Abstract
Unlocked nucleic acid (UNA) is an acyclic analog of RNA that can be introduced into RNA or DNA oligonucleotides. The increased flexibility conferred by the acyclic structure fundamentally affects the strength of base-pairing, createing opportunities for improved applications and new insights into molecular recognition. Here we test how UNA substitutions affect allele-selective inhibition of trinucleotide-repeat genes Huntingtin (HTT) and Ataxin-3 (ATX-3) expression. We find that the either the combination of mismatched bases and UNA substitutions or UNA substitutions alone can improve potency and selectivity. Inhibition is potent and selectivities of > 40-fold for inhibiting mutant versus wild-type expression can be achieved. Surprisingly, even though UNA preserves the potential for complete base-pairing, the introduction of UNA substitutions at central positions within fully complementary duplexes leads to >19-fold selectivity. Like mismatched bases, the introduction of central UNA bases disrupts the potential for cleavage of substrate by Argonaute 2 (AGO2) during gene silencing. UNA-substituted duplexes are as effective as other strategies for allele-selective silencing of trinucleotide repeat disease genes. Modulation of AGO2 activity by the introduction of UNA substitutions demonstrates that backbone flexibility is as important as base-pairing for catalysis of fully complementary duplex substrates. UNA can be used to tailor RNA silencing for optimal properties and allele-selective action.
Introduction
Synthetic nucleic acids are widely used for research, medical diagnosis, and drug development. Nucleic acids are promising approach for clinical therapy1 and a systemically administered antisense oligonucleotide (ASO) has recently been approved by Food and Drug Administration (FDA) as a therapy for familial hypercholesterolemia.2 The clinical success of oligonucleotides after many years of slow progress has led to optimism that nucleic acids may become a major class of therapeutics.
Many different chemically modified nucleosides are available and their wide range of properties encourages development.3 Upon introduction into an oligonucleotide, each type of nucleoside has a unique potential to alter the oligonucleotide’s properties. When carefully chosen, the introduced nucleosides can tailor the function of the oligonucleotide for the demands of a given application. This engineering improves the robustness of protocols that use nucleic acids and the likelihood that they will be widely adopted.
Locked nucleic acid (LNA) nucleosides4, 5 have a methylene bridge connecting the 2′ ribose with the 4′ carbon. This bridge increases rigidity and reduces the entropic penalty paid upon binding a complementary sequence. The reduced entropic cost of binding leads to higher affinity, with LNA substitutions improving melting temperature (Tm) values by as much as 3–8°C per substitution.6 LNA has become one of the most successful modified nucleosides, with applications to clinical development of ASOs,7 as a research tool for inhibiting the action of miRNAs,8 and as molecular probes.9 Similar bridged nucleic acids (BNAs) that contain alternate ribose constraints add to the potential for using increased rigidity to improve applications.10, 11
In contrast to the increased rigidity of LNA nucleosides, unlocked nucleic acid (UNA) substitutions12–14 permit researchers to explore decreased rigidity as a variable for improving the function of oligonucleotides. UNA nucleosides are acyclic and lack a connection between the C2′ and C3′ atoms (Figure 1). In contrast to LNA, the introduction of UNA nucleosides allows a tailored decrease in the affinity of oligonucleotide binding. For duplex RNAs, UNA substitutions can reduce off-target gene silencing, possibly by destabilizing seed sequence interactions at off-target genes15,16 and can improve function in combination with other types of chemical modification.17 Acyclic oligonucleotides with 2′, 3′ secouridine substitutions have also been studied to investigate the interplay of backbone flexibility and enzyme activity.18 When introduced into antisense oligonucleotides the acyclic substitutions increased the rate of substrate cleavage by RNase H, suggesting that strategically-placed modifications can affect catalysis.
Figure 1.
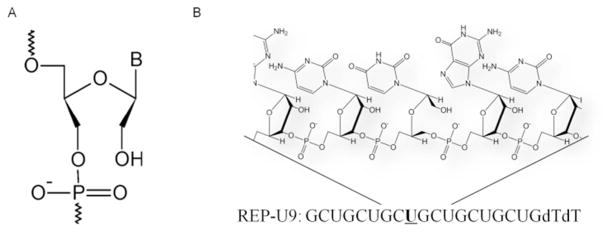
(A) UNA structure. (B) Representative chemical structure of UNA-modified RNA
In this report we test the hypothesis that UNA substitutions can improve allele-selective gene silencing of mutant huntingtin (HTT) and ataxin-3 (ATX-3). Mutant HTT causes Huntington’s disease (HD)19,20 and mutant ATX-3 causes Machado-Joseph disease (MJD).21,22 Relative to the wild-type HTT or ATX-3 genes, the mutated alleles contain longer CAG repeats within their mRNA coding regions. For HTT, the CAG repeats are close to the 5′ terminal. For ATX-3 mRNA, the repeats are located at its 3′ end. HD patients have an average of 45 CAG repeats while the number of wild-type HTT is almost always fewer than 26. For MJD, mean repeat length can vary from 73 to 80 repeats in different populations and in most unaffected individuals the CAG tract is less than 31 repeats. HD and MJD are representative of a large family of neuromuscular diseases caused by CAG expansions and anti-CAG therapeutic strategies have the potential to treat multiple pathologies.
Antisense oligonucleotide and duplex RNA inhibitors of HTT and ATX-3 expression have been intensively studied.23–33 We and others have previously shown that the allele-selective inhibition of gene expression can be achieved by duplex RNAs or single-stranded silencing RNAs (ss-siRNAs) containing central mismatches.34–42 These mismatches prevent argonaute 2 (AGO2) from cleaving the target mRNA and shift the mechanism of action towards one that resembles the mechanism of miRNAs. Allele-selectivity can also be achieved by duplexes containing abasic substitutions that, like mismatches, remove the potential for normal base-pairing.41
While mismatched and abasic duplexes provide a substantial pool of promising compounds for therapeutic discovery, meeting the challenges of clinical development of inhibition candidates will benefit from the identification of a wider number of potent and allele-selective agents. Exploring the limits for applying chemical modification to gene silencing by duplex RNA also provides insights into substrate recognition by AGO2 during catalysis.
Here we find that duplexes that contain both central mismatches and UNA substitutions possess improved potencies and selectivities. Even though UNA substitutions preserve base-paring with the target mRNA, UNA substitutions within fully complementary duplexes also yield allele-selective inhibition. These results expand the range of therapeutic leads for allele-selective inhibition of CAG repeat disease genes and introduce UNA as a strategy for tailoring the properties of allele-selective duplexes.
MATERIALS AND METHODS
RNA Synthesis
UNA-modified antisense RNAs and unmodified sense RNAs were synthesized and characterized using electrospray ionization mass spectrometry by Sigma Custom Products (The Woodlands, TX) and reconstituted in nuclease-free water. Double-stranded RNAs were prepared by mixing the two RNA strands and annealing them in 2.5X PBS solutions. 20 μM stock solutions were prepared for transfection in cell cultures.
Thermal denaturing by UV melt analysis
Thermal denaturation analysis of UNA-containing RNA duplexes was carried out using a CARY Varian model 3 UV-Vis spectrophotometer (Agilent Tech, Santa Clara, CA). In a 1-cm quartz cuvette, absorbance was monitored at 260 nm. UNA-modified antisense RNAs (1 μM) were mixed with equimolar sense RNA strand (5′-CAGCAGCAGCAGCAGCAGCdTdT-3′) in 0.1 M phosphate buffer (pH 7.4) and melted three times from 15 °C to 95°C at a ramp rate of 1°C/min. Melting temperature (Tm) was calculated using a CARY WinUV Thermal Application software using a baseline fitting method.
Cell culture and transfection
Patient-derived fibroblast cell lines GM04281 (HTT; 69 CAG repeats) and GM06151 (ATX-3; 74 CAG repeats) were obtained from the Coriell Institute (Camden, NJ). The fibroblasts were maintained at 37 °C and 5% CO2 in Minimal Essential Media Eagle (MEM) (Sigma, M4655) supplemented with 10% heat inactivated fetal bovine serum (Sigma) and 0.5% MEM nonessential amino acids (Sigma). Cells were plated at a density of 60,000 (HTT) or 70,000 (ATX-3) per well of a 6-well plate 48 h before transfection. siRNAs were transfected into cells with lipid RNAiMAX (Life Technologies) as previously described.33,34 Cells were typically harvested 3 days after transfection for quantitative PCR (qPCR) and RNA immunoprecipitation (RIP), or 4 days for protein assay.
Western blot and qPCR analysis
HTT and ATX-3 expression was analyzed by western blot analysis. SDS-PAGE was used to separate HTT isoforms as described previously34 and ATX-3 protein was separated by 4–20% acrylamide pre-cast gels (Bio-Rad). The primary antibodies were used: anti-HTT (MAB2166, Millipore), anti-ATX-3 (MAB5360, Millipore) and anti-β-actin (Sigma). Protein bands were quantified using ImageJ software. The percentage of inhibition was calculated as a relative value to a control sample. Dose fitting curve was generated using GraphPad Prism 6 program by the equation: , where y is percentage of inhibition and x is the siRNA concentration, n is the IC50 value, and m is the Hill coefficient value.
Total RNA was extracted using TRIzol (Life Technologies) and 2 μg of RNA was subjected to DNase I (Worthington Biochemical Corp.) treatment. cDNA was prepared using High Capacity cDNA Reverse Transcription Kit (Life Technologies). After an appropriate dilution of cDNA sample, qPCR was performed on a CFX96 real-time PCR system (Bio-Rad) using iTaq SYBR Green Supermix (Bio-rad). Data was normalized relative to levels of GAPDH mRNA. The following qPCR primer sets were used for HTT: F 5′–CGACAGCGAGTCAGTGAATG–3′/R 5′–ATCCTGAGCCTCTGATACTC–3′. GAPDH primers were obtained from Applied Biosystems. The qPCR cycles are as follows: 50°C for 2 min; 95°C for 3min; (95°C for 15s; 60°C for 1min) × 40 cycles.
RNA immunoprecipitation (RIP)
HTT fibroblast cells were seeded at 1400K in 150 cm2 dishes. Duplex RNAs were transfected with RNAiMAX on the next day. Cells were harvested 72 hrs later, and were lysed in a buffer (20 mM Tris.HCl pH7.4, 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40, 0.5 mM DTT, protease inhibitor (EDTA-free, Roche) and RNase inhibitor (Promega, 50 U/ml final)) with a volume about 3 times of the cell pellet size. The mixture was sat on ice for 10 min after thorough mixing. After centrifugation, the supernatant were isolated and stored at −80 °C. After 60 SL Protein A/G agarose Plus was washed with lysis buffer twice, beads were incubated with 4 SL of antibodies (anti-AGO2, 4G8, 011-22033, Wako; or normal mouse IgG, 12-371, Millipore) and cell lysate in lysis buffer at 4°C with gentle agitation for 3 hours.42 The beads were further washed with above lysis buffer three times. The beads were finally eluted with elution buffer (1% SDS, 20 mM NaHCO3 and RNase inhibitor). After proteinase K treatment, RNA extraction and precipitation, samples were treated with recombinant DNase I, followed by reverse transcription. The mRNA levels were quantified by qPCR. Results were normalized first by GAPDH levels and second by that of IgG.
In vitro cleavage assay
RNA substrate containing fragment of HTT exon1 with 17 CAG repeats was prepared as previously reported.44 This transcript was gel purified, dephosphorylated and 5′-phosphorylated with [γ-32P] ATP. Purified recombinant human Ago2 protein (a gift from Dr. Qinghua Liu) was pre-incubated with 5′-phosphorylated antisense RNA with or without UNA modification at room temperature for 1.5 h. Then the 5′-radiolabeled RNA substrate was added and the solution was further incubated at 37°C for 1.5h. The final reaction conditions are as follows: 50 nM 5′-phosphorylated antisense RNA, 10 U Superase-IN (Ambion), 50 mM Tris (pH 7.4), 2 mM MgCl2, 0.5 mM DTT, 0.25 mM ATP, 100 mM KCl and 50 mM NaCl. The reaction was stopped by adding 2% LiClO4 in acetone and RNA was precipitated by centrifuge. After washing with acetone, the RNA was reconstituted in 90% formaldehyde and 1X TBE with dye and separated with 12% acrylamide/7M urea gel.
RESULTS
Design and synthesis of UNA oligonucleotides
In our initial studies we observed that RNA duplexes that were fully complementary to CAG repeats were potent but non-allele selective inhibitors of HTT and ATX-3 expression.35,37 We subsequently observed that RNA duplexes containing centrally-mismatched bases were allele-selective inhibitors of expression for both genes.34,35,37 Our goal in this study was to test the effect of UNA substitutions (Figure 1) within duplexes that were fully complementary relative to the target mRNAs or into duplexes that contained a mismatched base at position 9 of antisense strand. These compounds test whether preserving base-pairing while “unlocking” the backbone of key nucleosides can affect allele-selectivity.
Effect of UNA substitutions on inhibition of HTT by fully complementary duplexes
We synthesized duplexes that were fully complementary to the CAG repeat and contained single UNA substitutions at positions 9, 10, or 11 from the 5′ termini of guide (antisense) strand (Figure 2A). We also synthesized a duplex that was substituted at positions 9, 10, and 11 with three UNA nucleosides. The UNA nucleosides were introduced by standard oligonucleotide coupling protocols.
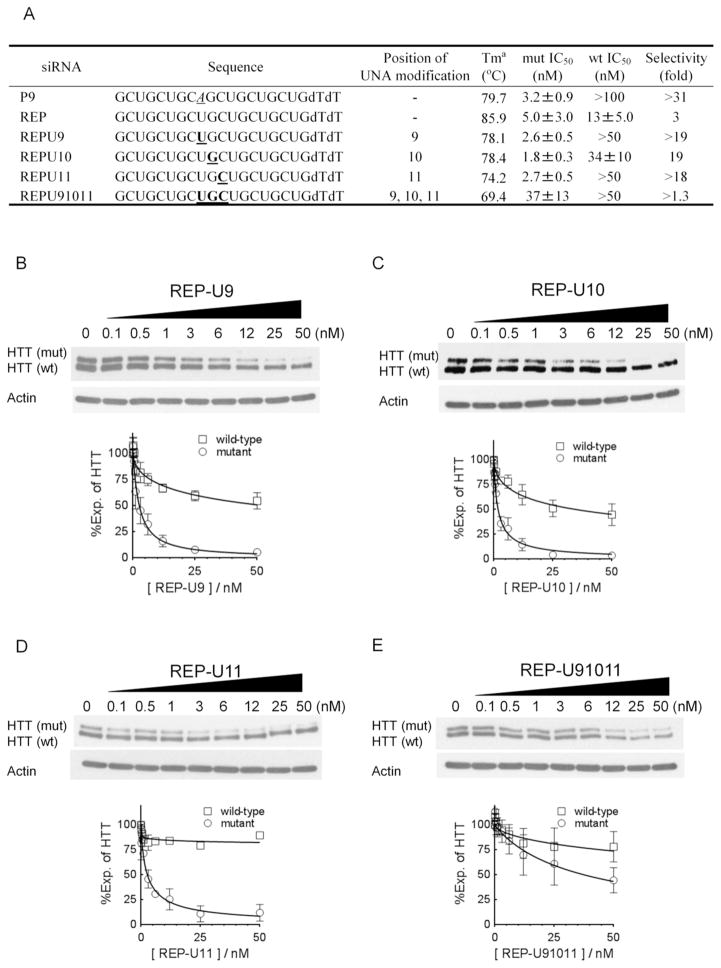
Figure 2.
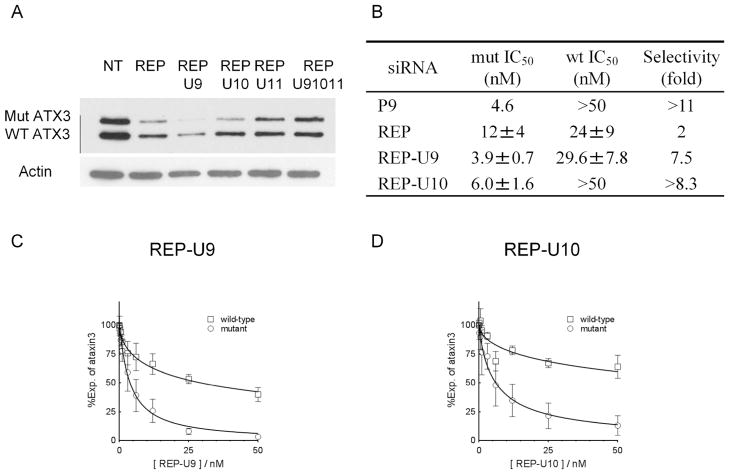
Effects of UNA substitutions on allele-selective inhibition of HTT by RNA duplexes that are fully complementary to the CAG repeat. Data shown are western analyses of HTT protein expression in GM04281 patient-derived fibroblast cells. (A) Sequences of duplex RNAs. REP is a duplex with two fully complementary strands that targets the CAG repeat and has no UNA substitutions. P9 is a duplex that has a mismatch at position 9 and no UNA substitution. For other RNAs, UNA nucleosides are boldface and underlined. Gels and dose response curves showing effect of adding (B) REP-U9, (C) REP-U10, (D) REP-U11, or (E) REP-U91011 at increasing concentrations. Dose curves are averaged data from three independent experiments.
We founds that a single UNA substitution reduced the melting temperature (Tm) of the duplex by about 8~12 °C compared with the fully complementary RNA duplex (REP). Three of the UNA-substituted duplexes (REPU9, REPU10, and REPU11) contained a single UNA and the Tm values of these RNAs were 1~2 °C lower than analogous RNAs that containing mismatched bases. For the duplex with three UNA substitutions (REPU91011), Tm drops 16.5 °C compared with duplex REP. This value is about 4 °C lower than that of the triply-mismatched analogue.
We transfected the RNA duplexes into patient-derived fibroblast cells using cationic lipid and monitored inhibition of HTT protein expression by western analysis. Duplexes containing single substitutions at positions 9, 10, or 11 were potent and selective inhibitors of mutant HTT expression, demonstrating that unlocking the ribose could yield good allele-selectivity even though the potential for full base-pairing is maintained. Potencies were 1.8–2.7 nM and selectivities for inhibition of mutant versus wild-type expression were > 18 fold.
By contrast to the potent and allele-selective duplexes with single UNA substitutions, duplex REPU91011 substituted with three UNA nucleobases was less potent and had low selectivity. REPU91011 had a 5–9°C lower Tm value than those of RNAs with one UNA substitution. This lowered ability to recognize a complementary target is a likely the cause of its reduced potency and selectivity.
Effect of UNA substitutions on inhibition of HTT by mismatched duplexes
We also introduced UNA substitutions into duplexes containing a mismatched base at position 9 (Figure 3A). We had previously observed that the introduction of centrally located mismatches yields high selectivities and potencies37. We reasoned that the combination of mismatches and UNA substitutions might yield improved compounds.
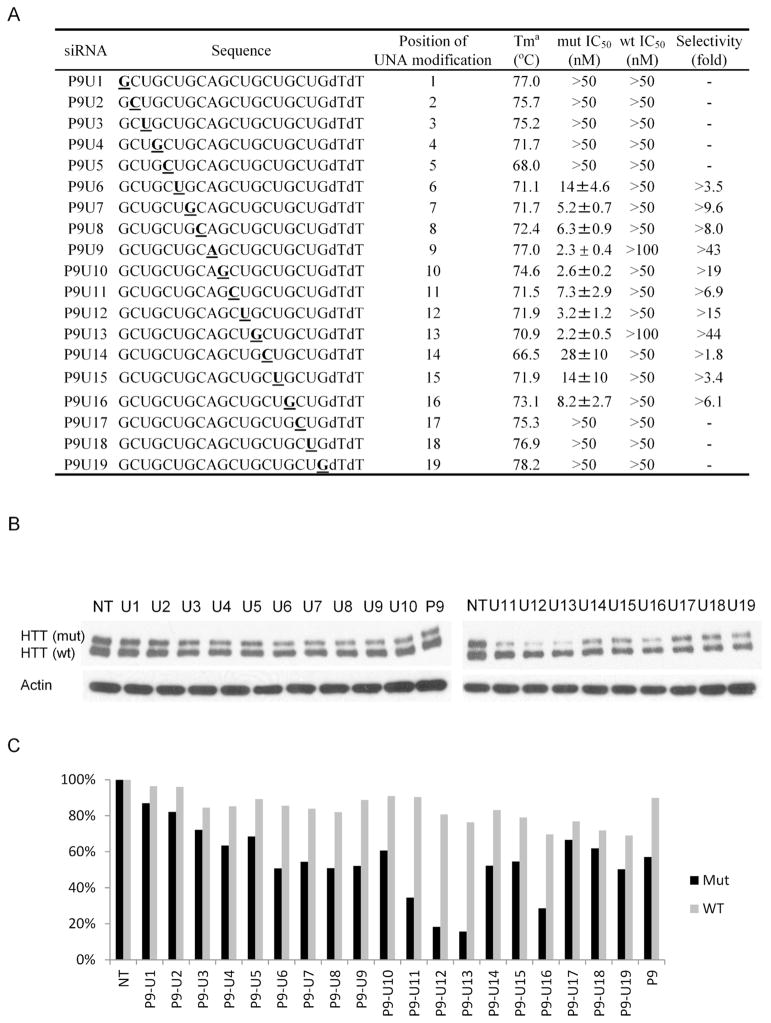
Figure 3.
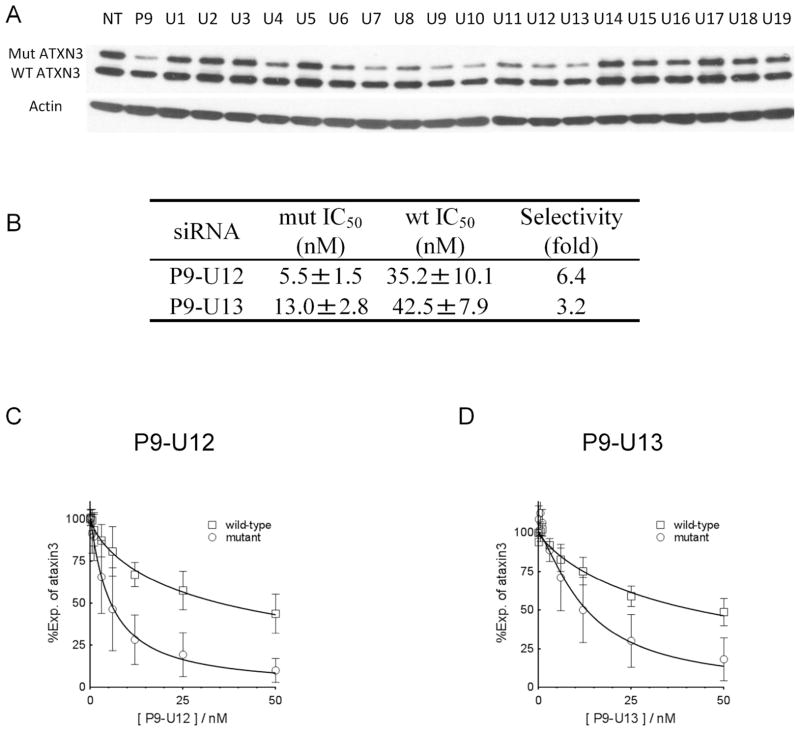
Effects of UNA substitutions within duplex RNAs that also have a mismatch at position 9 (P9). (A) Sequences of UNAs. UNA nucleosides are boldface and underlined. (B) Effect of 25 nM UNA on inhibition of HTT expression, analyzed by western analysis. (C) Quantitation of inhibition of wild-type or mutant HTT expression shown in part (B). NT: no treatment/no duplex added. The Tm value of the corresponding mismatched duplex at position 9 (P9) that lacks a UNA substitution is 79.7°C.
Introducing a UNA nucleoside blocked inhibition of both alleles when the substitution was at positions one through five (Figure 3BC, Supplementary Figures 1 and 2). This result is consistent with positions 1–8 acting as a critical seed sequence during RNAi. Increased flexibility can disrupt key seed interactions even if the potential for base-pairing is maintained.
Other substitutions were more compatible potent RNAi. When the substitution as at position 6, the IC50 for inhibition of the mutant allele was 14 nM and the potency dropped to under 10 nM for substitutions at positions 7–13. Two duplexes (P9U9, P9U13) had both outstanding potencies (~2 nM) and selectivities > 40 fold, demonstrating that the combination of UNA substitutions and mismatched bases has the potential to yield promising compounds. Moving the position of the UNA substitution of the UNA base by just one position in P9U14 reduced potency dramatically, emphasizing that UNA substitutions must be positioned carefully. For several UNA-substituted duplexes (e.g. P9-U9, P9-U11) we observed little or no inhibition of wild-type expression (Figure 4), making them excellent candidates for further investigation.
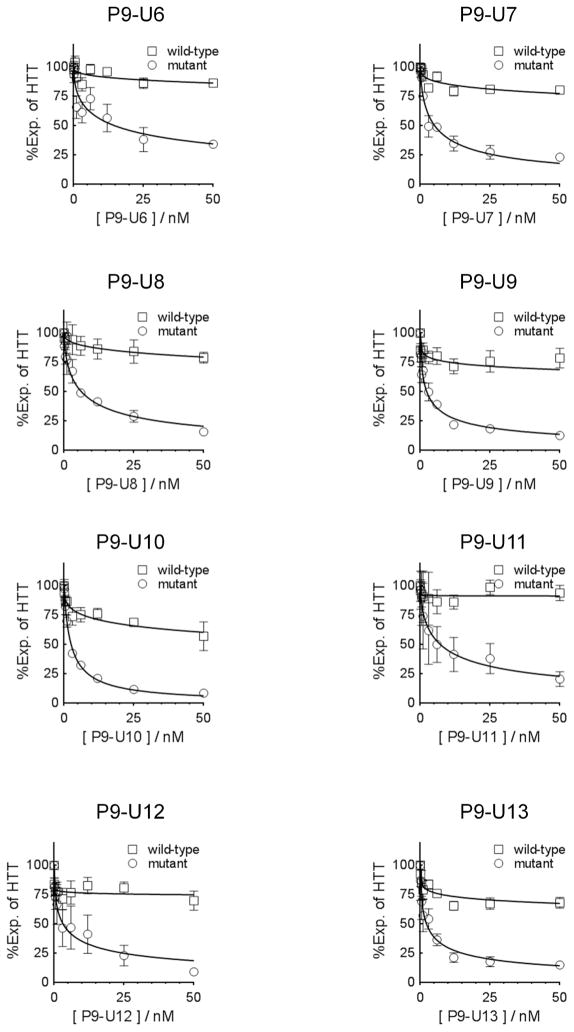
Figure 4.
Dose response data showing the effect of UNA substitutions on duplexes based on P9 siRNA containing a mismatched base at position 9 (P9) relative to the CAG repeat. Dose response curves for (A-H) P9-U6, P9-U7, P9-U8, P9-U9, P9-U10, P9-U11, P9-U12, and P9-U13 are shown. Dose curves are averaged data from three independent experiments and are based on western analysis of HTT inhibition in GM04281 patient-derived fibroblast cells.
Effect of UNA substitutions on cleavage of RNA targets during RNAi
Argonaute 2 (AGO2) is the catalytic engine of RNAi.45 When a duplex RNA is fully complementary to its RNA target, the anticipated outcome is that AGO2 will mediate recognition and cleavage of the target RNA. We had previously observed that a duplex RNA that is fully complementary to a CAG repeat will cause a modest reduction of RNA levels, while little or no reduction is observed when the duplex contains centrally-located mismatches.
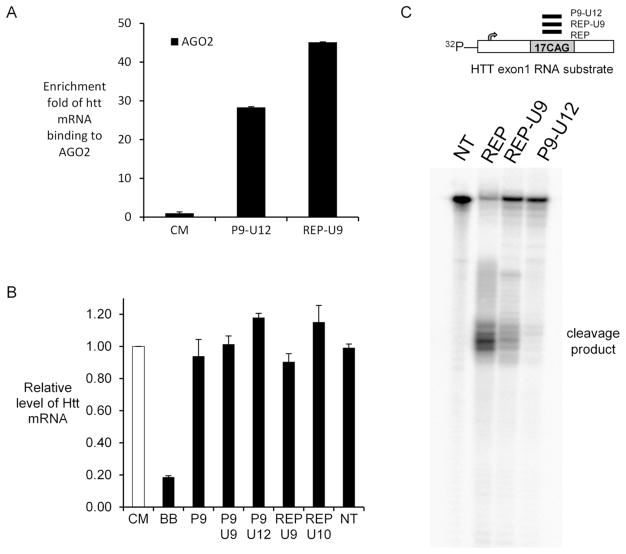
We tested whether UNA modified duplexes reduce HTT mRNA levels and whether recognition of UNAs involves the key RNAi factor AGO2. We transfected UNA duplexes into fibroblast cells and used RNA immunoprecipitation (RIP) with anti-AGO2 antibody to examine the recruitment of AGO2 to HTT mRNA. Effects on AGO2 recruitment were compared with a non-complementary control duplex (CM). Both UNA-modified duplexes recruited AGO2 to HTT mRNA (Figure 5A), consistent with functioning through RNAi.
Figure 5.
Effect of addition of UNAs on levels of HTT mRNA. (A) RNA immunoprecipitation (RIP) by use of an anti-AGO2 antibody to examine the association of AGO2/UNA-modified siRNA complex with HTT mRNA. (B) Quantitative PCR (qPCR) showing effect of UNA duplexes (25 nM) on levels of HTT RNA expression. (C) In vitro cleavage assay using RNA antisense strands and recombinant human AGO2 protein. NT: no treatment/no duplex added. BB is a duplex RNA that targets sequences of HTT mRNA outside the CAG repeat. CM is a control duplex lacking complementarity to HTT mRNA.
Next, we used quantitative PCR to test how UNA-substituted duplexes would affect the level of HTT mRNA (Figure 5B). Duplex RNA BB is a positive control siRNA targeting an outside region of CAG repeat in HTT mRNA and, as expected, it reduced HTT mRNA. We then tested four allele-selective potent UNA-modified duplexes and observed that all four duplexes caused little alteration of HTT mRNA level. These findings are consistent with the conclusion that UNA substitutions disrupt AGO2-mediated cleavage of target mRNA in a fashion similar to the introduction of central mismatches.
We further examined the effect of UNA substitutions on RNA cleavage through the RNAi pathway using a radiolabeled substrate HTT RNA with CAG repeats (Figure 5C). UNA-modified RNAs were individually incubated with AGO2 for a predetermined time to allow loading and formation of an AGO2/RNA complex. The complex was then mixed with the radiolabeled RNA substrate. The duplex RNAs substituted with UNA induced cleavage less than the fully complementary duplex, consistent with our qPCR data. Cleavage was almost undetectable when the duplex has both mismatch and UNA substitution at positions 9 and 12 position, respectively (P9-U12).
Taken together, data from RIP, qPCR, and in vitro cleavage assays show that the UNA substitution blocks cleavage of RNA substrates as effectively as introducing a mismatched base. These data suggest that allele-selective inhibition can be achieved without the need for cleavage of the target HTT mRNA and that these UNA-substituted duplexes function more like miRNAs than fully complementary siRNAs.
Effect of UNA substitutions on inhibition of ATX-3 expression by fully complementary and mismatch-containing duplexes
As noted above, several neurological diseases are caused by expanded CAG repeats and a single-anti-CAG compound may be able to treat several disease. To understand the potential of UNA-substituted duplexes to inhibit expression of other disease genes, we examined their ability to allele-selectively inhibit ATX-3 expression in patient-derived fibroblast cells.
Previously, we had observed that duplex RNAs that contained central mismatches could achieve allele-selective inhibition of ATX-3.36 The potencies and selectivities of ATX-3 modulation, however, were less than for inhibition of HTT expression and we identified fewer potent and selective duplexes. This result - that selectivity was different even though the CAG target sequence was found in both genes - suggested that the surrounding unique sequence of the HTT or ATX-3 genes contributed to selectivity and that ATX-3 was a more difficult target.
We tested UNA duplexes that were fully complementary to the CAG repeat and contained one or three UNA substitutions (Figure 2A). The three duplexes with one UNA substitution were allele-selective inhibitors (Figure 6). In contrast to inhibition of HTT expression, where inhibition by U9 or U11 had been more selective than inhibition by U10, for ATX-3 the highest selectivity was achieved by U10 with an IC50 value of 6.0 nM and a selectivity >8.3 fold.
Figure 6.
Effect of UNA substitutions on allele-selective inhibition of ATX-3 by RNA duplexes that are fully complementary to the CAG repeat. Western analyses of ATX-3 protein expression in GM06151 patient-derived fibroblast cells. (A) Effect of 25 nM UNA duplexes on inhibition of ATX-3 expression. (B) Summary of data. Dose response curves showing effect of adding (C) REP-U9 and (D) REP-U10 at increasing concentrations. NT: no treatment/no duplex added. Dose curves are averaged data from three independent experiments.
We also tested UNA duplexes that contained a single UNA substitution in combination with a mismatch at position 9 (Figure 3A). Several of these duplexes were allele-selective inhibitors of mutant HTT expression (Figure 7A) and P9-U12 and P9-U13 were chosen for further analysis (Figure 7B, C). P9-U12 possessed the best selectivity with an IC50 value of 5.5 nM and a selectivity of 6.4-fold.
Figure 7.
Effect of UNA substitutions on inhibition of ATX-3 expression. Duplexes were systematically substituted with UNA bases and contained mismatched base at position 9 (P9) relative to the CAG repeat target. (A) Effect of 25 nM UNA duplexes on inhibition of ATX-3 expression, analyzed by western analysis. (B) Summary of data. Dose response curves for inhibition of ATX-3 expression by (C) P9-U12 and (D) P9-U13. NT: no treatment/no duplex added. Dose response curves are averaged data from three independent experiments.
Discussion
Effects of the UNA substitution on AGO2 activity
RNA duplexes containing UNA modifications achieve allele-selective inhibition of HTT and ATX-3 expression. Both potency and selectivity are sensitive to shifting the position of UNA substitutions. Previously, we34,35,37–39 and others36 have shown that the introduction of a mismatch or abasic site that disrupt AGO2-mediated cleavage can lead to allele-selective anti-CAG duplexes. Allele-selective inhibition by UNA duplexes adds increased backbone flexibility as a new strategy for generating allele-selective duplexes.
Examination of the crystal structure data for human AGO2 in complex with RNA46 suggests that stacking is disrupted and kinks appears between bases 6–7, and 9–10. Furthermore, the ribose-phosphodiester backbone curves around bases 13 and 14. By introducing UNA residues at positions 9, 10, and 11, the flexibility of surrounding RNA backbone is increased. Increased flexibility may lead to slight displacement in strand position relative to catalytic residues, making the RNA strand a poorer substrate for cleavage by AGO2 mediated cleavage.
Trinucleotide repeats and the challenge of allele-selectivity
Genes that contain CAG trinucleotide repeats are responsible for up to nineteen different hereditary diseases. These diseases have severe consequences for patients and there are currently no curative treatments available. Useful drugs are urgently needed. Nucleic acids, with their potential to silence expression of disease genes, offer significant advantages as a strategy for therapeutic development.33
One approach to silencing trinucleotide repeat genes involves nonallele-selective antisense oligonucleotides or duplex RNAs.24,30,31 These compounds are active in vivo in mouse models of HD and may offer the most rapid route to human clinical trials. Reduced levels of the wild-type protein during nonallele-selective inhibition, however, may have adverse consequences.
Allele-selective strategies are a useful alternative in case trials with non-allele selective drugs encounter problems. Allele-selective inhibition can be achieved by antisense oligonucleotides23 or duplex RNAs25 that take advantage of single nucleotide polymorphisms to gain specificity for inhibiting expression of the mutant allele. While effective, the identity of SNPs varies between patients and it would be necessary to develop several drugs to treat most of the population for a disease like HD.26 By contrast, the expanded CAG repeat exists in all patients. Targeting the CAG repeat might not only be useful for all HD patients, but because the expanded CAG repeat is common among several diseases it is possible that one molecule might be able to treat several different hereditary pathologies.
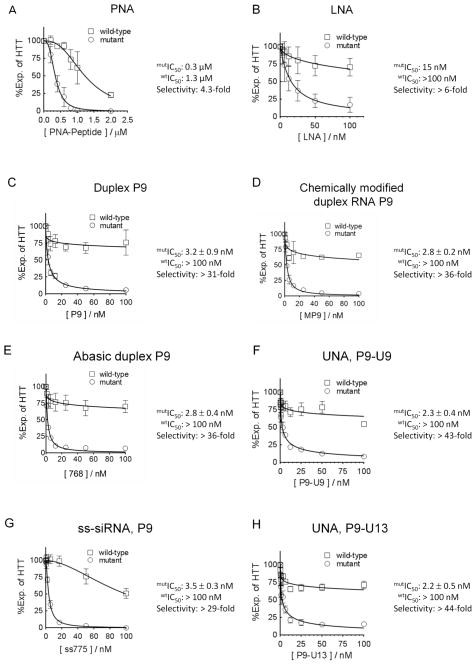
We have shown that several different types of synthetic oligomer can achieve allele-selective inhibition of HTT or ATX-3 expression (Figure 8). These oligomers include peptide nucleic acid (PNA),40 LNA or cEt bridged nucleic acid,40,44 mismatch-containing RNA duplexes with no chemical modifications,37 mismatched duplexes with extensive chemical modifications,40 mismatched duplexes with abasic substitutions,41 and single-stranded silencing RNAs.38,42 We find that potencies and selectivities for the best UNA match the best molecules from any other class of anti-CAG nucleic acid (Figure 8).
Figure 8.
Representative does response curves for inhibition of HTT expression by (A) PNA (peptide nucleic acid),39 (B) LNA (locked nucleic acid),40 (C) duplex siRNA with a central mismatch (P9),37 (D) chemically modified duplex siRNA with a central mismatch,41 (E) duplex siRNA with abasic substitution,41 (F) P9-U9, (G) single-stranded siRNA with a central mismatch,42 and (H) P9-U13. Dose response curves are averaged data from three independent experiments.
In contrast to previous work showing that the introduction of acyclic substitutions could enhance RNAse H activity,18 we observe that UNA substitutions block cleavage activity by AGO2. This difference emphasizes that the effect of UNA substitution will depend on context, include the properties of the enzyme involved in silencing and the exact placement of the UNA modifications. It is also worth noting that there are other acyclic modifications, like the 2′,3′-secouridine modifications used in the earlier study, and these might also provide a basis for allele-selective duplexes.
To be a drug, a nucleic acid inhibitor of mutant HTT expression will need to enter brain cells and be well-tolerated. This need to balance cellular uptake, potency, allele-selectivity, and toxicity presents a challenge for drug development. An encouraging outcome from our studies is that diverse nucleic acid chemistries are compatible with effective inhibition of mutant alleles. These chemistries, (UNA, abasic, ss-siRNA, 2′-modified) can be combined with each other or with mismatched bases at varied positions to create a large number of duplexes that are potent and selective. This pool of candidates then becomes a reservoir for testing compounds in animals to identify agents with optimal in vivo properties.
Supplementary Material
Acknowledgments
Funding Sources
Work in the Corey Laboratory was supported by the National Institutes of Health (NIGMS 73042), an award from the McKnight Foundation for Neuroscience, Cure Huntington’s Disease Initiative (CHDI) Inc. Foundation Inc., and the Robert A. Welch Foundation (I-1244); supported by a Young Investigator Award from the National Ataxia Foundation (to J.H.); supported by a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to Y.A.).
ABBREVIATIONS
- HTT
huntingtin
- ATX-3
ataxin-3
- UNA
unlocked nucleic acid
- AGO2
argonaute 2
- Tm
melting temperature
- HD
Huntington’s Disease
- MJD
Machado Joseph Disease
- RIP
RNA immunoprecipitation
Footnotes
Author Contributions
YA, JH, JL planned and executed experiments testing inhibition of HTT. CM and XQ designed and synthesized UNAs. DRC planned experiments and wrote the manuscript.
Representative gel images of the dose response curves for inhibition of HTT expression by UNA-modified P9 duplexes (Figure S1 and S2). This material is available free of charge via the Internet at HTTp://pubs.acs.org.
References
- 1.Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson JG. Management of familial hypercholesterolemia: a review of the recommendations from the national lipid association expert panel on familial hypercholesterolemia. J Managed Care Pharm. 2013;19:139–149. doi: 10.18553/jmcp.2013.19.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deleavey GF, Damha MJ. Designing Chemically Modified Oligonucleotides for Targeted Gene Silencing. Chem Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Obika S, Nanbu D, Hari Y, Andoh J-i, Morio K-i, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation. 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 5.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 6.Braasch DA, Liu YH, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundin KE, Hojland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J, Smith CIE. Biological Activity and Biotechnological Aspects of Locked Nucleic Acids. In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics. Vol. 82. 2013. pp. 47–107. [DOI] [PubMed] [Google Scholar]
- 8.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostergaard ME, Cheguru P, Papasani MR, Hill RA, Hrdlicka PJ. Glowing Locked Nucleic Acids: Brightly Fluorescent Probes for Detection of Nucleic Acids in Cells. J Am Chem Soc. 2010;132:14221–14228. doi: 10.1021/ja1057295. [DOI] [PubMed] [Google Scholar]
- 10.Yahara A, Shrestha AR, Yamamoto T, Hari Y, Osawa T, Yamaguchi M, Nishida M, Kodama T, Obika S. Amido-Bridged Nucleic Acids (AmNAs): Synthesis, Duplex Stability, Nuclease Resistance, and in Vitro Antisense Potency. Chem Bio Chem. 2012;13:2513–2516. doi: 10.1002/cbic.201200506. [DOI] [PubMed] [Google Scholar]
- 11.Prakash TP. An Overview of Sugar-Modified Oligonucleotides for Antisense Therapeutics. Chem Biodivers. 2011;8:1616–1641. doi: 10.1002/cbdv.201100081. [DOI] [PubMed] [Google Scholar]
- 12.Pasternak A, Wengel J. Unlocked nucleic acid - an RNA modification with broad potential. Org Biomol Chem. 2011;9:3591–3597. doi: 10.1039/c0ob01085e. [DOI] [PubMed] [Google Scholar]
- 13.Campbell MA, Wengel J. Locked vs. unlocked nucleic acids (LNA vs UNA): contrasting structures work towards common therapeutic goals. Chem Soc Rev. 2011;40:5680–5689. doi: 10.1039/c1cs15048k. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen P, Dreioe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: Introduction of three novel analogues and a summary. Biorg Med Chem. 1995;3:19–28. doi: 10.1016/0968-0896(94)00143-q. [DOI] [PubMed] [Google Scholar]
- 15.Vaish N, Chen F, Seth S, Fosnaugh K, Liu Y, Adami R, Brown T, Chen Y, Harvie P, Johns R, Severson G, Granger B, Charmley P, Houston M, Templin MV, Polisky B. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, Herdewijn P, Wengel J, Kjems J. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, Langklaer N, Wengel SL, Wengel J, Kjems J, Bramsen JB. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol BioSyst. 2010;6:862–870. doi: 10.1039/b918869j. [DOI] [PubMed] [Google Scholar]
- 18.Mangos MM, Min KL, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-deficient cleavage of RNA promoted by antisense DNA or 2F-ANA constructs containing acyclic nucleotide inserts. J Am Chem Soc. 2003;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 19.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 20.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 21.Costa Mdo C, Paulson HL. Toward understanding Machado-Joseph disease. Prog Neurobiol. 2012;97:239–257. doi: 10.1016/j.pneurobio.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulson HL. Dominantly inherited ataxias: Lessons learned from Machado Joseph disease/spinocerebellar atria type 3. Sem Neurol. 2007;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- 23.Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and Selective Antisense Oligonucleotides Targeting Single-Nucleotide Polymorphisms in the Huntington Disease Gene/Allele-Specific Silencing of Mutant Huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, Hung G, Bennett CF, Cleveland DW. Sustained Therapeutic Reversal of Huntington’s Disease by Transient Repression of Huntingtin Synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. Plos Genetics. 2006;2:1307–1318. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs Targeting Three SNPs May Provide Therapy for Three-Quarters of Huntington’s Disease Patients. Current Biology. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller VM, Xia HB, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper SQ, Staber PD, He XH, Eliason SL, Martins IH, Mao QW, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DWY, Zamore PD, Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, Deglon N. Sustained Effects of Nonallele-Specific Huntingtin Silencing. Annals of Neurology. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 31.Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific Silencing of Mutant and Wild-type Huntingtin Demonstrates Therapeutic Efficacy in Huntington’s Disease Mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alves S, Nascimento-Ferreira I, Dufour N, Hassig R, Auregan G, Nobrega C, Brouillet E, Hantraye P, Pedroso de Lima MC, Deglon N, de Almeida LP. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum Mol Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 33.Sah DWY, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Liu J, Yu D, Chu Y, Corey DR. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res. 2012;40:11270–11280. doi: 10.1093/nar/gks907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J, Gagnon KT, Liu J, Watts JK, Syeda-Nawaz J, Bennett CF, Swayze EE, Randolph J, Chattopadhyaya J, Corey DR. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiszer A, Mykowska A, Krzyzosiak WJ. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, Liu J, Corey DR. Allele-Selective Inhibition of Huntingtin Expression by Switching to an miRNA-like RNAi Mechanism. Chem Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Yu D, Aiba Y, Hannah P, Swayze EE, Lima WF, Hu J, Prakash TP, Corey DR. ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucl Acids Res. 2013 doi: 10.1093/nar/gkt693. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsui M, Corey DR. Allele-selective inhibition of trinucleotide repeat genes. Drug Discovery Today. 2012;17:443–450. doi: 10.1016/j.drudis.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Pendergraff H, Narayanannair KJ, Lackey JG, Kuchimanchi S, Rajeev KG, Manoharan M, Hu J, Corey DR. RNA duplexes with abasic substitutions are potent and allele-selective inhibitors of huntingtin and ataxin-3 expression. Nucleic Acids Res. 2013;41:8788–8801. doi: 10.1093/nar/gkt594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP, Corey DR. Single-Stranded RNAs Use RNAi to Potently and Allele-Selectively Inhibit Mutant Huntingtin Expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Hu J, Corey DR. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 2012;40:1240–1250. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, Montaillier C, Lemaitre M, Corey DR. Allele-Selective Inhibition of Mutant Huntingtin Expression with Antisense Oligonucleotides Targeting the Expanded CAG Repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 46.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The Structure of Human Argonaute-2 in Complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.