Abstract
Inspired by a multi-resolution community detection (MCD) based network segmentation method, we suggest an automatic method for segmenting fluorescence lifetime (FLT) imaging microscopy (FLIM) images of cells in a first pilot investigation on two selected images. The image processing problem is framed as identifying segments with respective average FLTs against the background in FLIM images. The proposed method segments a FLIM image for a given resolution of the network defined using image pixels as the nodes and similarity between the FLTs of the pixels as the edges. In the resulting segmentation, low network resolution leads to larger segments, and high network resolution leads to smaller segments. Further, using the proposed method, the mean-square error (MSE) in estimating the FLT segments in a FLIM image was found to consistently decrease with increasing resolution of the corresponding network. The MCD method appeared to perform better than a popular spectral clustering based method in performing FLIM image segmentation. At high resolution, the spectral segmentation method introduced noisy segments in its output, and it was unable to achieve a consistent decrease in MSE with increasing resolution.
Keywords: Fluorescence lifetime imaging microscopy, multi-resolution community detection, spectral clustering
1 Introduction
Fluorescence lifetime imaging microscopy (FLIM), a promising technique for imaging molecular processes, generates images using the characteristic fluorescence lifetimes (FLTs) from sub-cellular locations of biological samples (such as cells and thin tissue sections) that are treated with fluorescent contrast agents. The FLT is the average time a molecule resides in the excited state before returning to the ground state through fluorescence emission (Nothdurft et al., 2012). In this work, we propose a multi-resolution community detection (MCD) method based on graph partitioning theory (Fortunato, 2010) to automatically segment FLIM images of cells. MCD (Fortunato, 2010; Girvan & Newman, 2002; Hu et al., 2012; Newman, 2004; Ronhovde & Nussinov, 2009; Ronhovde & Nussinov, 2010) seeks to divide groups of nodes with dense interconnections among them and with more sparse connections with the other nodes in the network. It thus partitions a large physically interacting system into optimally decoupled communities. To explore the performance of the proposed method, we segmented donor FLTs in FLIM images of cells transfected with Förster resonance energy transfer (FRET) protein pairs (Orthaus et al., 2009).
Image segmentation plays a crucial role in medical imaging applications by enhancing the detection of biological structures of interest. Existing medical image segmentation methods (Pham et al., 1998) are typically based on spectral clustering, normalized cuts (Fortunato, 2010; Ng et al., 2002; Perona & Freeman, 1998; Scott & Longuet-Higgins, 1990; Shi & Malik, 2000), and the mixture of Gaussian distributions (MGD) (Dempster et al., 1977). FLIM is a relatively newer technique to the medical imaging community. Existing FLIM image analysis software packages (SPCImage, Becker-Hickl, Germany; SymPhoTime, PicoQuant, Germany; VistaVision, ISS Inc., Champaign, IL) deliver FLIM images and corresponding FLT histograms for data acquired by FLIM systems. The VistaVision software package further provides phasor histograms of FLIM images (Stringaria et al., 2011). Users are able to manually segment pixels corresponding to distinct FLTs in FLIM images based on their locations in the FLT histograms or the phasor histograms. However, to date there is no published method for automatically segmenting FLIM images.
The proposed MCD method performs automatic unsupervised segmentation of FLIM images for a given resolution of the network. The network is defined by image pixels as the nodes and similarity between the FLTs of the pixels as the edges. During this process, the input images are segmented starting from different initial states, and significant segments are determined in the final segmented images using information theoretic correlations. Low network resolution leads to larger segments, and high network resolution leads to smaller segments. The outcome is a segmented image containing distinct average FLTs in each of its segments.
We compared the performance of the proposed method with a popular spectral clustering method developed by Ng et al. in segmenting two selected FLIM images. Using the MCD method, the mean-square error (MSE) in estimating the FLT segments in a FLIM image was found to consistently decrease with increasing resolution of the network. In contrast, the spectral clustering method was unable to achieve a decrease in MSE in segmenting FLIM images with increasing resolution, and this method introduced noisy segments in its output at high resolution.
The study is presented as follows. In Section 2, FLIM imaging and its applications are discussed. In Section 3, the proposed MCD method for FLIM image segmentation is described. In Section 4, the performance of the proposed method in segmenting FLIM images of cells transfected with FRET protein pairs is explored in a first pilot investigation on two selected images, and this performance is compared with that attained using the spectral clustering method developed by Ng et al. We conclude in Section 5.
2 Fluorescence Lifetime Imaging Microscopy
FLIM is typically performed in the frequency or time domains. In the frequency domain, a sinusoidally modulated (0.1–1 GHz) light source illuminates the sample, and FLTs are measured by detecting and analyzing the amplitude and phase shift between the excitation light and fluorescence emission (Gadella et al., 1993). In the time domain, pulsed light illuminates the sample, and the time-course of fluorescence emission is detected and analyzed for FLTs. Imaging systems use either time-gated wide field image intensifiers (Elson et al., 2002) or time-resolved laser scanning point detection (Morgan et al., 1995). FLIM images depict FLTs of fluorophore molecules in each pixel emanating from the sample micro-environment. Applications of FLIM include imaging molecular signalling (Webb et al., 2008) and trafficking (Verveer et al., 2000), imaging the spatial concentration of intracellular ions (Lahn et al., 2011), assessing the intracellular environment (Kneen et al., 1998), characterizing tissue slices in vivo (Ushakov et al., 2011), and determining molecular interactions using FRET (Keese et al., 2010). However, the literature does not describe any automatic method for segmenting FLIM images. This void leaves the users to manually select regions with distinct FLTs in the FLIM images based on FLT histograms or phasor histograms computed by existing FLIM image analysis software packages. Enabling automatic segmentation of FLIM images will beneficially eliminate the manual intervention in analyzing FLIM data using these software packages.
3 Segmentation Using Multi-Resolution Community Detection
3.1 Potts Model Hamiltonian
To segment a FLIM image, we construct a network by using the image pixels as nodes and the absolute FLT difference between two pixels as the edge (weight) between the nodes formed by these pixels. The MCD method segments the nodes of the resulting network by minimizing a Potts model Hamiltonian,
| (1) |
The weight Wij denotes the absolute FLT difference between a pixel pair formed by the ith and jth ({i, j} ∈ {1, 2, …, N}) pixels in the input image with N pixels, and W denotes the background of Wij. The Heaviside function Θ(·) “turns on” or “turns off” the edge designation.
| (2) |
The parameter γ controls the resolution of the estimated segments. With decreasing γ, the minima of Eq. (1) leads to solutions with progressively lower intra-community edge densities, effectively “zooming out” toward larger segments. The Kronecker delta δ(·) is given by
| (3) |
In the above Hamiltonian, by virtue of the δ(σi, σj) term, each spin σi interacts only with other spins in its own segment. The spin σi (∀σi ∈ {1, 2, …, K}) defines the segment identity for the ith (i ∈ {1, 2, …, N}) pixel, and the algorithm optimizes it by minimizing the energy defined by Eq. (1). As such, the resulting model is local—a feature that enables high accuracy along with rapid convergence (Ronhovde & Nussinov, 2010). Thus, minimizing the Hamiltonian of Eq. (1) corresponds to identifying strongly connected segments of pixels.
3.2 Trials and Replicas
We review the notions of “trials” and “replicas” used in our community detection algorithms. Both of these notions pertain to the use of multiple identical copies of the same system which differ from one another by a permutation of the initial site indices. Thus, whenever the time evolution depends on sequentially ordered searches for energy lowering moves (as it does in our greedy algorithm), these copies may generally reach different local solutions. By the use of an ensemble of such identical copies, accurate results are attained, information theoretic correlations are determined (Appendix A) between the candidate solutions, and a detailed picture of the system is inferred from them.
In the definitions of “trials” and “replicas” given below, any given algorithm may be used to minimize the selected cost function. In our particular case, the Hamiltonian of Eq. 1 is minimized.
Trials: We use “trials” in our bare community detection algorithm. The algorithm is evaluated on the same problem T independent times. This may generally lead to different contending states that minimize Eq. 1. Out of these T trials, the lowest energy state is picked, and that state is used as the solution.
Replicas: We use both “trials” and “replicas” in our MCD algorithm. Each sequence of the above described T trials is termed a “replica.” The aforementioned T trials (and picking the solution that attains the lowest energy in the Hamiltonian of Eq. 1) are replicated R independent times. By examining information theoretic correlations between the R replicas, we infer which features of the contending solutions are well agreed on (and thus are likely to be correct), and on which features there is a large variance between the disparate contending solutions that may generally mark important physical boundaries. The information theoretic correlations are computed within the ensemble of R replicas. Specifically, the information theoretic extrema are a function of the resolution parameter, and generally correspond to more pertinent solutions that are locally stable to a continuous change of scale. In this way the important physical scales in the system are detected.
3.3 Community Detection
The community detection (CD) algorithm minimizes Eq. (1) in four steps (Ronhovde & Nussinov, 2010).
-
The pixels are partitioned based on a symmetric or fixedK initialization.
Symmetric initialization is used for unsupervised image segmentation, where each pixel forms its own segment; i.e., initially, there are K(0) = N segments. Here the algorithm does not know the number of segments, so the symmetric initialization provides the advantage of no bias towards a particular segment. The algorithm decides the number of segments K by means of the lowest energy solution. The method described here performs such unsupervised image segmentation.
Fixed K initialization is used in supervised image segmentation, where all pixels are divided into K segments using a random initial distribution. The community membership of an individual pixel is then changed to lower the solution energy, using the CD algorithm. Here the user decides the number of initial segments K based on the desired information. For instance, if only one target needs to be identified, K = 2 is enough to describe the target and background.
Each pixel is then placed in the segment that best lowers the energy of Eq. (1) based on the current state of the system.
This process is repeated for all pixels. The iteration is continued until no energy lowering moves are found after one full cycle through all pixels.
The above three steps are repeated forT trials, and the lowest energy is selected as the best solution. Different trials differ solely by the permuted pixel order of the initial state.
3.4 Multi-Resolution Community Detection
We illustrate below how the multi-resolution CD (MCD) algorithm (Ronhovde & Nussinov, 2009) works.
To begin applying the MCD algorithm, users first specify the number of replicas R at each resolution γ, the number of trials T per replica, and the starting and ending resolutions, γ0 and γf, respectively. See Section 3.2 for the definitions on “trial” and “replica.” We typically use 8 ≤ R ≤ 12 and 2 ≤ T ≤ 20. In the case of a symmetric initialized state of one pixel per community, the initial states of the replicas are generated by permuting the pixel labels. These permutations P simply reorder the pixel indices (1, 2, 3, …, i, …, N) → (P1, P2, …, PN) (with Pi the state of i under a permutation), and thus lead to a different initial state.
The algorithm starts from the initialization of the system, as described in item (1) of Section 3.3.
Eq. (1) is then minimized independently for all replicas at a resolutionγ = γi ∈ {γ0, γ1, …, γf−1, γf }, as described in Section 3.3.
For the entire range of the resolutions studied, the algorithm then calculates the average inter-replica information theoretic measures, such as IN and V, at each value of resolution γ. These measures determine the most pertinent scales of the systems in the following manner. Extrema in the average inter-replica information theoretic overlaps (Ronhovde & Nussinov, 2009) as a function of γ occur at particular set of the values of γ (which we denote here by { }). Community detection done at any one of these extremal values leads to an image segmentation (Hu et al., 2012) that is robust and locally insensitive to a change of resolution (i.e., to small variations in γ about ). These locally stable segmentations generally highlight prominent features of the image on all natural pertinent scales. Different levels of detail and resolutions can be determined by allowing the resolution parameter γ to assume different values when the Hamiltonian of Eq. (1) is minimized following the CD detection of Section 3.3. Higher values of lead to finer segmentations on smaller scales; coarser larger range features appear in the corresponding segmentation when smaller values of are inserted.
4 Results
This section describes selected examples of the proposed MCD method applied to FLIM images of cells transfected with FRET protein pairs (Orthaus et al., 2009) in a first pilot investigation on two images. A performance comparison between the MCD and a popular spectral clustering method developed by Ng et al. in segmenting FLIM images is also discussed.
4.1 Datasets
To explore the performance of the proposed MCD method, we employed two FLIM images of live 12V HC Red cells expressing a protein fused to EGFP (donor) and RFP (acceptor) separated by a short linker. The images were acquired using an Olympus FV1000 laser scanning microscope (LSM) equipped with the PicoQuant LSM Upgrade Kit for FLIM. Such a donor-acceptor fusion serves as a positive control for FRET. To capture images from cells transfected with EGFP-RFP fusion protein pairs, the samples received pulsed excitation at 470 nm with 40 MHz repetition. Photons were detected by a single channel SPAD (PicoQuant, Germany) set-up. A fluorescence bandpass filter (500–540 nm) limited the detection to the donor (EGFP) fluorescence only.
The FLIM image in Fig. 1A shows two cells with different average donor FLTs: a FRET cell and a cell where the acceptor RFP was irreversibly bleached, leading to a FLT shift from approximately 2.1 ns to 2.4 ns. In Fig. 1A, by carefully adjusting the colormap of the image, this shift of the average FLT is clearly distinguished by yellow and red, respectively. In the FRET cell (shown in yellow), half of the EGFP-RFP fusion proteins could adopt a proper conformation because complete maturation allowed for FRET (Orthaus et al., 2009). For the acceptor-bleached cell (shown in red), the situation was different. Only 15% of the EGFP molecules were quenched by energy transfer to some remaining acceptor molecules, whereas the majority of donor molecules (85%) could not undergo FRET anymore because an appropriate acceptor molecule was missing. In the similar image in Fig. 1B, an average donor FLT of 2.2 ns (yellow) was obtained in the cell corresponding to the quenched EGFP, and an average donor FLT of 2.9 ns (red) was found in the other cell, where the acceptor molecules were irreversibly destroyed.
Fig. 1.
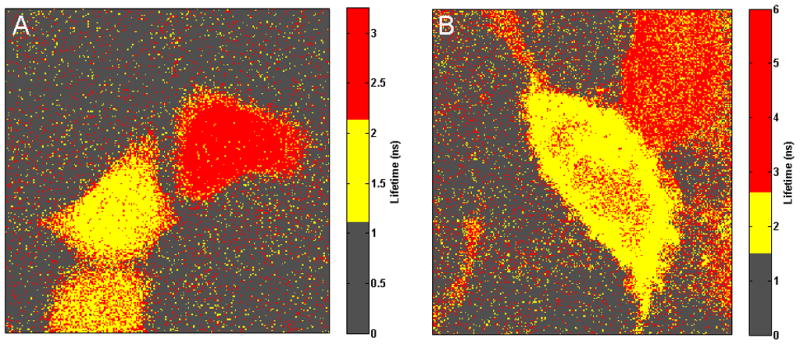
Fluorescence lifetime imaging microscopy images of live cells transfected with EGFP-RFP fusion Förster resonance energy transfer (FRET) protein pairs. False colors represent different segments. Images are depicted by carefully adjusting the respective colormaps. Color boundaries are specified based on the expected fluorescence lifetimes (FLT) in the FRET cell and the cell with no FRET. Ground-truths established based on these images are thus universal, and we used these images for evaluating the performance of the proposed segmentation method. (A–B) For the cells with EGFP-RFP fusion FRET pairs, two adjacent cells show distinct donor fluorescent lifetimes (FLTs). One of the cells is a FRET cell, and the other cell’s acceptor RFP was irreversibly bleached, leading to a FLT shift from the former.
4.2 Multiresolution Community Detection for Varying Resolution
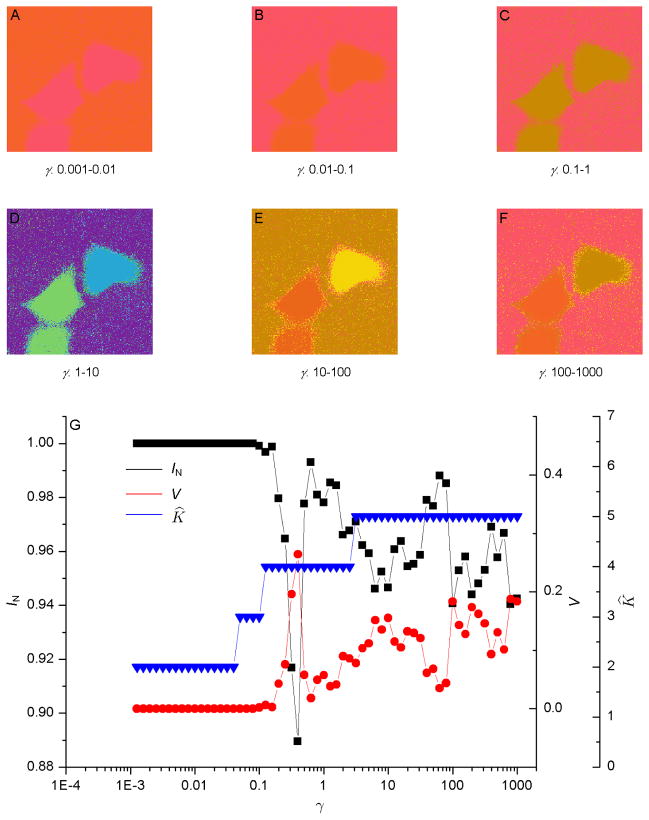
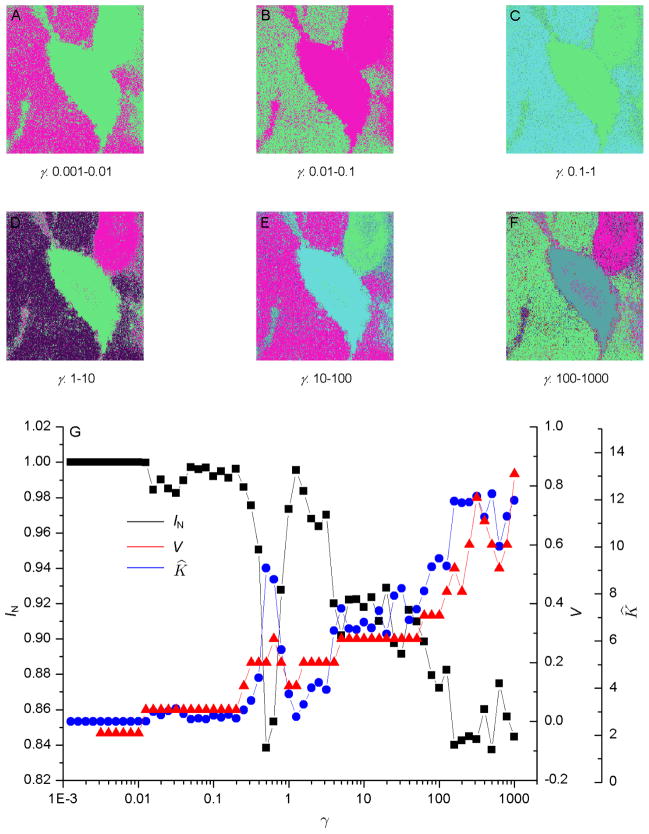
For the FLIM images shown in Fig. 1, we define the edge weight between two pixels as the absolute FLT difference between them. The MCD was applied to segment the resulting networks formed by the image pixels as nodes. Figs. 2G and 3G show plots of the respective information theoretic overlaps between the replicas of the MCD for the FLIM images in Fig. 1A & 1B, such as their normalized mutual information IN and variation of information V, together with the respective number of estimated segments K̂ as a function of the resolution parameter γ. With decreasing γ, the minima of Eq. (1) leads to solutions with progressively lower intra-segment edge densities, effectively “zooming out” toward larger segments. Natural network resolutions correspond to the values of γ for which the replicas exhibit extrema and plateau in the average of their information theoretic overlaps when expressed as a function of γ (Ronhovde & Nussinov, 2009). Recall that the independent solutions of the MCD method attained from different starting points were defined as replicas in Section 3.4.
Fig. 2.
(A–F) Segmentation of the fluorescence lifetime imaging microscopy image shown in Fig. 1A using the multi-resolution community detection (MCD) method for increasing resolution. False colors represent different segments. For resolution parameter γ > 1, two major segments, one representing the Förster resonance energy transfer cell and the other representing the unquenched cell, are clearly visible. (G) Information theoretic overlaps between the replicas of the MCD method, such as their normalized mutual information IN and variation of information V, together with the number of estimated segments K̂ as a function of γ. Natural network resolutions correspond to the values of γ for which the replicas exhibit extrema and plateau in the average of their information theoretic overlaps.
Fig. 3.
(A–F) Segmentation of the fluorescence lifetime imaging microscopy image shown in Fig. 1B using the multi-resolution community detection (MCD) method for increasing resolution. False colors represent different segments. The result obtained was identical to that achieved for Fig. 1. (G) Information theoretic overlaps between the replicas of the MCD method, together with the number of estimated segments K̂, as a function of γ.
Figs. 2A–2F and 3A–3F show the results of automatic image segmentation using our MCD algorithm at different resolutions for the two FLIM images shown in Fig. 1. The segments are depicted using false colors. As the resolution increases from Fig. 2A to 2F and from Fig. 3A to 3F, the images show more detailed segments. In Figs. 2D–2F and 3D–3F, two major segments, one representing the respective FRET cell and the other representing the respective unquenched cell, are clearly visible for γ > 1, in addition to the respective background. Thus, by using different resolutions γ, the MCD method was able to detect the segments at different scales. To generate the segmented images, we used eight replicas and one trial. Parallel implementation of the automatic segmentation in different resolutions will allow users to obtain segments without having any human intervention in the adjustment of the image colormap.
4.3 Spectral Clustering for Varying Resolution
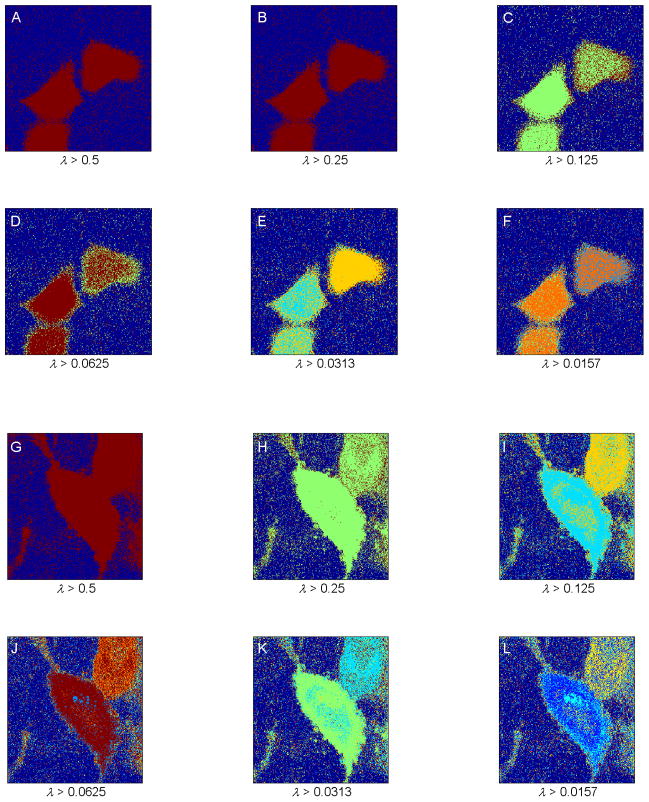
To compare the performance of the proposed MCD method in automatically segmenting FLIM images, we used a popular spectral clustering method developed by Ng et al. In brief, we first constructed a network with image pixels as nodes and took the edge weight between two nodes as the squared distance between the FLTs of the corresponding pixels smoothed by a Gaussian kernel. Then we normalized the affinity matrix formed by the resulting edge weights, and performed eigendecomposition of the resulting matrix. Eigenvectors corresponding to the eigenvalues λ ≥ α were chosen, these eigenvectors were normalized again, and then they were segmented along the rows to segment the network. As in the MCD method, α determines the resolution of the segmented images. Increasing α confines us to eigenvectors of smaller variation across the pixels, and decreasing α allows us to include eigenvectors of larger variation across the pixels. Consequently, increasing α effectively “zooms out” toward larger segments.
Figs. 4A–4F and 4G–4L show the performance of the spectral clustering method in segmenting the FLIM images shown in Fig. 1 for decreasing α. For Fig. 1A, the segmented images shown in Figs. 4A–4F using false colors depict increasing noise at high resolution, and none of these segmented images clearly depicts the two major expected segments. For the FLIM image shown in Fig. 1B, using false colors, the segmented images show the two respective cells as one segment in Figs. 4G and 4H in low resolution and as two expected segments in Figs. 4I and 4J in a slightly higher resolution. At the resolution limit, similar to the case of Fig. 1A, the segmented images for Fig. 1B exhibit a high amount of introduced noise. In comparison to the proposed MCD method, spectral clustering at high resolution thus was not able to provide the expected segments, and moreover introduced a high amount of noise in the segmented images.
Fig. 4.
(A–F) Segmentation of the fluorescence lifetime imaging microscopy (FLIM) image shown in Fig. 1A, using the spectral clustering method developed by Ng et al., 2002 for increasing resolution. False colors represent different segments. Spectral clustering was not able to provide the expected segments, and it introduced a high amount of noise in the segmented images. (G–L) Segmentation of the FLIM image shown in Fig. 1B, using the same spectral segmentation method for increasing resolution. False colors represent different segments. The segmented images show the two cells in a single segment in low resolution (G–H) and in two distinct segments in a slightly higher resolution (I–J). At the resolution limit, similar to the case for the image shown in Fig. 1A, the segmented images display a high amount of introduced noise.
4.4 Performance Comparison between Multi-Resolution Community Detection and Spectral Clustering
To quantitatively compare the MCD and spectral clustering methods, we evaluated mean-square errors (MSEs) in segmenting the major segments of the FLIM images shown in Fig. 1. To perform a fair comparison, the following procedure was used. The ground-truth FLTs of the cells in each FLIM image shown in Figs. 1A and 1B were computed to be the average FLTs in the yellow and red colored regions. The segmented images using the MCD method in Figs. 2A–2C and 3A–3C and those using the spectral clustering method in Figs. 4A–4D, 4F, and 4G–4H depict the two respective expected major segments as one segment. Consequently, the average FLT of this single segment was used as the estimated FLT for both respective cells in each segmented image. The segmented images using the MCD method (in Figs. 2D–2F and 3D–2F) and those using the spectral clustering method (in Fig. 4E and 4I–4L) depict the two respective expected major segments. Consequently, the average FLTs in these segments were used as the estimated FLTs for the two cells in each segmented image. The MSE in segmenting the FLIM images shown in Fig. 1 at each resolution was the squared distance between the estimated FLTs and the ground-truth FLTs of the respective cells.
Using the MCD method, the MSE in the estimation of average FLTs of the correct segments consistently decreases with increasing resolution, as seen in Fig. 5. As seen in Figs. 5–6, the MCD method yielded a lower MSE than the spectral clustering method in all its network resolutions for the FLIM image shown in Fig. 1A, and in its high network resolution region (γ > 10) for the FLIM image shown in Fig. 1B. The MSE in estimating average FLTs of the correct segments using the spectral clustering method does not consistently decrease with increasing resolution for the FLIM image shown in Fig. 1A. For Fig. 1B, the MSE in estimating average FLTs of the correct segments using the spectral clustering method shows first a decrease with increasing resolution. Recall that the resolution of the images segmented by the spectral clustering method is increased by decreasing α. Here, for Fig. 1B, the decrease in MSE in estimating the correct segments is apparent for λ > 0.0625. Decreasing α below 0.0625 introduces noisy segments in the output, and the MSE in estimating average FLTs of the correct segments becomes very high. Consequently, for clarity, such MSEs at limiting resolutions are not shown in Fig. 6B. In summary, in terms of MSE, the proposed MCD method appears to perform better than the spectral clustering method in automatically segmenting the FLIM images shown in Fig. 1.
Fig. 5.
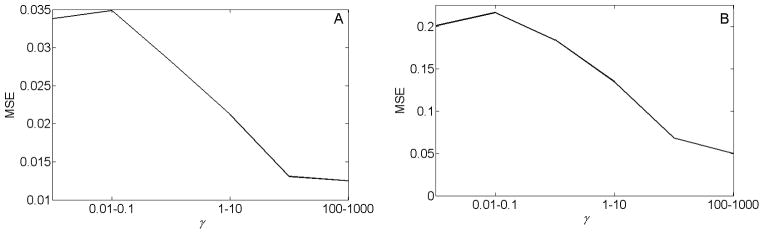
(A–B) The mean-square error (MSE) in estimating average fluorescence lifetimes of the correct segments, using the multi-resolution community detection method for images shown in Fig. 1A and Fig. 1B, respectively. The MSE consistently decreases with increasing resolution.
Fig. 6.
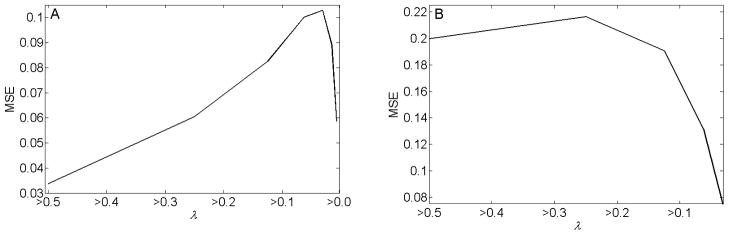
(A–B) The mean-square error (MSE) in estimating average fluorescence lifetimes of the correct segments, using the spectral clustering method developed by Ng et al. for the images shown in Fig. 1A and Fig. 1B, respectively. With the spectral clustering method, the MSE in estimating the average FLTs of the correct segments for the image shown in Fig. 1A does not consistently decrease with increasing resolution. For Fig. 1B, the MSE shows first a decrease with increasing resolution. Note that the resolution of the images segmented by the spectral clustering method is increased by decreasing α, where the eigenvectors corresponding to the eigenvalues λ ≥ α of the respective affinity matrix were chosen for the segmentation. Here the decrease in MSE in estimating the correct segments is apparent for λ > 0.0625. Decreasing α below 0.0625 increases the MSE to a very high value, and these MSE values are not depicted here for clarity.
5 Conclusion
We suggest a multi-resolution community detection (MCD) algorithm to automatically segment fluorescence lifetime imaging microscopy (FLIM) data. The proposed method is able to identify segments at different scales in the input FLIM images. In this study, it appears to perform better than a popular graph-based spectral clustering method developed by Ng et al. in segmenting two selected FLIM images. The MCD method was able to provide correct segments for FLIM images of cells transfected with EGFP-RFP fusion FRET protein pairs in our first pilot investigation on two selected images. The spectral clustering method was unable to provide such correct segments and introduced a high amount of noise in the segmented images, particularly, at high resolution. The MCD method offers lower mean-square errors in segmenting the FLIM images than that obtained using the spectral clustering method.
The MCD method for automatically segmenting FLIM images will avoid any manual selection of regions with distinct FLTs in the FLIM images based on FLT histograms or phasor histograms computed by existing FLIM image analysis software packages. Automatic segmentation of FLIM images will thus minimize error in analyzing FLIM data using such software.
Our study based on two images does not offer enough statistical power to conclude MCD method as a superior method in segmenting FLIM images than other image segmentation methods. Our study can however be considered as a first and promising pilot investigation in understanding how MCD performs in segmenting FLIM images in comparison with other graph based image segmentation methods. Our future work will use a large population of FLIM images of diverse types to establish a statistical significance in this context. On another note, the success of our bare MCD graph theory based method naturally suggests the possibility of yet more potent approaches which build on it. We will briefly propose and speculate on a possible extension involving the use of known prior information, and hope to explore this possibility in future work. We may apply expectation maximization (EM; Dempster et al., 1977) to a given image via a library of finite number of images of known tissue types to infer probabilities that different parts of the image will be locally similar to any of the previously known types. We may then use the similarity of these local probabilities to define weights in the graph and employ MCD. In this approach, the MCD will not invoke bare weights resulting from only local intensity strengths in an image (as we have in the current work). Rather, the MCD will use edge weights given by these probabilities (inferred via EM).
Software
The software package for the “multi-resolution community detection” algorithm (Ronhovde & Nussinov, 2009) that was used in this work is available at http://www.physics.wustl.edu/zohar/communitydetection/.
Acknowledgments
This work is supported by the National Science Foundation (NSF) under grant numbers NSF DMR-1106293, NSF 1066293 (Aspen Center for Physics) and by the National Institute of Health (NIH) under grant numbers NIH R01 EB008111, NIH R01 EB007276, NIH R33 CA123537, NIH U54 CA136398, and NIH HHSN268207000046C. We wish to acknowledge a discussion on EM with Prof. Veit Elser from Cornell University. The images shown in Fig. 1 were taken from the application note written by Orthaus et al., with kind permission from PicoQuant. We appreciate Prof. James Ballard’s (Director, Engineering Communication Center, Washington University in St. Louis) close reading of the manuscript.
Appendix
A Information Theoretic Measures
We use information theoretic measures to calculate correlations between community detection (CD) solutions. The CD method partitions N pixels for a replica r (∀r ∈ {1, 2, …, R}) into Kr segments, where segment k (k ∈ {1, 2, …, Kr}) consists of Nk pixels. The ratio Nk=N is the probability that a randomly selected pixel is found in the segment k (k ∈ {1, 2, …, Kr}).
The Shannon entropy (Hu et al., 2012) is
| (4) |
The mutual information I(r, s) between the replicas r and s ({r, s} ∈ {1, 2, …, R}) is
| (5) |
where Nk1k2 is the number of common pixels in the segment k1 (k1 ∈ {1, 2, …, Kr}) of replica r (r ∈ {1, 2, …, R}) and the segment k2 (k2 ∈ {1, 2, …, Ks}) of replica s (s ∈ {1, 2, …, R}).
The variation of information V (r, s) between the two segments r and s is
| (6) |
which has a range of 0 ≤ V (r, s) ≤ log2 N.
The normalized mutual information IN(r, s) is
| (7) |
with the obvious range of 0 ≤ IN(r, s) ≤ 1.
Higher IN(·) and lower V (·) values indicate better agreement between the compared segments.
References
- Dempster A, Laird N, Rubin D. Maximum likelihood from incomplete data via the expectation maximization algorithm. Journal of the Royal Statistical Society. 1977;39(1):1–38. [Google Scholar]
- Elson DS, Siegel J, Webb SED, et al. Wide-field fluorescence lifetime imaging with optical sectioning and spectral resolution applied to biological samples. Journal of Modern Optics. 2002;49(5–6):985–995. [Google Scholar]
- Fortunato S. Community detection in graphs. Physics Reports. 2010;486(3–5):75–174. [Google Scholar]
- Gadella TWJ, Jovin TM, Clegg RM. Fluorescence lifetime imaging microscopy (FLIM)– spatial-resolution of microstructures on the nanosecond time-scale. Biophysical Chemistry. 1993;48(2):221–239. [Google Scholar]
- Girvan M, Newman MEJ. Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the USA. 2002;99(12):7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Ronhovde P, Nussinov Z. Phase transition in random Potts systems and the community detection problem: Spin-glass type and dynamic perspectives. Philosophical Magazine. 2012;92(4):406–445. [Google Scholar]
- Hu D, Ronhovde P, Nussinov Z. A replica inference approach to unsupervised multiscale image segmentation. Physical Review E. 2012;85(1 Pt 2):016101. doi: 10.1103/PhysRevE.85.016101. [DOI] [PubMed] [Google Scholar]
- Keese M, Yagublu V, Schwenke K, Post S, Bastiaens P. Fluorescence lifetime imaging microscopy of chemotherapy-induced apoptosis resistance in a syngenic mouse tumor model. International Journal of Cancer. 2010;126(1):104–113. doi: 10.1002/ijc.24730. [DOI] [PubMed] [Google Scholar]
- Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intra-cellular pH indicator. Biophysical Journal. 1998;74(3):1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn M, Dosche C, Hille C. Two-photon microscopy and fluorescence lifetime imaging reveal stimulus-induced intracellular Na+ and Cl− changes in cockroach salivary acinar cells. American Journal of Physiology: Cell Physiology. 2011;300(6):C1323–C1336. doi: 10.1152/ajpcell.00320.2010. [DOI] [PubMed] [Google Scholar]
- Morgan CG, Murray JG, Mitchell AC. Photon-correlation system for fluorescence lifetime measurements. Review of Scientific Instruments. 1995;66:3744–3749. [Google Scholar]
- Newman MEJ. Detecting community structure in networks. European Physical Journal B. 2004;38(2):321–330. [Google Scholar]
- Ng A, Jordan M, Weiss Y. On spectral clustering: analysis and an algorithm. In: Dietterich T, Becker S, Ghahramani Z, editors. Advances in Neural Information Processing Systems. Vol. 14. MIT Press; 2002. pp. 849–856. [Google Scholar]
- Nothdurft R, Sarder P, Bloch S, Culver J, Achilefu S. Fluorescence lifetime imaging microscopy using near-infrared contrast agents. Journal of Microscopy. 2012;247(2):202–207. doi: 10.1111/j.1365-2818.2012.03634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthaus S, Buschmann V, Bülter A, et al. Application note by PicoQuant GmbH. Berlin, Germany: 2009. Quantitative in-vivo imaging of molecular distances using FLIM-FRET; pp. 1–7. [Google Scholar]
- Perona P, Freeman WT. In: Burkardt H, Neumann B, editors. A factorization approach to grouping; Proceedings of European Conference on Computer Vision.; 1998. pp. 655–670. [Google Scholar]
- Pham DL, Xu C, Prince JL. A survey of current methods in medical image segmentation. Annual Review of Biomedical Engineering. 1998;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- Ronhovde P, Nussinov Z. Multiresolution community detection for megascale networks by information-based replica correlations. Physical Review E. 2009;80(1 Pt 2):016109. doi: 10.1103/PhysRevE.80.016109. [DOI] [PubMed] [Google Scholar]
- Ronhovde P, Nussinov Z. Local resolution-limit-free Potts model for community detection. Physical Review E. 2010;81(1 Pt 2):046114. doi: 10.1103/PhysRevE.81.046114. [DOI] [PubMed] [Google Scholar]
- Scott GL, Longuet-Higgins HC. Feature grouping by relocalisation of eigenvectors of the proximity matrix. Proceedings of British Machine Vision Conference.; 1990. pp. 103–108. [Google Scholar]
- Shi J, Malik J. Normalized cuts and image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000;22(8):888–905. [Google Scholar]
- Stringaria C, Cinquinb A, Cinquinb O, et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proceedings of the National Academy of Sciences of the USA.; 2011. pp. 13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushakov DS, Caorsi V, Ibanez-Garcia D, et al. Response of rigor cross-bridges to stretch detected by fluorescence lifetime imaging microscopy of myosin essential light chain in skeletal muscle fibers. The Journal of biological chemistry. 2011;286(1):842–850. doi: 10.1074/jbc.M110.149526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer PJ, Wouters FS, Reynolds AR, Bastiaens P. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 2000;290(5496):567–1570. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- Webb SED, Roberts SK, Needham SR, et al. Single-molecule imaging and fluorescence lifetime imaging microscopy show different structures for high- and low-affinity epidermal growth factor receptors in A431 Cells. Biophysical Journal. 2008;94(3):803–819. doi: 10.1529/biophysj.107.112623. [DOI] [PMC free article] [PubMed] [Google Scholar]