Abstract
Microtubule binding protein Tau has been implicated in a wide range of neurodegenerative disorders collectively classified as tauopathies. Exon 10 of the human tau gene, which codes for a microtubule binding repeat region, is alternatively spliced to form Tau protein isoforms containing either four or three microtubule binding repeats, Tau4R and Tau3R, respectively. The levels of different Tau splicing isoforms are fine-tuned by alternative splicing with the ratio of Tau4R/Tau3R maintained approximately at one in adult neurons. Mutations that disrupt tau exon 10 splicing regulation cause an imbalance of different tau splicing isoforms and have been associated with tauopathy. To search for factors interacting with tau pre-messenger RNA (pre-mRNA) and regulating tau exon 10 alternative splicing, we performed a yeast RNA–protein interaction screen and identified polypyrimidine tract binding protein associated splicing factor (PSF) as a candidate tau exon 10 splicing regulator. UV crosslinking experiments show that PSF binds to the stem-loop structure at the 5′ splice site downstream of tau exon 10. This PSF-interacting RNA element is distinct from known PSF binding sites previously identified in other genes. Overexpression of PSF promotes tau exon 10 exclusion, whereas down-regulation of the endogenous PSF facilitates exon 10 inclusion. Immunostaining shows that PSF is expressed in the human brain regions affected by tauopathy. Our data reveal a new player in tau exon 10 alternative splicing regulation and uncover a previously unknown mechanism of PSF in regulating tau pre-mRNA splicing.
Keywords: Tau, Alternative splicing regulation, Tauopathy, RNA stem-loop secondary structure, Polypyrimidine tract binding protein associated splicing factor (PSF)
Introduction
Alternative splicing is one of the most powerful mechanisms for gene regulation and increasing proteomic diversity in metazoans (Black 2000; Matlin et al. 2005; Nilsen and Graveley 2010). The majority of mammalian genes undergo alternative pre-messenger RNA (pre-mRNA) splicing to generate distinct splicing isoforms. Alternative splicing gives rise to isoforms that differ in their biochemical/biophysical properties, subcellular localization, and posttranslational modifications. Numerous genes utilize alternative splicing to achieve spatial and/or temporal regulation of their expression. The fundamental mechanisms regulating alternative splicing consist of complex and dynamic interactions between trans-acting splicing regulators and cis-elements in the intronic or exonic regions of individual pre-mRNAs (Wu et al. 2004; Wu and Postashkin 2009; Nilsen and Graveley 2010). The critical importance of alternative splicing regulation is demonstrated by large numbers of disease-causing mutations that affect alternative pre-mRNA splicing (Krawczak et al. 1992; Wu et al. 2004; Cooper et al. 2009; Ward and Cooper 2010). Splicing mutations can occur either in cis altering sequence elements within the splicing substrate genes or in trans affecting splicing accessory factors (Wang and Cooper 2007; Solis et al. 2008).
Fronto-temporal lobar degeneration (FTLD) refers to a group of neurological disorders that share the common pathological features of frontal and temporal lobe degeneration, accompanied by the presence of abnormal protein inclusions. There are several subgroups of FTLDs, which are classified on the basis of the predominant protein species found in the neuronal inclusion bodies. FTLD-tau is the condition wherein aggregated tau is the primary component of the neuronal inclusions, forming neurofibrillary tangles (Neumann et al. 2009; MacKenzie et al. 2010). The diseases that share this pathological feature are also known as tauopathies. Apart from FTLD-tau, tauopathies include Alzheimer’s disease, Pick’s disease, corticobasal degeneration, and progressive supra-nuclear palsy (Lee et al. 2001; Goedert and Jakes 2005; Mackenzie and Rademakers 2007; Boeve and Hutton 2008; Iqbal et al. 2010; Medeiros et al. 2010). Tau is a neuronally expressed microtubule binding protein that promotes microtubule assembly. It plays an important role in neuronal integrity and axonal transport by regulating microtubule stability and dynamics (Mercken et al. 1995; Panda et al. 1995; Iqbal et al. 2009; Morfini et al. 2009). Genetic studies have identified more than 30 mutations in the human tau gene among FTLD-tau patients (Hutton et al. 1998; Spillantini et al. 1998; D’Souza and Schellenberg 2000; Grover et al. 1999; Jiang et al. 2000; Pickering-Brown et al. 2000; Spillantini et al. 2000; Goedert and Spillantini 2001; Pickering-Brown et al. 2002; Jiang et al. 2003; Andreadis 2005, 2006; Goedert and Jakes 2005; Kar et al. 2005; Liu and Gong 2008; Wang et al. 2004). In addition to the mutations that alter the peptide sequence of Tau protein, many mutations have been found that affect alternative splicing of tau pre-mRNA, including both intronic and exonic ones (Lee et al. 2001; Goedert and Jakes 2005; Liu and Gong 2008; Colombo et al. 2009; Wolfe 2009).
The human tau gene contains 16 exons, of which exons 2, 3, and 10 are alternatively spliced to generate six isoforms, three containing exon 10 and three lacking exon 10 (Neve et al. 1986; Goedert et al. 1989a; Himmler 1989; Andreadis et al. 1992; Montejo de Garcini et al. 1992; Collet et al. 1997; Wei and Andreadis 1998; Andreadis 2005; Kar et al. 2005). Tau exons 9, 10, 11, and 12 encode four tandem microtubule binding repeat sequences (Goedert et al. 1989b; Goedert and Jakes 1990; Andreadis et al. 1992). Alternative splicing of exon 10 gives rise to two classes of tau isoforms: Tau4R (containing exon 10), which contains four microtubule binding repeats, and Tau3R (lacking exon 10), which contains three microtubule binding repeats. Tau4R and Tau3R isoforms differ in their binding affinity to microtubules (Lee et al. 1989; Goedert and Jakes 1990; Goode et al. 2000; Utton et al. 2002; Tomoo et al. 2005; Konzack et al. 2007; LeBoeuf et al. 2008). In the adult human brain, alternative splicing of tau exon 10 is tightly regulated to maintain the ratio of Tau4R to Tau3R at approximately one, whereas tau is expressed as almost exclusively Tau3R isoform in the fetal brain (Neve et al. 1986; Lichtenberg-Kraag and Mandelkow 1990; Andreadis et al. 1992; Gao et al. 2000). Disruption of this delicate balance in the Tau4R/ Tau3R ratio is associated with tauopathies (Hutton et al. 1998; Hasegawa et al. 1998; Grover et al. 1999; Hasegawa et al. 1999; Spillantini et al. 2000; Connell et al. 2001; Cairns et al. 2007; Mackenzie et al. 2010). Studies over the last decade have revealed both cis-acting elements and transacting regulatory proteins that regulate tau exon 10 alternative splicing (Hartmann et al. 2001; Broderick et al. 2004; Kondo et al. 2004; Glatz et al. 2006; Kar et al. 2006; Wu et al. 2006; Dawson et al. 2007; Gao et al. 2007; Zhou et al. 2008; Wang et al. 2010). The RNA sequence at the exon 10–intron 10 boundary of tau pre-mRNA is predicted to form a stem-loop structure with 6 bp stem and six nucleotide loop (Hutton et al. 1998; Grover et al. 1999; Varani et al. 1999; Jiang et al. 2000; Varani et al. 2000; Kar et al. 2005; Donahue et al. 2006). Previous studies show that this stem-loop structure may influence the binding of U1snRNP (U1 small nuclear ribonucleoprotein) to the 5′ splice site (5′ss) of exon 10 (Varani et al. 1999; Jiang et al. 2000; Varani et al. 2000; Kalbfuss et al. 2001; Jiang et al. 2003; Donahue et al. 2006). A number of FTLD-tau-associated mutations are clustered in this exon 10–intron 10 boundary region. Interestingly, most of the mutations in the stem-loop region are associated with increased formation of Tau4R isoforms, suggesting that these mutations lead to inclusion of exon 10 by disrupting the stem-loop structure. However, splicing factors that interact with this region and regulate tau exon 10 alternative splicing have not been fully characterized.
In this study, we carried out a yeast RNA–protein interaction screen and identified PSF as one of factors interacting with the tau pre-mRNA in this stem-loop region. Using molecular and biochemical assays, we characterized the interaction between PSF and tau pre-mRNA and examined the role of PSF in regulation of tau exon 10 splicing. Our experiments demonstrate that PSF interacts with the stem-loop structure and acts to promote tau exon 10 skipping. Finally, the expression of PSF in the human brain is consistent with its involvement in tauopathy.
Materials and Methods
Yeast Three-Hybrid Screening for RNA–Protein Interaction
We used the RNA–protein Hybrid Hunter system (Invitrogen) to screen for proteins interacting with the tau exon 10 5′ splice site stem-loop structure. The bait plasmid was constructed by fusing the 5′ splice site stem-loop region of 96 nucleotides to the MS2 binding site in the pRH5′ plasmid. The bait plasmid was transformed into L40uraMS2 yeast strain that carried the reporters HIS3 and LacZ, downstream of four or eight LexA binding sites, respectively. L40uraMS2 yeast strain carries the gene encoding LexA DNA binding domain–MS2 coat protein fusion. We confirmed that the bait we constructed did not activate reporter genes by itself in the L40 strain. The bait expressing yeast strain was transformed with a human fetal brain complementary DNA (cDNA) library that expresses cDNAs as proteins fused to B42 transcription activation domain. cDNA clones that activated the reporter genes were isolated and sequenced.
Plasmids
The wild-type and DDPAC tau Ex9–11 and Ex10–11 minigene constructs have been described previously (Jiang et al. 2000, 2003). Mammalian expression plasmid for PSF-GFP and bacterial expression construct for PSF-GST were a kind gift of Dr. James Patton, Vanderbilt University.
Transfection and RT-PCR Assay
HEK293 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (Gemini) and 1% penicillin-streptomycin (Gibco). For overexpression experiments, 2×105 cells/well were seeded onto six-well plates 24 h before transfection. Cells were transfected with 1 μg wild-type or DDPAC minigene and 3 μg expression vector using a modified calcium phosphate precipitation method (Jiang et al. 1998, PNAS). For small-interfering (siRNA) experiments, 250 pmol of either control or PSF specific siRNA (Ambion) were transfected using Lipofectamine 2000 following the manufacturer’s instructions. For RT-PCR, cells were harvested 48 h after transfection, and RNA was extracted using the RNAspin Mini RNA isolation kit (GE). After RT-PCR amplification, cDNAwas used for PCR using tau-specific primers (Jiang et al. 1998) in the presence of [α-32P]dCTP. The levels of Bcl-xl/xs, GAPDH, and β-actin were detected using corresponding specific primers. PCR products were separated on a 6% polyacrylamide gel, and the bands were quantified using a BAS 5000 Phosphor-imager (Fuji).
Antibodies and Western Blots
For detecting PSF expression, HEK293 cells expressing the corresponding mini-genes with PSF protein or control GFP were lysed in lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM DTT, 1× protease inhibitor (Roche)]. Equal amount of cell lysates were run on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), then transferred onto 0.22 μm Nitrocellulose membrane (Biorad) using a semi-dry transfer apparatus (Biorad). Membranes were blocked with 10% non-fat dry milk in PBST (phosphate-buffered saline, pH 7.4, 0.1% Tween-20), incubated overnight with corresponding primary antibody diluted in PBST containing 3% bovine serum albumin, washed, incubated with appropriate secondary antibody, and developed with ECL Plus Reagent (GE-Amersham). Antibodies used are as follows: anti-PSF (B92, Sigma), anti-GFP (B-34, Covance), and anti-beta actin (C-11, Santa Cruz).
RNA–Protein Interaction Assay
Uniformly labeled tau wild-type or DDPAC pre-mRNA transcripts were prepared by in vitro transcription in presence of [α-32P]UTP using T7 polymerase from linearized DNA templates as described previously (Jiang et al. 2000). For preparing radioactively labeled oligomer, a labeling reaction was done with [γ-32P] ATP by PNK. The RNA probes were used for UV crosslinking using cell lysates. Briefly, 50 μl HEK293 cell lysates overexpressing PSF or control protein were incubated with ~20 fmol of RNA probe for 2 h on ice. The samples were irradiated on ice under a 254-nm UV light for 10 min using a Stratalinker 2000 UV crosslinker (Stratagene) and subjected to RNase A (5 mg/ml, Sigma) digestion at 30°C for 30 min. End-labeled oligomer transcripts were not subjected to RNase digestion. For immunoprecipitation, anti-GFP antibody bound to protein A/G beads were added to the samples and incubated at 4°C for 2 h. Following precipitation, the beads were washed with cold buffer (20 mM Tris, 150 mM NaCl, 0.1% NP-40, pH 7.4), the RNA bound proteins eluted in sample buffer, and resolved on SDS-PAGE followed by autoradiography.
Oligonucleotide Directed RNase H Cleavage Assay
[α-32P]UTP-labeled wild-type and mutant RNA transcripts were incubated at 37°C for 60 min in standard splicing buffer in the presence of 0.5 U of RNase H (USB) in 25-μl reaction mixtures with 200 fmol of an oligonucleotide (5′-GAAGGTACTCACACTGCC-3′) complementary to the exon 10 splice donor site. The reaction also contained either purified PSF or GST protein (for details, see Jiang et al. 2000). The recombinant PSF-GST protein was purified from Escherichia coli using standard purification protocol (Patton et al. 1993). Cleaved RNA products were then separated on a 6% polyacrylamide–8 M urea gel. The cleavage ratios were measured with a PhosphorImager (Fuji).
Immunostaining
Human brain samples from autopsy material were fixed in formalin and paraffin-embedded. Six-micrometre-thick sections from the frontal and hippocampal regions were used for immunohistochemical staining. The Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) staining kit for mouse monoclonal antibody was used to perform immunostaining with the mouse anti-PSF antibody (B 92, Sigma-Aldrich). Antigen retrieval was carried out in the presence of 1 mM EDTA, pH 9.0. Primary antibody was used at a 1:1,000 dilution. Immunostaining was carried out following the instruction manual, color development was performed with 3,3′-diaminobenzidine, and counterstaining was with hematoxylene.
Results
Identification of PSF as a Protein Binding to Tau Pre-mRNA in a Yeast RNA–Protein Interaction Screen
To understand molecular mechanisms regulating alternative splicing of tau exon 10, we employed a yeast RNA–protein interaction screen approach to search for proteins interacting with tau pre-mRNA in the region flanking the 5′ splice site, where a number of FTLD-tau associated mutations are located. A 96-nucleotide RNA corresponding to the last 40 nucleotides of tau exon 10 followed by 56 nucleotide of the downstream intronic sequence was used as bait. This tau RNA fragment was fused with the RNA fragment containing phage MS2 binding site to make the plasmid expressing MS2-tau-pre-mRNA bait. The bait tau RNA expressing plasmid was transformed into yeast strain L40MS2 expressing transcription activator protein containing LexA DNA binding domain–MS2 coat protein. We confirmed that expression of MS2-tau-pre-mRNA bait by itself does not activate reporter genes. A human fetal brain cDNA library expressing cDNAs as proteins fused to B42 transcription activation domain was transformed into the yeast expressing MS2-tau-pre-mRNA bait together with reporter genes His3 and LacZ under the control of the promoter containing LexA binding sites (Fig. 1). A number of cDNA clones were isolated encoding proteins that activated the reporter genes in a manner dependent on the expression of the tau pre-mRNA. In this study, we focus on one of the candidate tau pre-mRNA interacting proteins identified in this screen (PSF). The remaining cDNA clones are being further characterized, and their detailed characterization will be described in separate reports.
Fig. 1.
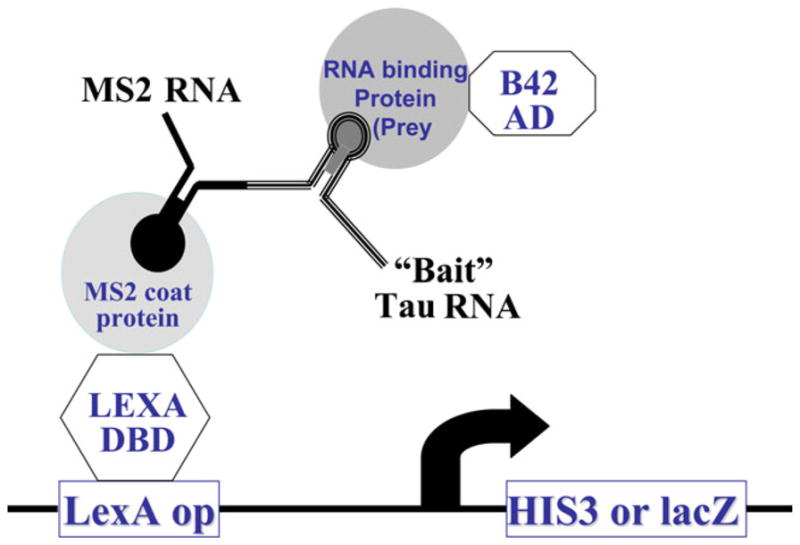
Schematic representation of the yeast RNA–protein interaction screen. As described in “Materials and Methods,” the bait plasmid was constructed by fusing the 5′ splice site stem-loop region of exon 10 nucleotides to the MS2 binding site in the pRH5′ plasmid. The bait plasmid was transformed into L40uraMS2 yeast strain that carried the reporters HIS3 and LacZ and the LexA DNA binding domain-MS2 coat protein fusion. The cDNA library consists of proteins fused to the B42 transcription activation domain. When there is an interaction between RNA binding protein and the bait RNA, transcription is activated, then the LacZ and His3 reporters are expressed
PSF Binds to Tau Pre-mRNA at the Stem-Loop Region at the Exon 10–Intron 10 Junction
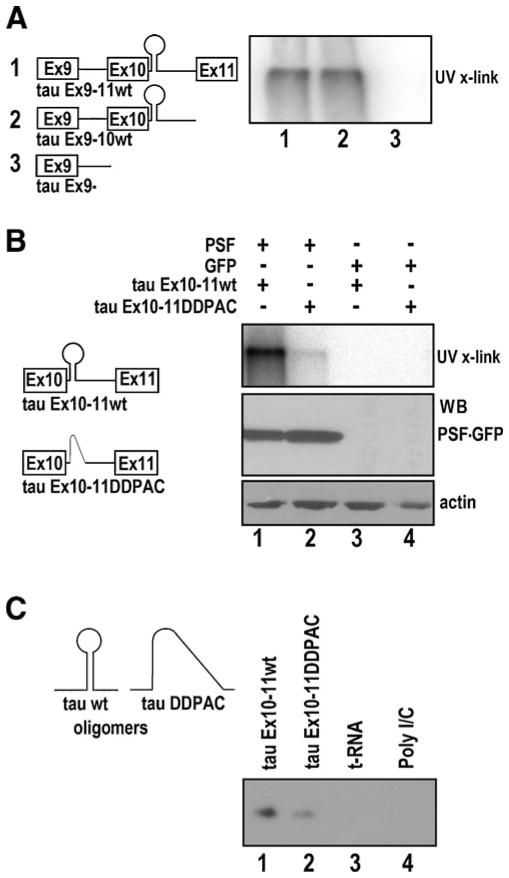
We tested whether PSF interacted with tau pre-mRNA under splicing conditions. As illustrated in Fig. 2, a series of tau pre-mRNA transcripts were prepared containing either the intact tau exon 9–exon10–exon 11 (tau Ex9–11) or truncated sequences with wild-type sequence (tau Ex9–11wt) or mutant sequence, which contained a single nucleotide mutation (C to T) named disinhibition–dementia–parkinsonism–amyotrophy complex (DDPAC) at +14 position of tau intron 10 (Hutton et al. 1998; Jiang et al. 2000) (tau Ex9–11DDPAC). These tau pre-mRNA transcripts were radio-labeled by in vitro transcription reaction as described in our previous studies (Jiang et al. 2000, 2003) and used in UV crosslinking experiments. As shown in Fig. 2A, different tau pre-mRNA transcripts were UV-irradiated following incubation under splicing conditions with cell lysates prepared from HEK293 cells overexpressing PSF tagged with GFP. RNAs cross-linked to PSF were immunoprecipitated using a specific anti-GFP antibody and separated on SDS-PAGE following RNase digestion. UV crosslinking signals were detected by autoradiography. PSF interaction with tau pre-mRNAwas detected when tau Ex9–11 wt (Fig. 2A, transcript 1, lane1) or tau exon9–10 together with the downstream intronic sequence (tau Ex9–10wt; Fig. 2A, transcript 2, lane 2) were used. However, when exon 10 with the intron 10 sequence was deleted (tau Ex9 wt; Fig. 2A, transcript 3), the UV crosslinking signal of PSF was no longer detectable (Fig. 2A, lane3). We then compared the wild-type and DDPAC mutant tau splicing substrates containing only exon 10–11 region. Interestingly, the wild-type (tau Ex10–11wt) transcript showed a strong UV crosslinking signal, but tau DDPAC (tau Ex10–11DDPAC) transcript had a significantly reduced signal (Fig. 2B, compare lanes 1 and 2). The crosslinking signal was specific because no signal was detected when the reactions were carried out using the control cell lysates expressing GFP vector alone (Fig. 2B, lanes 3 and 4). To further define the region of PSF binding on the tau pre-mRNA, a 48-nucleotide tau RNA oligomer containing the stem-loop region with either wild-type sequence or DDPAC mutation was used in the UV cross-linking experiments. Consistent with above results, the wild-type tau RNA oligomer showed a stronger UV crosslinking signal to PSF than the mutant DDPAC oligomer (Fig. 2C, compare lanes 4 and 3), suggesting that PSF preferentially binds to the stem-loop region. To rule out the possibility that PSF binds non-specifically to any RNAs with stem-loop secondary structures, we performed UV crosslinking experiments with Poly I/C and transfer RNA (tRNA), two molecules that have been predicted to form double stranded secondary structures in solution (Grant et al. 1968). PSF did not show any detectable binding to either tRNA or Poly I/C (Fig. 2C, lanes 3 and 4), indicating that PSF protein does not interact non-specifically with RNA containing double-stranded stem-loop structures. These results show that PSF specifically binds to the stem-loop region of tau pre-mRNA.
Fig. 2.
PSF interacts with tau Exon 10 stem-loop region. A HEK293 cells were transfected with PSF-GFP plasmid, and the lysates were used for UV crosslinking experiment with different tau wild-type (wt) transcripts (transcripts 1, 2, and 3). Following UV crosslinking and RNase treatment, the reaction products were analyzed by SDS-PAGE (lanes 1–3) and autoradiography. B Radiolabeled tauEx10–11 wt or DDPAC transcripts (illustrated on left) were incubated with HEK293 cell lysates overexpressing PSF-GFP or control GFP and UV cross-linked. After RNase treatment, anti-GFP antibody was used for immunoprecipitation and the products subjected to SDS-PAGE (lanes 1–4) followed by autoradiography. Middle and lower panels show levels of PSF-GFP and actin, respectively, as detected by immunoblotting. C UV crosslinking experiments using cell lysates (similar to B above) were carried out with a radiolabeled 48 nucleotide oligomer (illustrated on left). Binding of wild type (lane 1) was compared with DDPAC (lane 2) oligomers as well as to transfer RNA (tRNA, lane 3) and Poly I/C (lane 4)
PSF Regulates tau Exon 10 Splicing
To investigate the significance of PSF binding to the stem-loop region of tau pre-mRNA exon 10, we examined the effect on exon 10 splicing. HEK293 cells were co-transfected with tau exon 9–10–11 minigene (tau Ex9–11wt, Jiang et al. 2000) and either vector control or PSF expression plasmid. RNAwas extracted 48 h after transfection and levels of exon 10 alternative splicing products were analyzed by RT-PCR (Fig. 3A), using specific primers, as reported previously (Jiang et al. 2000). Overexpression of PSF led to a significant increase in exclusion of tau exon 10 as compared to the vector control, resulting in increased levels of exon 10-lacking tau splicing products (E10−; Fig. 3A, top left panel, compare lanes 1 and 2).
Fig. 3.
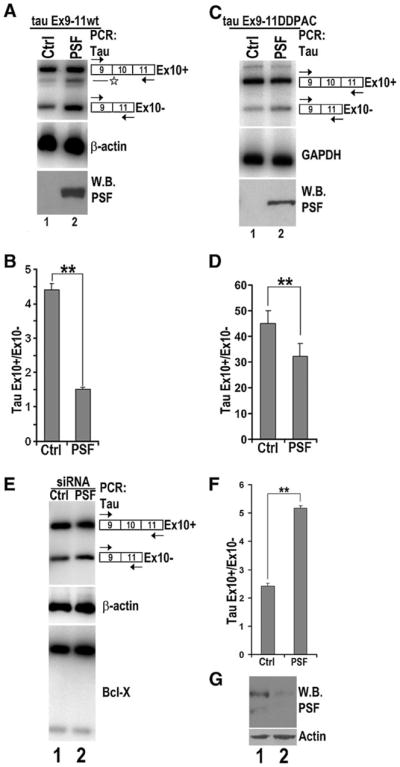
PSF regulates tau exon 10 exclusion. A–D The corresponding plasmids expressing either the vector control (lane 1) or PSF (lane 2) were co-transfected with the tau9–11 wild-type (tau Ex9–11wt) or tau Ex9–11 DDPAC minigene into HEK 293 cells. A, C tau exon 10 (tau Ex10) containing products were detected using RT-PCR, with β-actin or GAPDH transcripts in the corresponding groups showing comparable amounts of RNA used in each reaction. The levels of PSF protein in the corresponding reactions as detected by immunoblotting using anti-GFP antibody is shown in the bottom panels. Asterisk indicates background band in A. B, D Quantification of tau exon 10 splicing (ratio of tauEx10+/ Ex10− levels) as shown in A and C. The graph is average of five independent experiments. E The tau Ex9–11wt minigene was co-transfected into HEK293 cells together with either non-specific control RNAi (lane 1) or PSF-specific RNAi (lane 2). Alternative splicing products were detected by specific primers using RT-PCR. β-Actin levels were used as the internal control. Alternative splicing of the endogenous Bcl-x was not affected by decrease in PSF levels. F Quantification of tau exon 10 splicing isoforms as shown in E. The graph shows the average of six independent experiments. G Endogenous levels of PSF detected by anti-PSF antibody with actin levels used as loading control
Next, we tested whether PSF expression had a similar effect on the DDPAC mutant splicing as observed with tau wild-type minigene. After co-transfection of either vector control or PSF expression plasmid and tau exon 9–10–11 DDPAC minigene (tau Ex9–11 DDPAC, Jiang et al. 2000) in HEK293 cells, RNA was extracted, and cDNA levels were analyzed to determine the levels of tau transcripts containing or lacking exon 10. PSF overexpression resulted in a decrease in splicing products containing exon10 (E10+) in the DDPAC mutant background (Fig. 3C, top right panel, compare lanes 1 and 2).
To investigate the effect of down-regulating PSF on tau exon 10 alternative splicing, we used siRNA to decrease the endogenous levels of PSF in HEK293 cells. PSF-specific siRNA or a non-specific control siRNAwere co-transfected together with the tau Ex9–11wt minigene into HEK293 cells. After two rounds of siRNA transfection, levels of alternatively spliced tau mRNA products containing or lacking exon 10 was examined by RT-PCR. Western blotting using anti-PSF-specific antibody confirmed that specific siRNA transfection led to a significant reduction in the endogenous PSF (see Fig. 3G, top panel). Specific down-regulation of the endogenous PSF in HEK293 cells led to increased levels of products containing exon 10 (E10+), accompanied by a comparable decrease in the levels of product lacking exon 10 (E10−) (Fig. 3E, top panel, lane 2). The effect of PSF knockdown was specific for tau pre-mRNA splicing because alternative splicing of Bcl-x was not affected (Fig. 3E, bottom panel). Together with the PSF overexpression data, these results show that PSF regulates the inclusion of exon10 of tau pre-mRNA.
Oligonucleotide-Directed RNase H Cleavage Experiments Suggest that PSF Stabilizes the 5′ Splice Site Stem-Loop Structure
In our screen, PSF was identified as a protein interacting with the stem-loop region at exon 10 of tau pre-mRNA. To understand its mechanism of action, we examined whether PSF binding affected this secondary structure. Our group had previously reported an oligonucleotide directed RNase H cleavage assay to probe the secondary structure of this region using a DNA oligomer that is complementary to the 5′ splice site of the tau exon 10 (Jiang et al. 2000; Kar et al. 2011). This RNase H cleavage assay was carried out using either tau Ex10–11wt or tau Ex10–11DDPAC transcripts (as in Fig. 2B), and the splicing efficiency of these transcripts was measured following co-transfection with the PSF plasmid as described above for the tau Ex9–11wt minigene. The ratio of spliced products and unspliced pre-mRNAwas calculated to determine whether the effect of PSF on tau Ex10–11 transcript was comparable to the effect seen on tau Ex9–11 minigenes. PSF overexpression led to decreased levels of tau Ex10–11 mRNA, hence suppressing the splicing of exon 10 (see Fig. 4A and B). In the RNaseH cleavage assay, radiolabeled tau wt or tau DDPAC transcripts were incubated with control GST or purified PSF protein and then subjected to RNaseH treatment. Our results showed a decrease in the RNAase H cleavage products (indicative of the formation of DNA-RNA hybrid and thus the openness in the conformation of the stem-loop structure) in the reactions containing the purified PSF protein (Fig. 5A, left panel, compare lanes 1 and 3) as compared to the control reaction. This suggests that in the presence of PSF the stem-loop structure is less accessible for binding to the DNA oligonucleotide. To further identify the region responsible for PSF binding, we used a shorter oligomer RNA transcript corresponding to the stem-loop structure (as used in Fig. 2C). Consistent with our results using the longer tau pre-mRNA transcript, a reduction of cleavage of the tau wild-type oligomer RNA transcript was observed in the presence of PSF protein, suggesting that PSF interacts with tau pre-mRNA stem-loop structure in the region spanning the intron10–exon10 junction to ~48nucleotides downstream of exon 10. We also detected a concentration-dependent suppression of RNaseH cleavage in the presence of increasing amounts of PSF protein (data not shown, quantification shown in Fig. 5). This observation further supports the involvement of PSF protein in regulating the conformation of the tau stem-loop structure. We predicted tau DDPAC mutant to be more susceptible to RNaseH cleavage because of the destabilization of the stem-loop structure and the increased accessibility of the 5′ splice site to U1snRNP by the point mutation inside the stem structure. Consistent with our prediction, the DDPAC mutant RNA transcript showed an increased RNase H cleavage (Fig. 5A, right panel), suggesting that reduced PSF binding correlated with reduced protection from RNaseH cleavage. These results suggest that PSF may interact with the 5′ splice site stem-loop structure to prevent it from switching to a more open conformation that would allow an increased U1snRNP binding to the 5′ splice site.
Fig. 4.
PSF overexpression affects exon 10 splicing. A tau exon 10–11 minigene plasmid was co-transfected along with vector control (lane 1) or PSF (lane 2) overexpression plasmids into HEK293 cells, cDNA extracted 48 h post-transfection and levels of mRNA analyzed by PCR with levels of GAPDH used as internal control. Western blot analysis of corresponding lysates using anti-GFP antibody show levels of PSF in the lysates used for the experiments. B Quantification of ratio of tau mRNA/pre-mRNA. Graph shows the average of three independent experiments
Fig. 5.

Oligonucleotide directed RNaseH cleavage assay shows that PSF protects the 5′ splice site of tau exon 10. A Radiolabeled tau wild-type (left panel) or DDPAC mutant (right panel) pre-mRNA transcripts containing exon 10 and intron 10 were incubated with RNase H and the oligonucleotide complementary to the 5′ splice site at 37°C for 20 min under splicing conditions in the presence of purified PSF (lanes 2 and 3) or control GST protein (lanes 1). After incubation, RNA was isolated, separated on denaturing gel and detected by autoradiography. Lane 2 (0 min) in both the panels contain corresponding input RNA transcripts. Tau RNA transcript and its RNase H cleavage products are illustrated in the diagram between the two panels in A, with the arrowhead indicating the cleavage site and the horizontal bar below exon10–intron 10 boundary representing the DNA oligomer that is complementary to the 5′ splice site. Bottom panel represents the quantification of data from lanes 1 and 3 in each panel, of cleavage of tau RNA as detected by autoradiography. Data are an average of four independent experiments. B RNase H cleavage assay performed with the 48 nucleotide oligomer under same conditions as described in A but with increasing amounts of purified PSF protein. Cleavage products were analyzed on denaturing gel followed by autoradiography, and efficiency of cleavage was determined by calculating the ratio of total cleavage products to the corresponding total transcript input. Top panel represents a schematic representation of the stem-loop structure with the arrowhead showing the cleavage site
Expression of PSF in Mammalian Brain
PSF expression has been reported in the mammalian nervous system (Chanas-Sacre et al. 1999). In the mouse brain, PSF is expressed widely across all regions. PSF expression is under developmental control in the neural tissue, and PSF immunoreactivity has been detected in both neurons and oligodendrocytes (Chanas-Sacre et al. 1999). To determine whether PSF is expressed in neurons where tau exon 10 alternative splicing is important, we examined PSF expression in the human brains by immunohistochemical staining. Our results show that PSF is expressed at a high level in the hippocampus and the cortex (Fig. 6A, B) and that PSF expression appears higher in the fetal than in the adult hippocampus (Fig. 6C). Therefore, PSF is expressed in the human brain regions affected by tauopathy and developmental changes in PSF expression appear to be consistent with the role of PSF in regulating tau exon 10 alternative splicing in humans.
Fig. 6.
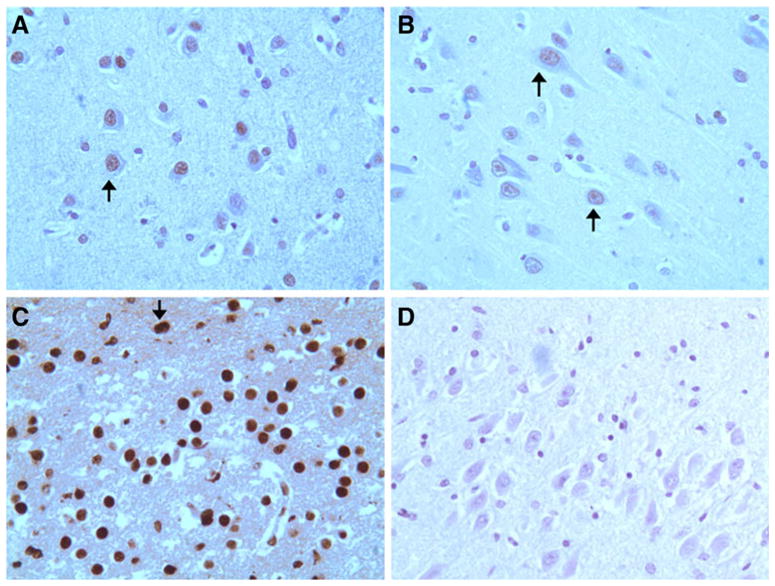
PSF is expressed in the brain regions affected by FTLD-tau. Human brain sections were stained using a specific anti-PSF antibody. A–C Staining of the adult frontal cortex (A), adult hippocampus (B), and fetal brain (C). D Negative control, staining of the hippocampal section in the absence of the primary antibody. PSF immunostaining was detected in the frontal cortex neurons (A) and hippocampal neurons (B, C). The arrows in A and B mark the pyramidal neurons. The pattern of staining is strong nuclear with fetal neurons displaying stronger signal. Stronger signals were detected in the fetal hippocampal neurons than that in the adult hippocampal neurons under the same staining conditions. All sections shown are of high magnification (×600)
Discussion
Genetic analyses of FTLD-tau patients have revealed a number of mutations near the 5′ splice site of exon 10 and the downstream intronic region in the human tau gene. Several studies have been conducted to characterize this region to elucidate molecular mechanisms of these splicing mutations and their effects on tau exon 10 alternative splicing (Wei et al. 2000; Hartmann et al. 2001; Broderick et al. 2004; Kondo et al. 2004; Glatz et al. 2006; Kar et al. 2006; Wu et al. 2006; Dawson et al. 2007; Gao et al. 2007; Zhou et al. 2008; Wang et al. 2010; Kar et al. 2011). These studies have uncovered several transacting proteins that interact with exonic or intronic cis-elements to regulate tau exon 10 alternative splicing. Interestingly, a significant fraction of tauopathy cases with increased Tau4R levels show no detectable mutation in the tau gene. It is possible that in such cases, increased expression or function of trans-acting factors that promote tau exon 10 inclusion play a role in the development of tauopathy.
Using a yeast RNA–protein interaction screen and biochemical assays, we identified PSF as a candidate regulator of tau exon 10 alternative splicing. PSF is a multifunctional nuclear protein that has been implicated in several cellular processes, including DNA recombination, transcription, pre-mRNA splicing (Patton et al. 1993), 3′ end processing (Kaneko et al. 2007), translation (Straub et al. 1998; Akhmedov and Lopez 2000; Urban et al. 2000; Mathur et al. 2001; Zhang and Carmichael 2001; Sewer et al. 2002; Shav-Tal and Zipori 2002; Cobbold et al. 2008), and control of circadian rhythm in mice (Duong et al. 2011). It has been proposed that PSF plays a role in tumor suppression (Wang et al. 2009). PSF has also been shown to be essential for cell survival and neuronal development in zebra fish (Lowery et al. 2007).
Our data show that PSF binds to the stem-loop structure at the exon 10–intron10 boundary of the tau pre-mRNA transcript. PSF was initially identified as a spliceosome associated factor (Patton et al. 1993). PSF is associated with the pre-spliceosome complex as well as spliceosomal snRNPs (Gozani et al. 1994; Peng et al. 2002). PSF also binds to U5snRNP and the 5′ splice site (Peng et al. 2002; Kameoka et al. 2004). Previous studies have reported the binding of PSF to different RNA sequences including AU-rich elements (Buxade et al. 2008), a stem-loop structure in hepatitis delta virus (Greco-Stewart et al. 2006), an exonic splicing suppressor sequence element in CD45 exon 4 (Melton et al. 2007), as well as other sequence elements in transcription promoter region (Yang et al. 2007). Furthermore, the binding of PSF to the stem 1b of U5snRNA has been shown to be dependent on both a specific sequence and a stem structure (Peng et al. 2002). In our studies, the wild-type tau RNA showed stronger PSF UV-crosslinking signal than the tau DDPAC mutant, suggesting that a more stable stem-loop structure may be necessary for efficient binding of PSF (Fig. 2). This reduced PSF binding to the DDPAC mutant tau RNA was accompanied by a reduction in the ability of PSF in suppressing exon 10 inclusion (Fig. 3). On the other hand, no UV-crosslinking signal was detected between PSF and Poly I/C or tRNA (Fig. 2C), indicating that stem-loop structures alone are not sufficient to confer PSF binding.
To better understand the mechanism of PSF in regulating tau exon 10 splicing, we investigated the effect of PSF binding on the stem-loop structure using oligonucleotide directed RNaseH cleavage assay. Our experiments show that the binding of PSF to the stem-loop structure is correlated with reduction of the association of U1snRNP to the 5′ splicing and with the exclusion of tau exon 10. Destabilization of this stem structure by the DDPAC mutation allows more U1snRNP binding to the 5′ splice site, leading to increased inclusion of exon 10 in tau transcripts. Previous studies have shown that different RNA secondary structures may influence interaction of the 5′ splice site with U1snRNP and play a role in regulating alternative splicing (Blanchette and Chabot 1997; Buratti and Baralle 2004; Eperon et al. 1988; Zychlinski et al. 2009). A stem-loop secondary structure has been reported in the 5′ splice site of SMN2 exon 7 that regulates SMN2 exon 7 alternative splicing (Singh et al. 2007). Other genes in which specific RNA secondary structures are associated with regulation of alternative exon inclusion include dystrophin (Matsuo et al. 1992) and the β-tropomyosin gene (Clouet d’Orval et al. 1991; Libri et al. 1991; Sirand-Pugnet et al. 1995).
PSF expression in the mouse brain is the highest around embryonic stage 16 (E16) and gradually decreases in the adulthood. In the developing mouse brain, PSF expression is high in the post-mitotic differentiating neurons in the cortex and cerebellum, whereas in the adult brain, PSF expression appears highest in the olfactory bulb and the hippocampus (Chanas-Sacre et al. 1999). Interestingly, these are the brain regions that are frequently affected in tauopathy patients. Our immunohistochemical staining of the human fetal and adult brain sections showed higher levels of PSF in fetal hippocampus than that in the adult brain. This PSF expression pattern is consistent with the potential role of PSF in suppressing tau exon 10 inclusion in vivo. Previous work by our group shows that another regulatory factor, SRp54, is expressed at a high level in the fetal brain (Wu et al. 2006). These observations suggest that tau alternative splicing pattern may be a result of the complex interplay of different splicing factors whose expressions are controlled in a temporal and spatial manner. Although PSF expression is well documented in nervous system, neuronal specific splicing substrates of PSF had not been reported previously. Our study identifies tau pre-mRNA as a neuronally expressed alternative splicing substrate for PSF. A recent study reported an upregulation of PSF and the splicing factor SRp20 in mice during hippocampal memory formation (Antunes-Martins et al. 2007). Interestingly, SRp20 overexpression leads to increased exclusion of exon 10 not only in wild-type tau minigene but also in FTDP-17 mutants (Yu et al. 2004). It will be interesting to study how these splicing regulators act to affect learning and memory and whether their expression is altered in tauopathy patients.
In summary, we report the identification of PSF as a previously unknown alternative-splicing regulator of tau gene expression. This is the first report of PSF in regulating alternative splicing of a neuron-specific gene. PSF acts to repress the inclusion of tau exon 10 and gives rise to 3R isoform (Fig. 7). PSF functions by stabilizing the stem-loop structure at the 5′ splice site and blocking the accessibility of U1snRNP to the splice site. We also show that PSF is expressed in the brain regions that are affected in FTDP patients. The observation that PSF has a moderate effect on the splicing of tau DDPAC mutant that lacks a regular stem-loop secondary structure can be exploited to develop novel therapeutic approaches for FTDP patients.
Fig. 7.
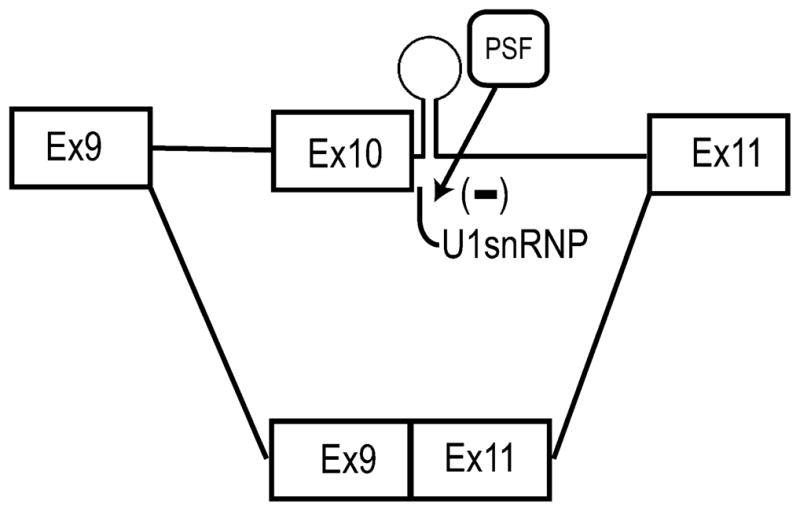
A model for the role of PSF in regulating tau exon 10 splicing. A schematic representation of tau exon 10 splicing regulation by PSF. Under normal conditions the closed stem-loop configuration is in a dynamic flux with the open configuration. PSF binds to and stabilizes the stem-loop structure at the 5′ splice site of exon 10 thereby facilitating the closed configuration. This prevents the association of U1snRNP with the 5′ splice site leading to exclusion of exon 10 from tau transcripts
Acknowledgments
We are grateful for generous help from Dr. James Patton (Vanderbilt University) and Dr. Douglas Black (UCLA) for various reagents. We thank members of the Wu laboratory for helpful discussions, critical reading and suggestions. This work was supported by National Institutes of Health (R01 AG033004 and R56NS074763 to JYW).
Contributor Information
Payal Ray, Department of Neurology, Lurie Cancer Center, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Amar Kar, Department of Neurology, Lurie Cancer Center, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Kazuo Fushimi, Department of Neurology, Lurie Cancer Center, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Necat Havlioglu, Department of Pathology, Saint Louis University, St. Louis, MO, USA.
Xiaoping Chen, Department of Neurology, Lurie Cancer Center, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Jane Y. Wu, Email: jane-wu@northwestern.edu, Department of Neurology, Lurie Cancer Center, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA
References
- Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28(16):3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta. 2005;1739(2–3):91–103. doi: 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Andreadis A. Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog Mol Subcell Biol. 2006;44:89–107. doi: 10.1007/978-3-540-34449-0_5. [DOI] [PubMed] [Google Scholar]
- Andreadis A, Brown WM, Kosik KS. Structure and novel exons of the human tau gene. Biochemistry. 1992;31(43):10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- Antunes-Martins A, Mizuno K, Irvine EE, Lepicard EM, Giese KP. Sex-dependent up-regulation of two splicing factors, Psf and Srp20, during hippocampal memory formation. Learn Mem. 2007;14:693–702. doi: 10.1101/lm.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103(3):367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNPA1 alternative exon 7B. RNA. 1997;3(4):405–419. [PMC free article] [PubMed] [Google Scholar]
- Boeve BF, Hutton M. Refining frontotemporal dementia with parkinsonism linked to chromosome 17: introducing FTDP-17 (MAPT) and FTDP-17 (PGRN) Arch Neurol. 2008;65(4):460–464. doi: 10.1001/archneur.65.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick J, Wang J, Andreadis A. Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene. 2004;331:107–114. doi: 10.1016/j.gene.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24(24):10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxade M, Morrice N, Krebs DL, Proud CG. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J Biol Chem. 2008;283(1):57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, White CL, 3rd, Schneider JA, Grinberg LT, Halliday G, Duyckaerts C, Lowe JS, Holm IE, Tolnay M, Okamoto K, Yokoo H, Murayama S, Woulfe J, Munoz DG, Dickson DW, Ince PG, Trojanowski JQ, Mann DM. Consortium for frontotemporal lobar degeneration. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the consortium for frontotemporal lobar degeneration. Acta Neuropathol. 2007;114(1):5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas-Sacre G, Mazy-Servais C, Wattiez R, Pirard S, Rogister B, Patton JG, Belachew S, Malgrange B, Moonen G, Leprince P. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J Neurosci Res. 1999;57(1):62–73. doi: 10.1002/(SICI)1097-4547(19990701)57:1<62::AID-JNR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Clouet d’Orval B, d’Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252 (5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28(1):40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Fehrat L, Pollard H, Ribas de Pouplana L, Charton G, Bernard A, Moreau J, Ben-Ari Y, Khrestchatisky M. Developmentally regulated alternative splicing of mRNAs encoding N-terminal tau variants in the rat hippocampus: structural and functional implications. Eur J Neurosci. 1997;9:2723–2733. doi: 10.1111/j.1460-9568.1997.tb01701.x. [DOI] [PubMed] [Google Scholar]
- Colombo R, Tavian D, Baker MC, Richardson AM, Snowden JS, Neary D, Mann DM, Pickering-Brown SM. Recent origin and spread of a common Welsh MAPT splice mutation causing frontotemporal lobar degeneration. Neurogenetics. 2009;10(4):313–318. doi: 10.1007/s10048-009-0189-x. [DOI] [PubMed] [Google Scholar]
- Connell JW, Gibb GM, Betts JC, Blackstock WP, Gallo J, Lovestone S, Hutton M, Anderton BH. Effects of FTDP-17 mutations on the in vitro phosphorylation of tau by glycogen synthase kinase 3beta identified by mass spectrometry demonstrate certain mutations exert long-range conformational changes. FEBS Lett. 2001;493:40–44. doi: 10.1016/s0014-5793(01)02267-0. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136 (4):777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson HN, Cantillana V, Chen L, Vitek MP. The tau N279K exon 10 splicing mutation recapitulates frontotemporal dementia and parkinsonism linked to chromosome 17 tauopathy in a mouse model. J Neurosci. 2007;27(34):9155–9168. doi: 10.1523/JNEUROSCI.5492-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J Biol Chem. 2006;281(33):23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza I, Schellenberg GD. Determinants of 4-repeat tau expression. Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J Biol Chem. 2000;275(23):17700–17709. doi: 10.1074/jbc.M909470199. [DOI] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332 (6036):1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Gao QS, Memmott J, Lafyatis R, Stamm S, Screaton G, Andreadis A. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J Neurochem. 2000;74(2):490–500. doi: 10.1046/j.1471-4159.2000.740490.x. [DOI] [PubMed] [Google Scholar]
- Gao L, Wang J, Wang Y, Andreadis A. SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations. Mol Cell Neurosci. 2007;34(1):48–58. doi: 10.1016/j.mcn.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Moller HJ, Andreadis A, Henkel K, Stamm S. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer’s disease. J Neurochem. 2006;96(3):635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- Goedert M, Jakes R. Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990;9(13):4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta. 2005;1739(2–3):240–250. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG. Tau gene mutations and neurodegeneration. Biochem Soc Symp. 2001;(67):59–71. doi: 10.1042/bss0670059. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989a;3(4):519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 1989b;8(2):393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Chau M, Denis PE, Feinstein SC. Structural and functional differences between 3-repeat and 4-repeat tau isoforms. Implications for normal tau function and the onset of neurodegenetative disease. J Biol Chem. 2000;275:38182–38189. doi: 10.1074/jbc.M007489200. [DOI] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13(14):3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RC, Harwood SJ, Wells RD. The synthesis and characterization of poly d (I-C) poly d (I-C) J Am Chem Soc. 1968;90(16):4474–4476. doi: 10.1021/ja01018a060. [DOI] [PubMed] [Google Scholar]
- Greco-Stewart VS, Thibault CS, Pelchat M. Binding of the polypyrimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA. Virology. 2006;356(1–2):35–44. doi: 10.1016/j.virol.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, Hutton M. 5′ Splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J Biol Chem. 1999;274(21):15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Rujescu D, Giannakouros T, Nikolakaki E, Goedert M, Mandelkow EM, Gao QS, Andreadis A, Stamm S. Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol Cell Neurosci. 2001;18(1):80–90. doi: 10.1006/mcne.2001.1000. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Smith MJ, Goedert M. Tau proteins with FTDP-17 mutations have a reduced ability to promote microtubule assembly. FEBS Lett. 1998;437:207–210. doi: 10.1016/s0014-5793(98)01217-4. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Smith MJ, Iijima M, Tabira T, Goedert M. FTDP-17 mutations N279K and S305N in tau produce increased splicing of exon 10. FEBS Lett. 1999;443:93–96. doi: 10.1016/s0014-5793(98)01696-2. [DOI] [PubMed] [Google Scholar]
- Himmler A. Structure of the bovine tau gene: alternatively spliced transcripts generate a protein family. Mol Cell Biol. 1989;9:1389–1396. doi: 10.1128/mcb.9.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118(1):53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Grundke-Iqbal I. Tau in Alzheimer Disease and related Tauopathies. Curr Alzheimer Res. 2010;7:656–664. doi: 10.2174/156720510793611592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci USA. 1998;95(16):9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol Cell Biol. 2000;20 (11):4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Tang H, Havlioglu N, Zhang X, Stamm S, Yan R, Wu JY. Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 beta. J Biol Chem. 2003;278(21):18997–19007. doi: 10.1074/jbc.M301800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfuss B, Mason SA, Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J Biol Chem. 2001;276(46):42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM. P54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23(8):1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21(14):1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Kuo D, He R, Zhou J, Wu JY. Tau alternative splicing and frontotemporal dementia. Alzheimer Dis Assoc Disord. 2005;19 (Suppl 1):S29–S36. doi: 10.1097/01.wad.0000183082.76820.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Havlioglu N, Tarn WY, Wu JY. RBM4 interacts with an intronic element and stimulates tau exon 10 inclusion. J Biol Chem. 2006;281(34):24479–24488. doi: 10.1074/jbc.M603971200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A, Fushimi K, Zhou Xh, Ray P, Shi C, Chen Xp, Liu Zr, Chen S, Wu JY. RNA Helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol Cell Biol. 2011;31:1812–1821. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- Konzack S, Thies E, Marx A, Mandelkow EM, Mandelkow E. Swimming against the tide: mobility of the microtubule-associated protein tau in neurons. J Neurosci. 2007;27:9916–9927. doi: 10.1523/JNEUROSCI.0927-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90(1–2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- LeBoeuf AC, Levy SF, Gaylord M, Bhattacharya A, Singh AK, Jordan MA, Wilson L, Feinstein SC. FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an “alternative core” model for normal and pathological Tau action. J Biol Chem. 2008;283:36406–36415. doi: 10.1074/jbc.M803519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Neve RL, Kosik KS. The microtubule binding domain of tau protein. Neuron. 1989;2:1615–1624. doi: 10.1016/0896-6273(89)90050-0. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Libri D, Piseri A, Fiszman MY. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lichtenberg-Kraag B, Mandelkow EM. Isoforms of tau protein from mammalian brain and avian erythrocytes: structure, self-assembly, and elasticity. J Struct Biol. 1990;105:46–53. doi: 10.1016/1047-8477(90)90097-v. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236(5):1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenet Nov. 2007;8(4):237–248. doi: 10.1007/s10048-007-0102-4. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, Mann DM. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119(1):1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21(7):2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Nishio H, Kitoh Y, Francke U, Nakamura H. Partial deletion of a dystrophin gene leads to exon skipping and to loss of an intra-exon hairpin structure from the predicted mRNA precursor. Biochem Biophys Res Commun. 1992;182(2):495–500. doi: 10.1016/0006-291x(92)91759-j. [DOI] [PubMed] [Google Scholar]
- Medeiros R, Baglietto-Vargas D, Laferla FM. The role of tau in Alzheimer’s disease and related disorders. CNS Neurosci Ther. 2010 doi: 10.1111/j.1755-5949.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27(19):6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercken M, Fischer I, Kosik KS, Nixon RA. Three distinct axonal transport rates for tau, tubulin, and microtubule-associated proteins: evidence for dynamic interactions of tau with micro-tubules in vivo. J Neurosci. 1995;15(12):8259–8267. doi: 10.1523/JNEUROSCI.15-12-08259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montejo de Garcini E, Corrochano L, Wischik CM, Diaz Nido J, Correas I, Avila J. Differentiation of neuroblastoma cells correlates with an altered splicing pattern of tau RNA. FEBS Lett. 1992;299:10–14. doi: 10.1016/0014-5793(92)80088-x. [DOI] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, Brown RH, Jr, Brown H, Tiwari A, Hayward L, Edgar J, Nave KA, Garberrn J, Atagi Y, Song Y, Pigino G, Brady ST. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29(41):12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Tolnay M, Mackenzie IR. The molecular basis of frontotemporal dementia. Expert Rev Mol Med. 2009;11:e23. doi: 10.1017/S1462399409001136. [DOI] [PubMed] [Google Scholar]
- Neve RL, Harris P, Kosik KS, Kurnit DM, Donlon TA. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 1986;387(3):271–280. doi: 10.1016/0169-328x(86)90033-1. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Goode BL, Feinstein SC, Wilson L. Kinetic stabilization of microtubule dynamics at steady state by tau and microtubule-binding domains of tau. Biochemistry. 1995;34 (35):11117–11127. doi: 10.1021/bi00035a017. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7(3):393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8(10):1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering-Brown S, Baker M, Yen SH, Liu WK, Hasegawa M, Cairns N, Lantos PL, Rossor M, Iwatsubo T, Davies Y, Allsop D, Furlong R, Owen F, Hardy J, Mann D, Hutton M. Pick’s disease is associated with mutations in the tau gene. Ann Neurol. 2000;48(6):859–867. [PubMed] [Google Scholar]
- Pickering-Brown SM, Richardson AM, Snowden JS, McDonagh AM, Burns A, Braude W, Baker M, Liu WK, Yen SH, Hardy J, Hutton M, Davies Y, Allsop D, Craufurd D, Neary D, Mann DM. Inherited frontotemporal dementia in nine British families associated with intronic mutations in the tau gene. Brain. 2002;125(Pt 4):732–751. doi: 10.1093/brain/awf069. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Nguyen VQ, Huang CJ, Tucker PW, Kagawa N, Waterman MR. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology. 2002;143(4):1280–1290. doi: 10.1210/endo.143.4.8748. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 2002;531(2):109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35(2):371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirand-Pugnet P, Durosay P, Clouet d’Orval BC, Brody E, Marie J. Beta-tropomyosin pre-mRNA folding around a muscle-specific exon interferes with several steps of spliceosome assembly. J Mol Biol. 1995;251(5):591–602. doi: 10.1006/jmbi.1995.0458. [DOI] [PubMed] [Google Scholar]
- Solis AS, Shariat N, Patton JG. Splicing fidelity, enhancers, and disease. Front Biosci. 2008;13:1926–1942. doi: 10.2741/2812. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Van Swieten JC, Goedert M. Tau gene mutations in frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) Neurogenetics. 2000;2(4):193–205. doi: 10.1007/pl00022972. [DOI] [PubMed] [Google Scholar]
- Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F. The RNA-splicing factor PSF/ p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273(41):26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- Tomoo K, Yao TM, Minoura K, Hiraoka S, Sumida M, Taniguchi T, Ishida T. Possible role of each repeat structure of the microtubule-binding domain of the tau protein in in vitro aggregation. J Biochem. 2005;138:413–423. doi: 10.1093/jb/mvi142. [DOI] [PubMed] [Google Scholar]
- Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14(6):774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- Utton MA, Connell J, Asuni AA, van Slegtenhorst M, Hutton M, de Silva R, Lees AJ, Miller CC, Anderton BH. The slow axonal transport of the microtubule-associated protein tau and the transport rates of different isoforms and mutants in cultured neurons. J Neurosci. 2002;22:6394–6400. doi: 10.1523/JNEUROSCI.22-15-06394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc Natl Acad Sci USA. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Spillantini MG, Goedert M, Varani G. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Res. 2000;28:710–719. doi: 10.1093/nar/28.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8 (10):749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang J, Gao QS, Wang Y, Lafyatis R, Stamm S, Andreadis A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J Neurochem. 2004;88(5):1078–1090. doi: 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- Wang G, Cui Y, Zhang G, Garen A, Song X. Regulation of protooncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci. 2009;106(39):16794–16798. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao L, Tse SW, Andreadis A. Heterogeneous nuclear ribonucleoprotein E3 modestly activates splicing of tau exon 10 via its proximal downstream intron, a hotspot for frontotemporal dementia mutations. Gene. 2010;451(1–2):23–31. doi: 10.1016/j.gene.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220(2):152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ML, Andreadis A. Splicing of a regulated exon reveals additional complexity in the axonal microtubule-associated protein tau. J Neurochem. 1998;70:1346–1356. doi: 10.1046/j.1471-4159.1998.70041346.x. [DOI] [PubMed] [Google Scholar]
- Wei ML, Memmott J, Screaton G, Andreadis A. The splicing determinants of a regulated exon in the axonal MAP tau reside within the exon and in its upstream intron. Brain Res Mol Brain Res. 2000;80(2):207–218. doi: 10.1016/s0169-328x(00)00137-6. [DOI] [PubMed] [Google Scholar]
- Wolfe MS. Tau mutations in neurodegenerative diseases. J Biol Chem. 2009;284(10):6021–6025. doi: 10.1074/jbc.R800013200. [DOI] [PubMed] [Google Scholar]
- Wu JY, Postashkin JA. Alternative splicing in the nervous system. In: Squire LR, editor. Encyclopedia of neuroscience. Vol. 1. Academic/Elsevier; Oxford: 2009. pp. 245–251. [Google Scholar]
- Wu JY, Yuan L, Havlioglu N. Alternatively spliced genes. In: Meyers RA, editor. Encyclopedia of molecular cell biology and molecular medicine. 2. Vol. 1. Wiley-VCH; Chichester: 2004. pp. 125–177. [Google Scholar]
- Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol Cell Biol. 2006;26(18):6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sui Y, Xiong S, Liour SS, Phillips AC, Ko L. Switched alternative splicing of oncogene CoAA during embryonal carcinoma stem cell differentiation. Nucleic Acids Res. 2007;35 (6):1919–1932. doi: 10.1093/nar/gkl1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Guo J, Zhou J. A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J Neurochem. 2004;90(1):164–172. doi: 10.1111/j.1471-4159.2004.02477.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106 (4):465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yu Q, Zou T. Alternative splicing of exon 10 in the tau gene as a target for treatment of tauopathies. BMC Neurosci. 2008;9 (Suppl 2):S10. doi: 10.1186/1471-2202-9-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D, Erkelenz S, et al. Limited complementarity between U1 snRNA and a retroviral 5′ splice site permits its attenuation via RNA secondary structure. Nucleic Acids Res. 2009;37 (22):7429–7440. doi: 10.1093/nar/gkp694. [DOI] [PMC free article] [PubMed] [Google Scholar]