly, it demonstrates that using biomimetic strategies which emulate viruses, such as size, symmetry, and simultaneous intracellular delivery of antigen and activator to DCs, can be an effective strategy for eliciting immune response.
Methods
Materials
All buffer reagents were purchased from Fisher Scientific, unless otherwise noted. The oligodeoxynucleotide Toll-like receptor 9 ligand CpG 1826 (5’-tccatgacgttcctgacgtt-3’) (CpG) was synthesized with a phosphorothioated backbone and 5’ benzaldehyde modification by TriLink Biotechnologies. The CpG 1826 oligonucleotide with a 5’ Alexa Fluor 488 modification was synthesized by Integrated DNA Technologies. The MHC I immunodominant peptide SIINFEKL (OVA257–264) was synthesized with an N-terminal cysteine by Genscript. All cell culture media was comprised of RPMI 1640 (Mediatech) supplemented with 10% heatinactivated fetal bovine serum (Gibco), 1 mM sodium pyruvate (Hyclone), 2 mM L-glutamine (Lonza), 100 units/ml penicillin (Hyclone), 100 µg/ml streptomycin (Hyclone), 50 µM 2-mercaptoethanol (Fisher), 0.1 mM non-essential amino acids (Lonza) (complete RPMI media). NP-40 and chlorophenol red β–galactoside were from Sigma.
Cell Lines
B3Z, a CD8 T cell hybridoma containing a T cell receptor specific for the MHC I ovalbumin epitope SIINFEKL in the context of H-2Kb, was kindly provided by Prof. Nilabh Shastri (University of California, Berkeley). Cells were maintained in complete RPMI media at less than 7 × 105 cells/ml.50
E2 Nanocapsule Preparation
The D381C E2 protein (E2) was prepared as previously described.13 D381C is an E2 mutant with a non-native cysteine introduced to the internal cavity of the nanoparticle for site-directed functionalization. Briefly, proteins were expressed in E. coli, cells were lysed, and soluble cell lysates were applied to a HiPrep Q Sepharose anion exchange column (GE Healthcare) followed by a Superose 6 (GE Healthcare) size exclusion column for purification. The purified proteins were analyzed by dynamic light scattering (Zetasizer Nano ZS, Malvern) for size measurements. Electrospray ionization mass spectrometry and SDS-PAGE were performed for molecular weight and purity confirmation. Final protein preparations were stored in 50 mM potassium phosphate at pH 7.4 with 100 mM NaCl at 4°C for short-term and −80°C for long-term storage.
Lipopolysaccharide (LPS), a component of gram-negative bacterial cell walls, is recognized by Toll-like receptor 4 expressing immune cells (e.g. DCs), causing potentially unwanted immune activation. Residual LPS was removed following the method described by Aida and Pabst.51 Briefly, Triton X-114 (Sigma) was added to the purified protein at 1% (v/v), chilled to 4°C, vortexed vigorously, and heated to 37°C. The mixture was then centrifuged at 18,000 × g and 37°C for 30 seconds, and the protein-containing aqueous portion was separated from the detergent. This total process repeated ≥ 8 times. Residual Triton was removed with detergent removal spin columns (Pierce). LPS levels were below 8 EU (0.8 ng) per milligram of E2 protein (LAL ToxinSensor gel clot assay, Genscript), significantly lower than levels that activate DCs in our assays.
CpG and Peptide Conjugation
For CpG conjugation (Figure 1B), the cysteines of the E2 internal cavity were first reduced with 10-fold excess of TCEP (Pierce) for 30 minutes followed by incubation with the N-(β-maleimidopropionic acid) hydrazide (BMPH) linker (Pierce) at a 10-fold excess for 2 hr at room temperature (RT). Unreacted linker was removed using Zeba Spin Desalting columns with a 40 kDa cutoff (Pierce). The aldehyde-modified CpG 1826 was added at 5-fold excess over protein monomer, incubated overnight at RT, and excess CpG removed by desalting spin columns. Conjugation was estimated by SDS-PAGE and measured by band intensity analysis with the NIH ImageJ software normalized to protein concentration measured with the BCA assay (Pierce). Conjugation measurements are given as an average number of CpG molecules per E2 particle (n=3). For CpG acid-hydrolysis assays, the conjugated-E2 nanocapsules were dialyzed against 50 mM potassium phosphate with 100 mM NaCl at either pH 7.4 (negative control) or pH 5 using drop dialysis membranes (Millipore) for 60–90 minutes. The nanoparticles were then removed from dialysis, incubated at 37°C for an additional 1 hr, and examined by SDS-PAGE.
For peptide conjugation (Figure 1C), the E2 protein was first incubated with sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC, Pierce) at a 20-fold excess to protein monomer for 30 minutes at room temperature followed by removal of unreacted linker by desalting spin columns. SMCC-functionalized E2 was combined with a 10-fold excess to protein monomer of the CSIINFEKL (TCEP reduced) peptide for 2 hr at RT. Excess peptide was removed by desalting spin columns. Conjugation of the peptide to the E2 protein was assessed by SDS-PAGE, and the number of peptides attached per particle was measured by the difference in free thiol concentration (i.e. unreacted peptides) between a conjugation reaction with and without the SMCC cross-linker (non-specific loss of free thiols over the incubation time). Free thiol concentration was determined using Ellman’s assay (Pierce), following manufacturer’s instructions. Conjugation measurements are given as an average ± standard deviation of CSIINFEKL peptides per E2 particle (n=3).
For particles to which both peptide and CpGs were attached, the reaction schemes for the individual components were carried out as described above. The CpG oligonucleotide was conjugated first, followed by peptide conjugation with extent of conjugation of both components assessed by SDS-PAGE. Transmission electron micrographs of 2% uranyl acetate stained nanoparticles on Cu 150 mesh Formvar-carbon coated grids were obtained on a JEM1200EX (JEOL) with a BioScan600W digital camera (Gatan).
Bone Marrow-Derived Dendritic Cells (BMDCs)
Discarded tissue from C57Bl/6 wild-type mice was kindly provided by the laboratories of Prof. Paolo Casali and Prof. Wendy Liu at the University of California, Irvine. Bone marrow-derived dendritic cells (BMDCs) were prepared following the method described by Lutz et al.52 Briefly, the femurs and tibias were rinsed in 70% ethanol, epiphyses removed, and the marrow flushed. Cells were broken up to a single cell suspension and applied to a 70 µm cell strainer (Fisher). Red blood cells were depleted with ACK lysing buffer (Lonza), followed by washing with PBS. The marrow cells were plated at 2 × 105 cells/ml (10 ml total) on sterile bacteriological Petri dishes (Fisher) in complete RPMI media supplemented with 20 ng/ml murine recombinant GM-CSF (eBioscience) (DC media). Cells were maintained at 37°C and 5% CO2, and 10 ml fresh DC media was added on day 3. On day 6, 50% of the media was replaced, and the non-adherent cells were pelleted and added back to the plates. Loosely and non-adherent cells were collected and used as immature BMDCs on day 8.
BMDC Activation
Immature BMDCs (iDCs) harvested on day 8 were plated at 5 × 105 cells/well in 24-well plates and allowed to settle overnight. The E2 nanocapsule, unbound CpG, CpG-conjugated nanocapsules, or 100 ng/ml LPS (positive control) were added and incubated with cells for 24 hours at 37°C. After collecting DCs by gentle pipetting, surface expression of CD11c, MHC II, and CD86 was assessed by labeling with fluorescently-tagged monoclonal antibodies (eBioscience for FITC-anti-CD11c and FITC-anti-CD86 and Biolegend for PE-anti-MHC II). Cells were analyzed by flow cytometry, collecting 5×104 events per sample, using the Accuri C6 (BD Biosciences). The data is reported as fold-increase in percent positive cells (n ≥ 4 independent experiments), relative to iDCs. Representative gating can be found in Supporting Information Figure S2.
Antigen Display and B3Z Assays
BMDCs harvested on day 8 were plated at 2.5 × 105 per well in 48 well plates and allowed to settle overnight. The E2 nanocapsule, SIINFEKL peptide (with or without unbound CpG), the SIINFEKL-conjugated E2 (S-E2, with or without unbound CpG), or the CpG and SIINFEKL double conjugated E2 (CpG-S-E2) nanocapsule were added for 18 hours. Cell surface display of the SIINFEKL epitope in the context of H-2kb was labeled with PE-tagged monoclonal antibody 25-D1.16 (Biolegend) and measured with flow cytometry (collecting 5 × 104 events per sample). The data is presented as MFI (n ≥ 4 independent experiments) relative to DCs only (non-specific antibody labeling of BMDCs).
For the T cell activation assays, BMDCs harvested on day 8 were plated at 1 × 105 cells/well in a 96-well plate and allowed to settle overnight. The E2 nanocapsule, SIINFEKL peptide (with or without unbound CpG), S-E2 (with or without unbound CpG), or CpG-S-E2 were added to the BMDCs, washed away after 1 hr, and the B3Z CD8 T cells added at 1 × 105 cells/well for an additional 12 or 48 hr. The B3Z cells are activated by the SIINFEKL epitope in the context of H-2kb, which can be measured by lacZ activity.50 Cells were washed with PBS and incubated with 100 µl Z buffer (100 mM 2-mercaptoethanol, 9 mM MgCl2, 0.125% NP-40, and 0.15 mM chlorophenol red β–galactoside) for 4 hours at 37°C. Following incubation, 50 µl of stop buffer (300 mM glycine and 15 mM EDTA in water) was added and absorbance at 570 nm was measured. The data is presented as an average absorbance relative to unbound SIINFEKL peptide (n ≥ 4 independent experiments, unless otherwise noted).
Statistical Analysis
Statistical analyses were carried out using InStat version 3.10. Data is reported as mean ± standard deviation (SD) of at least four independent experiments (unless otherwise noted), with the value of a single independent experiment being the average of at least two replicates for that set. Statistical significance was determined by performing a one-way analysis of variance (ANOVA) followed by a Bonferroni post-test over pairs within the group. P-values less than 0.05 were considered significant.
Supplementary Material
Acknowledgements
We thank the laboratories of Prof. Paolo Casali and Prof. Wendy Liu at UCI for providing excess animal tissue and the laboratory of Prof. Aaron Esser-Kahn for use of their flow cytometer. We thank Prof. Nilabh Shastri at UC Berkeley for providing the B3Z CD8 T cell hybridoma cell line. DLS and mass spectrometry were carried out at the UCI Laser Spectroscopy Facility and the UCI Mass Spectrometry Facility, respectively. We are also grateful to Dr. Sergey Ryazantsev for assistance with obtaining TEM images in the EMi Laboratory at UCLA. This work was supported by NIH (R21 EB010161), in part by the National Cancer Institute of NIH (P30CA062203), and the University of California Cancer Research Coordinating Committee.
Footnotes
Supporting Information Available:
Additional data for DLS of the E2, CpG-E2, S-E2, and CpG-S-E2 particles, representative dot plot flow cytometry data for DC activation and SIINFEKL display, DC uptake of fluorescent CpG and CpG-E2, and relative MFI DC-activation data for the CpG-S-E2 particle. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Aly HA. Cancer Therapy and Vaccination. J. Immunol. Methods. 2012;382:1–23. doi: 10.1016/j.jim.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Klebanoff CA, Acquavella N, Yu ZY, Restifo NP. Therapeutic Cancer Vaccines: Are We There Yet? Immunol. Rev. 2011;239:27–44. doi: 10.1111/j.1600-065X.2010.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plummer EM, Manchester M. Viral Nanoparticles and Virus-Like Particles: Platforms for Contemporary Vaccine Design. Wiley Interdiscip. Rev.: Nanomed. and Nanobiotechnol. 2011;3:174–196. doi: 10.1002/wnan.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kushnir N, Streatfield SJ, Yusibov V. Virus-Like Particles as a Highly Efficient Vaccine Platform: Diversity of Targets and Production Systems and Advances in Clinical Development. Vaccine. 2012;31:58–83. doi: 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pokorski JK, Steinmetz NF. The Art of Engineering Viral Nanoparticles. Mol. Pharmaceutics. 2011;8:29–43. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchida M, Klem MT, Allen M, Suci P, Flenniken M, Gillitzer E, Varpness Z, Liepold LO, Young M, Douglas T. Biological Containers: Protein Cages as Multifunctional Nanoplatforms. Adv. Mater. 2007;19:1025–1042. [Google Scholar]
- 7.Bachmann MF, Jennings GT. Vaccine Delivery: A Matter of Size, Geometry, Kinetics and Molecular Patterns. Nat. Rev. Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 8.Dalmau M, Lim S, Wang SW. Design of a pH-Dependent Molecular Switch in a Caged Protein Platform. Nano Lett. 2009;9:160–166. doi: 10.1021/nl8027069. [DOI] [PubMed] [Google Scholar]
- 9.Molino NM, Bilotkach K, Fraser DA, Ren D, Wang SW. Cell Uptake and Complement Responses Toward Polymer-Functionalized Protein Nanocapsules. Biomacromolecules. 2012;13:974–981. doi: 10.1021/bm300083e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren DM, Dalmau M, Randall A, Shindel MM, Baldi P, Wang SW. Biomimetic Design of Protein Nanomaterials for Hydrophobic Molecular Transport. Adv. Funct. Mater. 2012;22:3170–3180. doi: 10.1002/adfm.201200052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren DM, Kratz F, Wang SW. Protein Nanocapsules Containing Doxorubicin as a pH-Responsive Delivery System. Small. 2011;7:1051–1060. doi: 10.1002/smll.201002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grgacic EV, Anderson DA. Virus-Like Particles: Passport to Immune Recognition. Methods. 2006;40:60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmau M, Lim S, Chen HC, Ruiz C, Wang SW. Thermostability and Molecular Encapsulation Within an Engineered Caged Protein Scaffold. Biotechnol. Bioeng. 2008;101:654–664. doi: 10.1002/bit.21988. [DOI] [PubMed] [Google Scholar]
- 14.Dalmau M, Lim S, Wang SW. pH-Triggered Disassembly in a Caged Protein Complex. Biomacromolecules. 2009;10:3199–3206. doi: 10.1021/bm900674v. [DOI] [PubMed] [Google Scholar]
- 15.Domingo GJ, Chauhan HJ, Lessard IAD, Fuller C, Perham RN. Self-Assembly and Catalytic Activity of the Pyruvate Dehydrogenase Multienzyme Complex from Bacillus stearothermophilus. Eur. J. Biochem. 1999;266:1136–1146. doi: 10.1046/j.1432-1327.1999.00966.x. [DOI] [PubMed] [Google Scholar]
- 16.Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. Principles of Quasi-Equivalence and Euclidean Geometry Govern the Assembly of Cubic and Dodecahedral Cores of Pyruvate Dehydrogenase Complexes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milne JLS, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu XW, Brooks BR, Perham RN, Henderson R, Subramaniam S. Molecular Architecture and Mechanism of an Icosahedral Pyruvate Dehydrogenase Complex: A Multifunctional Catalytic Machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy ST, Rehor A, Schmoekel HG, Hubbell JA, Swartz MA. In Vivo Targeting of Dendritic Cells in Lymph Nodes with Poly(Propylene Sulfide) Nanoparticles. J. Controlled Release. 2006;112:26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Reddy ST, van der Vlies AJ, Simeoni E, O'Neil CP, Swartz MA, Hubbell JA. Exploiting Lymphatic Transport and Complement Activation in Nanoparticle Vaccines. Tissue Eng. Part A. 2008;14:734–735. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 20.Caivano A, Doria-Rose NA, Buelow B, Sartorius R, Trovato M, D'Apice L, Domingo GJ, Sutton WF, Haigwood NL, De Berardinis P. HIV-1 Gag p17 Presented as Virus-Like Particles on the E2 Scaffold from Geobacillus stearothermophilus Induces Sustained Humoral and Cellular Immune Responses in the Absence of IFNγ Production by CD4+ T Cells. Virology. 2010;407:296–305. doi: 10.1016/j.virol.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaworski JP, Krebs SJ, Trovato M, Kovarik DN, Brower Z, Sutton WF, Waagmeester G, Sartorius R, D'Apice L, Caivano A, et al. Co-Immunization with Multimeric Scaffolds and DNA Rapidly Induces Potent Autologous HIV-1 Neutralizing Antibodies and CD8(+) T Cells. PLOS One. 2012;7:e31464. doi: 10.1371/journal.pone.0031464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apetoh L, Locher C, Ghiringhelli F, Kroemer G, Zitvogel L. Harnessing Dendritic Cells in Cancer. Semin. Immunol. 2011;23:42–49. doi: 10.1016/j.smim.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Tacken PJ, de Vries IJ, Torensma R, Figdor CG. Dendritic-Cell Immunotherapy: From Ex Vivo Loading to In Vivo Targeting. Nat. Rev. Immunol. 2007;7:790–802. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]
- 24.Ueno H, Klechevsky E, Schmitt N, Ni L, Flamar AL, Zurawski S, Zurawski G, Palucka K, Banchereau J, Oh S. Targeting Human Dendritic Cell Subsets for Improved Vaccines. Semin. Immunol. 2011;23:21–27. doi: 10.1016/j.smim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joffre OP, Segura E, Savina A, Amigorena S. Cross-Presentation by Dendritic Cells. Nat. Rev. Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 26.Reddy ST, Swartz MA, Hubbell JA. Targeting Dendritic Cells with Biomaterials: Developing the Next Generation of Vaccines. Trends Immunol. 2006;27:573–579. doi: 10.1016/j.it.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Joshi MD, Unger WJ, Storm G, van Kooyk Y, Mastrobattista E. Targeting Tumor Antigens to Dendritic Cells Using Particulate Carriers. J. Controlled Release. 2012;161:25–37. doi: 10.1016/j.jconrel.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Bourquin C, Anz D, Zwiorek K, Lanz AL, Fuchs S, Weigel S, Wurzenberger C, von der Borch P, Golic M, Moder S, et al. Targeting CpG Oligonucleotides to the Lymph Node by Nanoparticles Elicits Efficient Antitumoral Immunity. J. Immunol. 2008;181:2990–2998. doi: 10.4049/jimmunol.181.5.2990. [DOI] [PubMed] [Google Scholar]
- 29.Burgdorf S, Scholz C, Kautz A, Tampe R, Kurts C. Spatial and Mechanistic Separation of Cross-Presentation and Endogenous Antigen Presentation. Nat. Immunol. 2008;9:558–566. doi: 10.1038/ni.1601. [DOI] [PubMed] [Google Scholar]
- 30.Nierkens S, den Brok MH, Sutmuller RPM, Grauer OM, Bennink E, Morgan ME, Figdor CG, Ruers TJM, Adema GJ. In Vivo Colocalization of Antigen and Cytidyl Guanosyl Within Dendritic Cells Is Associated with the Efficacy of Cancer Immunotherapy. Cancer Res. 2008;68:5390–5396. doi: 10.1158/0008-5472.CAN-07-6023. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamachari Y, Geary SM, Lemke CD, Salem AK. Nanoparticle Delivery Systems in Cancer Vaccines. Pharm. Res. 2011;28:215–236. doi: 10.1007/s11095-010-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peek LJ, Middaugh CR, Berkland C. Nanotechnology in Vaccine Delivery. Adv. Drug Delivery Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson NS, Behrens GMN, Lundie RJ, Smith CM, Waithman J, Young L, Forehan SP, Mount A, Steptoe RJ, Shortman KD, et al. Systemic Activation of Dendritic Cells by Toll-Like Receptor Ligands or Malaria Infection Impairs Cross-Presentation and Antiviral Immunity. Nat. Immunol. 2006;7:165–172. doi: 10.1038/ni1300. [DOI] [PubMed] [Google Scholar]
- 34.Storni T, Ruedl C, Schwarz K, Schwendener RA, Renner WA, Bachmann MF. Nonmethylated CG Motifs Packaged into Virus-Like Particles Induce Protective Cytotoxic T Cell Responses in the Absence of Systemic Side Effects. J. Immunol. 2004;172:1777–1785. doi: 10.4049/jimmunol.172.3.1777. [DOI] [PubMed] [Google Scholar]
- 35.Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-Based Immune Adjuvants. Vaccine. 2011;29:3341–3355. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez A, Regnault A, Kleijmeer M, Ricciardi-Castagnoli P, Amigorena S. Selective Transport of Internalized Antigens to the Cytosol for MHC Class I Presentation in Dendritic Cells. Nat. Cell Biol. 1999;1:362–368. doi: 10.1038/14058. [DOI] [PubMed] [Google Scholar]
- 37.Zwiorek K, Bourquin C, Battiany J, Winter G, Endres S, Hartmann G, Coester C. Delivery by Cationic Gelatin Nanoparticles Strongly Increases the Immunostimulatory Effects of CpG Oligonucleotides. Pharm. Res. 2008;25:551–562. doi: 10.1007/s11095-007-9410-5. [DOI] [PubMed] [Google Scholar]
- 38.Roman BS, Irache JM, Gomez S, Tsapis N, Gamazo C, Espuelas MS. Co-Encapsulation of an Antigen and CpG Oligonucleotides into PLGA Microparticles by TROMS Technology. Eur. J. Pharm. Biopharm. 2008;70:98–108. doi: 10.1016/j.ejpb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Tacken PJ, Zeelenberg IS, Cruz LJ, van Hout-Kuijer MA, van de Glind G, Fokkink RG, Lambeck AJA, Figdor CG. Targeted Delivery of TLR Ligands to Human and Mouse Dendritic Cells Strongly Enhances Adjuvanticity. Blood. 2011;118:6836–6844. doi: 10.1182/blood-2011-07-367615. [DOI] [PubMed] [Google Scholar]
- 40.Sioud M, Leirdal M. Design of Nuclease Resistant Protein Kinase Cα DNA Enzymes with Potential Therapeutic Application. J. Mol. Biol. 2000;296:937–947. doi: 10.1006/jmbi.2000.3491. [DOI] [PubMed] [Google Scholar]
- 41.Peacey M, Wilson S, Baird MA, Ward VK. Versatile RHDV Virus-Like Particles: Incorporation of Antigens by Genetic Modification and Chemical Conjugation. Biotechnol. Bioeng. 2007;98:968–977. doi: 10.1002/bit.21518. [DOI] [PubMed] [Google Scholar]
- 42.Molinari P, Crespo MI, Gravisaco MJ, Taboga O, Moron G. Baculovirus Capsid Display Potentiates OVA Cytotoxic and Innate Immune Responses. PLOS One. 2011;6:e24108. doi: 10.1371/journal.pone.0024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Tongchusak S, Mizukami Y, Kang YJ, Ioji T, Touma M, Reinhold B, Keskin DB, Reinherz EL, Sasada T. Induction of Anti-Tumor Cytotoxic T Cell Responses Through PLGA-Nanoparticle Mediated Antigen Delivery. Biomaterials. 2011;32:3666–3678. doi: 10.1016/j.biomaterials.2011.01.067. [DOI] [PubMed] [Google Scholar]
- 44.Wu CCN, Lee JD, Raz E, Corr M, Carson DA. Necessity of Oligonucleotide Aggregation for Toll-Like Receptor 9 Activation. J. Biol. Chem. 2004;279:33071–33078. doi: 10.1074/jbc.M311662200. [DOI] [PubMed] [Google Scholar]
- 45.Hirosue S, Kourtis IC, van der Vlies AJ, Hubbell JA, Swartz MA. Antigen Delivery to Dendritic Cells by Poly(Propylene Sulfide) Nanoparticles with Disulfide Conjugated Peptides: Cross-Presentation and T Cell Activation. Vaccine. 2010;28:7897–906. doi: 10.1016/j.vaccine.2010.09.077. [DOI] [PubMed] [Google Scholar]
- 46.Shen H, Ackerman AL, Cody V, Giodini A, Hinson ER, Cresswell P, Edelson RL, Saltzman WM, Hanlon DJ. Enhanced and Prolonged Cross-Presentation Following Endosomal Escape of Exogenous Antigens Encapsulated in Biodegradable Nanoparticles. Immunology. 2006;117:78–88. doi: 10.1111/j.1365-2567.2005.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baldwin AD, Kiick KL. Tunable Degradation of Maleimide-Thiol Adducts in Reducing Environments. Bioconjugate Chem. 2011;22:1946–1953. doi: 10.1021/bc200148v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datta SK, Redecke V, Prilliman KR, Takabayashi K, Corr M, Tallant T, DiDonato J, Dziarski R, Akira S, Schoenberger SP, et al. A Subset of Toll-Like Receptor Ligands Induces Cross-Presentation by Bone Marrow-Derived Dendritic Cells. J. Immunol. 2003;170:4102–4110. doi: 10.4049/jimmunol.170.8.4102. [DOI] [PubMed] [Google Scholar]
- 49.den Brok MH, Sutmuller RP, Nierkens S, Bennink EJ, Toonen LW, Figdor CG, Ruers TJ, Adema GJ. Synergy Between In Situ Cryoablation and TLR9 Stimulation Results in a Highly Effective In Vivo Dendritic Cell Vaccine. Cancer Res. 2006;66:7285–7292. doi: 10.1158/0008-5472.CAN-06-0206. [DOI] [PubMed] [Google Scholar]
- 50.Sanderson S, Shastri N. LacZ Inducible, Antigen/MHC-Specific T Cell Hybrids. Int. Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 51.Aida Y, Pabst MJ. Removal of Endotoxin from Protein Solutions by Phase Separation Using Triton X-114. J. Immunol. Methods. 1990;132:191–195. doi: 10.1016/0022-1759(90)90029-u. [DOI] [PubMed] [Google Scholar]
- 52.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An Advanced Culture Method for Generating Large Quantities of Highly Pure Dendritic Cells from Mouse Bone Marrow. J. Immunol. Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.