Abstract
Identifying genes associated with cancer development is typically accomplished by comparing mean expression values in normal and tumor tissues, which identifies differentially expressed (DE) genes. Interindividual variation (IV) in gene expression is indirectly included in DE gene identification because given the same absolute differences in means, genes with lower variance tend to have lower P values. We explored the direct use of IV in gene expression to identify candidate genes associated with cancer development. We focused on prostate (PCa) and lung (LC) cancers and compared IV in the expression level of genes shown to be cancer related with that in all other genes in the human genome. Compared with all those other genes, cancer-related genes tended to have greater IV in normal tissues and a greater increase in IV during the transition from normal to tumorous tissue. Genes without significantly different mean expression values between tumor and normal tissues but with greater IV in tumor than in normal tissue (note: the DE-based approach completely ignores those genes) had stronger associations with clinically important features like Gleason score in PCa or tumor histology in LC than all other genes were. Our results suggest that analyzing IV in gene expression level is useful in identifying novel candidate genes associated with cancer development.
Keywords: Gene expression; interindividual variation in gene expression, prostate cancer, lung cancer
1. Background
Genome-wide profiling of gene expression is frequently used to identify cancer-related genes. The traditional approach compares the mean expression in tumor and normal tissues and identifies differentially expressed (DE) genes, which are usually considered candidate genes associated with cancer initiation and/or progression.1-3 Interindividual variation (IV) in the expression level, which is usually estimated as variance, is used indirectly in such analyses as part of the corresponding statistical test.
A number of studies suggest that IV in gene expression in tumor and in normal tissue of the type in which the tumor originated plays a crucial role in cancer heterogeneity at the level of clinical features. This is evident from studies conducted on breast cancer,4 for example, as well as other types of cancer.5 A number of genes with strong IV in expression level, e.g., HER-2, ER, and p53, have been shown to play an essential role in breast cancer initiation and progression.6,7 This suggests that identification of genes with strong IV in expression level will be useful in the detection of cancer-related genes. No studies on the link between the variation in gene expression and a gene’s probability of being cancer related have yet been conducted. With this study, we aimed to fill the gap between the analysis of IV in gene expression and the identification of candidate DE genes. Using lung and prostate cancer (LC and PCa) as examples, we demonstrated that taking into account the IV in gene expression may help identify novel candidate genes that are missed by the classical approach of analyzing the DE genes.
2. Methods
A relatively large sample is required to obtain a reliable estimate of IV. To meet this requirement, we used the publicly available gene expression data from the two largest LC and PCa studies included in the Gene Expression Omnibus (GEO) database. The LC data came from the study by Hou et al.,8 and the PCa data, from the study by Chandran et al.9 Table 1 briefly summarizes those datasets.
Table 1.
Summary of the studies used in our analysis
| Cancer type | GEO ID | No. of adjacent normal tissues |
No. of tumor tissues |
No. of probes |
|---|---|---|---|---|
| Lung | 19188 | 65 | 91 | 54,675 |
| Prostate | 6919 | 63 | 66 | 12,553 |
To identify PCa- and LC-related genes, we used the KnowledgeNet approach,10 which combines literature mining with gene-classification data from the Gene Ontology database.11 For functional annotation, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID).12 DAVID tests the null hypothesis that genes are uniformly distributed across pathways and biologic functions. The resulting P values characterize the strength of the statistical evidence for clustering: the lower the P value, the stronger the evidence that the genes are overrepresented in a specific pathway.
Most comparisons were made between KnowledgeNet-identified cancer-related genes and all other genes in the dataset. To test for tissue specificity for each type of cancer, we separately compared LC- and PCa-related genes with all other genes in the dataset. Correlation analysis was used to test for a relationship between Gleason score and IV. To assess an association between IV and histologic type of lung cancer, we used ANOVA. Student’s t test was used to compare mean expression values. Log-transformed and normalized expression values were used. Because there was no significant correlation between variance and mean expression values in the processed gene expression data, we used variance in the gene expression as a measure of IV. For each probe, we computed the ratio between the variance in the tumor and that in normal tissue separately for cancer-related and all other genes. We used SAS software (SAS Institute, Inc., Cary, North Carolina, USA) for performing the statistical analyses.
3. Results
3.1. Cancer-related genes have higher IV in normal tissues than all the other genes have
We identified 200 genes related to LC and 205 related to PCa (see the Appendix for a complete list). Some overlap exists between LC and PCa genes: there are only 167 unique LC genes and 162 unique PCa genes.
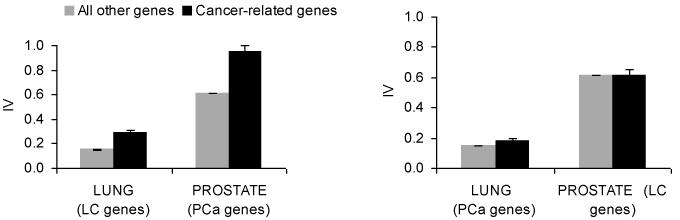
We found that compared with all other genes, LC-related genes had a higher IV in normal lung tissue but not in normal prostate tissue. Likewise, the PCa-related genes had a higher IV in normal prostate tissue but not in normal lung tissue (Figure 1).
Figure 1.
(Left) Interindividual variance (IV) in normal lung (or prostate) tissue for the lung (or prostate) cancer-related and all other genes. (Right) IV in the adjacent normal lung/prostate tissue for the prostate/lung cancer-related and all other genes when tissue type is different from gene type.
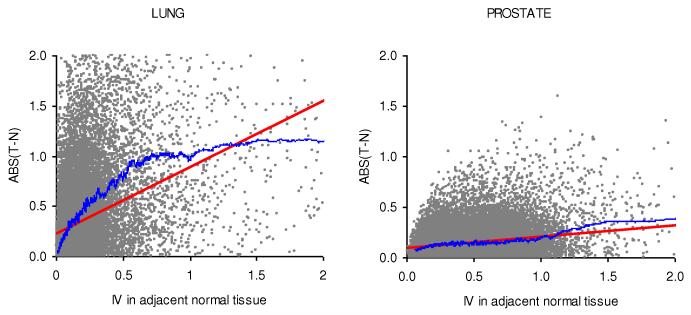
In addition, we assessed whether the genes with higher IV in normal tissue were expressed differently in tumorous and adjacent normal tissues. We estimated the correlation between the IV in normal tissue and the absolute difference in gene expression between tumor and adjacent normal tissues (Figure 2). The correlation between those two variables was positive for both LC (R = 0.43, N = 54,675, P << 10−6) and PCa (R = 0.26, N = 37,690, P << 10−6). The observed correlations can not be explained by the effect of sampling from a population with a higher variance. Indeed, aside from the correlation between the IV in normal tissue and absolute differences in expression levels, we have also noted a positive correlation between the IV in normal tissue and absolute values of t-statistics for both LC (R = 0.23, N = 54,675, P << 10−6) and PCa (R = 0.06, N = 37,690, P << 10−6). These positive correlations are counterintuitive because if we assume the same level of differentiation, e.g. the same level of fold change for high and low IV genes the absolute values of t-statistics are expected to be lower (not higher as we have observed) for genes with higher IV.
Figure 2.
An association between IV in adjacent normal tissue and absolute differences in the expression levels between normal tissue (N) and tumor (T). Each dot represents a probe. The red line is a linear regression curve, and the blue line is a moving average computed for the 250 closest probes in terms of variance. There is a positive correlation between IV and absolute differences in the expression levels between N and T in both lung (left) and prostate (right) cancers.
3.2. Genes with the highest IV in normal tissues cluster in a small number of functional categories
We performed functional annotation of the top 5% of the genes with the highest IV. The analysis was done separately for normal lung and normal prostate tissues. The top 5% was used because our previous analyses indicated that this percentage is optimal in terms of robustness of clustering and in the proportion of false positives included in the annotation list.13,14 For LC genes, the top functional categories were “extracellular region,” “inflammation,” “angiogenesis,” “chemotaxis,” and “cell adhesion,” whereas for the PCa genes, they were “actin cytoskeleton” and “cell adhesion.” It is interesting that we previously identified those same functions by analyzing the genes that are expressed differently in normal versus tumorous tissue.13-15 The overlap at the functional level was partially driven by the overlap at the gene level, though at the functional level it was more prominent, similarly as it was found in our previous study.15 .
3.3. IV is higher in tumor than it is in adjacent normal tissue
Overall, the IV in gene expression in tumorous tissue was higher than it was in adjacent normal tissue: for lung cancer, the mean ratio between variance in tumor and that in normal tissue was 3.29 ± 0.04, and for prostate cancer, it was 1.28 ± 0.01. In both cases, the mean ratio was greater than 1, which is to be expected under the null hypothesis.
3.4. For the cancer-related genes, IV increases more than it does in the other genes
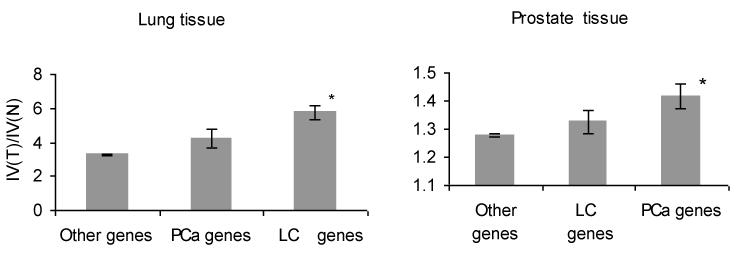
For the LC-related genes, the ratio of the IV between lung tumor and adjacent normal tissue was 5.76 ± 0.82. All other (not LC-related) genes showed a smaller ratio: 3.28 ± 0.04. The increase is tissue specific: for the PCa-related genes, the ratio of the IV between lung tumor and normal lung was 4.21 ± 0.56, which was not significantly different for all other genes in the dataset: t test = 1.66, N = 37,690, P < 0.21 (Figure 3, left).
Figure 3.
Ratios of IVs between tumor and adjacent normal tissues: LC-related, PCa-related, and all other genes. Left panel shows lung, and the right panel shows prostate tissues. Note that the ratio for PCa genes in lung tissue seems to be slightly elevated, most likely due to overlap between PCa- and LC-related genes. The same is true for LC genes in prostate tissue.
For the PCa-related genes, the mean ratio of the IV between tumorous and adjacent normal prostate tissue was 1.41 ± 0.06, which is significantly higher than the mean ratio for all other genes (1.24 ± 0.01; t test = 2.17, N = 37,690, P < 0.01). Again, the difference between those two ratios was tissue specific: the mean ratio for LC-related genes in prostate tissue was 1.33 ± 0.05, which was not statistically significant from that for all other genes: t test = 0.66, N = 37,690, P < 0.84 (Figure 3, right).
3.5. Cancer-related genes can be identified by analyzing the IV
We took the top 1% of the genes that had the highest increase in IV in prostate tumor compared with that in the adjacent normal tissues. From among those genes, we identified a subset of 96 that had no significant differences in mean expression level between normal and tumorous tissues (i.e., P >0.05). It is important to note that those genes are ignored by a traditional analysis that compares mean expression values.
To assess whether the IV can be used to identify cancer-related genes, we estimated the correlation between the expression level of those 96 genes in tumor and the Gleason score (GS) in PCa patients. GS is a key clinical characteristic that is associated with PCa progression and patients’ survival.16 We found that the absolute value of the Spearman’s correlation coefficient (ρ) was 0.14 ± 0.01 for the 96 genes, which was significantly higher than that for other genes in the human genome: ρ = 0.10 ± 0.01, P < 0.001. The top genes we identified as being strongly associated with GS are CD74, EEF1A1, HLA-F, MAPK12, NFYC, RCL1, RPL9, RPS23, RPS3A, TFDP1, TREM2, and ZNF789.
A similar approach was used to identify LC-related genes. We took 107 genes with the highest increase in IV and no significant difference in mean expression between normal and tumorous tissues. Those genes were more likely than all other genes in the dataset to be differently expressed in different histologic types of LC: adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma. The average F statistic was 10.4 ± 0.9 for the LC-related genes with increased IV in expression and 4.3 ± 0.1 for the average gene. The top genes we identified as having a different level of expression in different histologic types of LC are DCX, CADPS, DLK1, GRIA2, HESRG, KRTDAP, MTMR7, SEZ6L, STXBP5L, and TPTE.
4. Discussion
Our results showed that (i) cancer-related genes have greater IV in normal tissues and (ii) there is a greater increase in IV in the transition from normal to tumorous tissue than there is for other (non–cancer-related) genes. We believe that tumor heterogeneity may explain both these observations. Ample evidence exists to show that both LC17-19 and PCa20-22 are heterogeneous at the gene expression level. This may underlie both the increased IV in the expression of cancer-related genes and the increased IV in gene expression in the transition from normal to tumorous tissue. Indeed, to be able to influence cancer risk, a gene must be important for cancer development and also must have substantial IV in its expression level. Different tumors may “use” different genes for progression. If, for example, some tumors are driven by increased expression of gene A and other tumors by increased expression of gene B, then the IV in expression will be increased in tumor samples for both genes.
It is more difficult, however, to explain why an elevated IV in normal tissue is higher in cancer-associated genes than it is in all other genes in the human genome. In our preliminary analysis (results not shown), we found that a significant fraction of genes in normal prostate tissue show a bimodal distribution in gene expression. For example, the distribution of the expression of the KLK3 gene, which is crucial for prostate tumorigenesis, is bimodal in normal prostate tissue, with non-overlapping low and high expression variants. In tumor samples, however, only the high expression variant is present. This suggests that normal tissue with a high level of KLK3 expression is more likely to develop tumor than is that with a low level of its expression. Bimodal distribution is also a reason for the high variance of KLK3 expression in normal prostate tissue. In general, we believe that selection for this kind of preexisting variation in gene expression during carcinogenesis may cause differences in gene expression between normal and cancerous tissues and result in the observed association.
Our findings that interindividual heterogeneity at the gene expression level is higher in tumors than in normal tissue suggest that there are multiple paths from the normal to tumorous gene expression patterns. This observation does not contradict clonal expansion hypothesis that assumes a survival of meanest (most aggressive) from originally heterogeneous cell population. Our results simply suggest that there are many ways to be “mean” and different tumors are “mean” in different ways, which is demonstrated by the analysis at the gene expression level.
Overall, we found that the association between IV and ABS(T-N) was stronger for LC than it was for PCa (Figure 2). One possible explanation for the differences may be differences in tumor biology. Prostate tumors are usually diagnosed by screening, and many of them are slow-growing tumors, allowing the use of a watchful waiting strategy in many cases.23 Lung cancer, however, is typically diagnosed through symptoms and is often incurable after its detection.24 So it is possible that for PCa we are in fact comparing the gene expression in normal tissue with that in early stages of tumorigenesis, whereas in the case of LC, we are comparing gene expression in normal tissue and advanced tumors. This idea is supported by our observation of a stronger correlation between the IV in adjacent normal tissue and the absolute differences in expression levels between primary normal and metastatic prostate tumors (correlation coefficient ρ = 0.39, N = 37,690, P << 10−6), which is significantly higher than the correlation between the IV in normal tissue and the absolute difference in gene expression between tumor and adjacent normal tissues (Figure 2, R = 0.26, N = 37,690, P << 10−6).
Our results also indicate that a more effective approach than is currently used for identifying cancer-related genes will include both the traditional approach of comparing the mean gene expression levels and an analysis of the IV. The key remaining question is how best to combine these approaches.
5. Conclusions
The results of this analysis suggest that when combined with the traditional, mean-based approach to identifying cancer-related genes, the IV-based approach can facilitate the detection of cancer-related genes that are missed by the traditional approach.
Acknowledgments
This study was supported by the David H. Koch Center for Applied Research of Genitourinary Cancers; National Institutes of Health Prostate SPORE grant 1 P50 CA140388-01; and NIH grants R01 CA149462 and R01 AR055258 (both to O.Y.G.). It is also supported in part by the NIH through MD Anderson’s Cancer Center Support Grant, 5 P30 CA16672 and by NSFC (No. 31100958).
Appendix A
List of the prostate cancer (PCa)– and lung cancer (LC)–related genes identified by the KnowledgeNet approach, including their EntrezGene numbers.
| Cancer | Gene Symbol | EntrezGene | confidence score(SD) |
|---|---|---|---|
| PCa | AR | 367 | 2.663 |
| PCa | KLK3 | 354 | 0.785 |
| PCa | CDKN1B | 1027 | 0.493 |
| PCa | AMACR | 23600 | 0.478 |
| PCa | IGFBP3 | 3486 | 0.464 |
| PCa | PTEN | 5728 | 0.401 |
| PCa | TP53 | 7157 | 0.387 |
| PCa | NOS3 | 4846 | 0.385 |
| PCa | CDH1 | 999 | 0.382 |
| PCa | SRD5A2 | 6716 | 0.362 |
| PCa | ELAC2 | 60528 | 0.326 |
| PCa | EGFR | 1956 | 0.311 |
| PCa | BCL2 | 596 | 0.304 |
| PCa | TGFBI | 7045 | 0.301 |
| PCa | NKX3-1 | 4824 | 0.277 |
| PCa | IL6 | 3569 | 0.258 |
| PCa | GSTP1 | 2950 | 0.249 |
| PCa | IGF1 | 3479 | 0.245 |
| PCa | GDF15 | 9518 | 0.207 |
| PCa | VEGFA | 7422 | 0.186 |
| PCa | MAPK8 | 5599 | 0.181 |
| PCa | VDR | 7421 | 0.178 |
| PCa | CDKN1A | 1026 | 0.174 |
| PCa | ESR2 | 2100 | 0.166 |
| PCa | TRPS1 | 7227 | 0.165 |
| PCa | PTGS2 | 5743 | 0.161 |
| PCa | MSH2 | 4436 | 0.157 |
| PCa | MSR1 | 4481 | 0.156 |
| PCa | SDC1 | 6382 | 0.154 |
| PCa | ACPP | 55 | 0.153 |
| PCa | SKP2 | 6502 | 0.15 |
| PCa | CD82 | 3732 | 0.148 |
| PCa | KLK11 | 11012 | 0.146 |
| PCa | ITGB3 | 3690 | 0.145 |
| PCa | PPARG | 5468 | 0.142 |
| PCa | ERBB3 | 2065 | 0.138 |
| PCa | MET | 4233 | 0.138 |
| PCa | MTA1 | 9112 | 0.138 |
| PCa | PCA3 | 50652 | 0.138 |
| PCa | LEP | 3952 | 0.137 |
| PCa | PSCA | 8000 | 0.137 |
| PCa | PRKCE | 5581 | 0.135 |
| PCa | BMP5 | 653 | 0.134 |
| PCa | HIF1A | 3091 | 0.134 |
| PCa | SMAD4 | 4089 | 0.132 |
| PCa | ERBB2 | 2064 | 0.131 |
| PCa | STAT3 | 6774 | 0.128 |
| PCa | JUND | 3727 | 0.127 |
| PCa | FOLH1 | 2346 | 0.125 |
| PCa | STEAP1 | 26872 | 0.125 |
| PCa | BMP2 | 650 | 0.124 |
| PCa | ALOX15B | 247 | 0.123 |
| PCa | ID1 | 3397 | 0.122 |
| PCa | MMP9 | 4318 | 0.122 |
| PCa | CXCL12 | 6387 | 0.12 |
| PCa | FGF8 | 2253 | 0.12 |
| PCa | PTHLH | 5744 | 0.118 |
| PCa | RNF14 | 9604 | 0.118 |
| PCa | XRCC1 | 7515 | 0.117 |
| PCa | KLK2 | 3817 | 0.115 |
| PCa | TIMP1 | 7076 | 0.113 |
| PCa | ALOX12 | 239 | 0.112 |
| PCa | SLC30A4 | 7782 | 0.111 |
| PCa | OR51E2 | 81285 | 0.11 |
| PCa | GSK3B | 2932 | 0.108 |
| PCa | ITGAV | 3685 | 0.108 |
| PCa | RCBTB2 | 1102 | 0.107 |
| PCa | NAT2 | 10 | 0.106 |
| PCa | CHEK2 | 11200 | 0.105 |
| PCa | KLK10 | 5655 | 0.105 |
| PCa | PRKCA | 5578 | 0.104 |
| PCa | MAP2K5 | 5607 | 0.102 |
| PCa | ANP32C | 23520 | 0.101 |
| PCa | CCND2 | 894 | 0.101 |
| PCa | GSTM1 | 2944 | 0.099 |
| PCa | SRD5A1 | 6715 | 0.098 |
| PCa | RNASEL | 6041 | 0.097 |
| PCa | CARM1 | 10498 | 0.096 |
| PCa | RXRA | 6256 | 0.096 |
| PCa | CHGA | 1113 | 0.094 |
| PCa | PIM1 | 5292 | 0.094 |
| PCa | CCND1 | 595 | 0.092 |
| PCa | ANP32D | 23519 | 0.091 |
| PCa | BAX | 581 | 0.09 |
| PCa | ENG | 2022 | 0.09 |
| PCa | NRP1 | 8829 | 0.09 |
| PCa | EZH2 | 2146 | 0.088 |
| PCa | FLT4 | 2324 | 0.088 |
| PCa | KLK14 | 43847 | 0.088 |
| PCa | NFKB1 | 4790 | 0.088 |
| PCa | BCL2L1 | 598 | 0.087 |
| PCa | HIP1 | 3092 | 0.087 |
| PCa | REPS2 | 9185 | 0.087 |
| PCa | KLK4 | 9622 | 0.086 |
| PCa | SSTR2 | 6752 | 0.084 |
| PCa | HGF | 3082 | 0.083 |
| PCa | HOXC8 | 3224 | 0.083 |
| PCa | IGFBP7 | 3490 | 0.083 |
| PCa | IL8 | 3576 | 0.083 |
| PCa | NCOR2 | 9612 | 0.083 |
| PCa | DAB2IP | 153090 | 0.082 |
| PCa | TMPRSS2 | 7113 | 0.082 |
| PCa | CYP1A1 | 1543 | 0.081 |
| PCa | GAGE1 | 2543 | 0.081 |
| PCa | GAGE12I | 26748 | 0.081 |
| PCa | GAGE2C | 2574 | 0.081 |
| PCa | GAGE2E | 26749 | 0.081 |
| PCa | GAGE3 | 2575 | 0.081 |
| PCa | GAGE4 | 2577 | 0.081 |
| PCa | GAGE5 | 2576 | 0.081 |
| PCa | GAGE6 | 2578 | 0.081 |
| PCa | GAGE7 | 2579 | 0.081 |
| PCa | PAGE1 | 8712 | 0.081 |
| PCa | CFLAR | 8837 | 0.079 |
| PCa | IGFBP2 | 3485 | 0.079 |
| PCa | ITGA6 | 3655 | 0.079 |
| PCa | NCOA3 | 8202 | 0.079 |
| PCa | CAV1 | 857 | 0.078 |
| PCa | LIMK1 | 3984 | 0.077 |
| PCa | ESR1 | 2099 | 0.076 |
| PCa | FASN | 2194 | 0.076 |
| PCa | MMP14 | 4323 | 0.076 |
| PCa | MMP2 | 4313 | 0.076 |
| PCa | STEAP2 | 261729 | 0.076 |
| PCa | TERT | 7015 | 0.076 |
| PCa | CLU | 1191 | 0.075 |
| PCa | RASSF1 | 11186 | 0.075 |
| PCa | C15orf21 | 283651 | 0.074 |
| PCa | MMP26 | 56547 | 0.074 |
| PCa | SULT2B1 | 6820 | 0.074 |
| PCa | ALOX5 | 240 | 0.073 |
| PCa | TRPV6 | 55503 | 0.073 |
| PCa | ITGA3 | 3675 | 0.072 |
| PCa | CTAG1B | 1485 | 0.071 |
| PCa | GRN | 2896 | 0.071 |
| PCa | PNN | 5411 | 0.071 |
| PCa | PRKD1 | 5587 | 0.071 |
| PCa | SERPINB5 | 5268 | 0.071 |
| PCa | SFN | 2810 | 0.07 |
| PCa | GHRH | 2691 | 0.069 |
| PCa | TNFSF10 | 8743 | 0.069 |
| PCa | ALOX15 | 246 | 0.068 |
| PCa | MCAM | 4162 | 0.068 |
| PCa | SPDEF | 25803 | 0.067 |
| PCa | SSTR1 | 6751 | 0.067 |
| PCa | SSTR3 | 6753 | 0.067 |
| PCa | ST7 | 7982 | 0.067 |
| PCa | TIMP2 | 7077 | 0.066 |
| PCa | ZNF185 | 7739 | 0.066 |
| PCa | GHRHR | 2692 | 0.065 |
| PCa | KLK13 | 26085 | 0.065 |
| PCa | KLK15 | 55554 | 0.065 |
| PCa | SFRP4 | 6424 | 0.065 |
| PCa | CDC25A | 993 | 0.064 |
| PCa | CDKN2A | 1029 | 0.064 |
| PCa | LSM1 | 27257 | 0.063 |
| PCa | PCAP | 7834 | 0.063 |
| PCa | SREBF1 | 6720 | 0.063 |
| PCa | SREBF2 | 6721 | 0.063 |
| PCa | TRIM68 | 55128 | 0.063 |
| PCa | BTG2 | 7832 | 0.062 |
| PCa | CASP8 | 841 | 0.062 |
| PCa | EEF1A1 | 1915 | 0.062 |
| PCa | MED15 | 51586 | 0.062 |
| PCa | OGG1 | 4968 | 0.062 |
| PCa | RARRES1 | 5918 | 0.062 |
| PCa | APOE | 348 | 0.061 |
| PCa | CYP27B1 | 1594 | 0.061 |
| PCa | HPN | 3249 | 0.061 |
| PCa | PPFIA2 | 8499 | 0.061 |
| PCa | TEGT | 7009 | 0.061 |
| PCa | CPA4 | 51200 | 0.06 |
| PCa | EPHA2 | 1969 | 0.06 |
| PCa | IGFBP1 | 3484 | 0.06 |
| PCa | PROS1 | 5627 | 0.06 |
| PCa | EIF3H | 8667 | 0.059 |
| PCa | SLC43A1 | 8501 | 0.059 |
| PCa | AKT1 | 207 | 0.058 |
| PCa | FXYD3 | 5349 | 0.058 |
| PCa | KLF6 | 1316 | 0.058 |
| PCa | TNFRSF11B | 4982 | 0.058 |
| PCa | ITGB4 | 3691 | 0.057 |
| PCa | PLK1 | 5347 | 0.057 |
| PCa | RORA | 6095 | 0.057 |
| PCa | WFDC1 | 58189 | 0.057 |
| PCa | CSMD1 | 64478 | 0.056 |
| PCa | NUDC | 10726 | 0.056 |
| PCa | PMEPA1 | 56937 | 0.055 |
| PCa | TGFB1I1 | 7041 | 0.055 |
| PCa | CXCR4 | 7852 | 0.054 |
| PCa | PAWR | 5074 | 0.054 |
| PCa | NCOA4 | 8031 | 0.053 |
| PCa | ADAMTS13 | 11093 | 0.052 |
| PCa | CSRP2 | 1466 | 0.052 |
| PCa | GJA1 | 2697 | 0.052 |
| PCa | GJB1 | 2705 | 0.052 |
| PCa | IL10 | 3586 | 0.052 |
| PCa | PARP1 | 142 | 0.052 |
| PCa | PDZD2 | 23037 | 0.052 |
| PCa | SEMG1 | 6406 | 0.052 |
| PCa | FLT1 | 2321 | 0.051 |
| PCa | MT3 | 4504 | 0.051 |
| PCa | TPTE2 | 93492 | 0.051 |
| PCa | VIM | 7431 | 0.051 |
| PCa | FGF1 | 2246 | 0.05 |
| LC | EGFR | 1956 | 2.69 |
| LC | GSTM1 | 2944 | 0.857 |
| LC | SKP2 | 6502 | 0.722 |
| LC | TP53 | 7157 | 0.684 |
| LC | CXCR4 | 7852 | 0.673 |
| LC | GSTP1 | 2950 | 0.619 |
| LC | CYP1A1 | 1543 | 0.568 |
| LC | ERBB2 | 2064 | 0.533 |
| LC | RASSF1 | 11186 | 0.462 |
| LC | CADM1 | 23705 | 0.445 |
| LC | MPO | 4353 | 0.404 |
| LC | PTGS2 | 5743 | 0.343 |
| LC | CDKN2A | 1029 | 0.343 |
| LC | IGFBP3 | 3486 | 0.329 |
| LC | KRAS | 3845 | 0.306 |
| LC | IL1B | 3553 | 0.305 |
| LC | GSTT1 | 2952 | 0.29 |
| LC | BIRC3 | 330 | 0.287 |
| LC | BIRC2 | 329 | 0.286 |
| LC | MMP2 | 4313 | 0.244 |
| LC | XIAP | 331 | 0.235 |
| LC | KRT8 | 3856 | 0.229 |
| LC | FHIT | 2272 | 0.229 |
| LC | VEGFA | 7422 | 0.22 |
| LC | BCL2 | 596 | 0.219 |
| LC | OGG1 | 4968 | 0.217 |
| LC | CYP2A13 | 1553 | 0.21 |
| LC | PLAUR | 5329 | 0.205 |
| LC | PLAU | 5328 | 0.205 |
| LC | LGALS3 | 3958 | 0.205 |
| LC | CDH1 | 999 | 0.2 |
| LC | FASN | 2194 | 0.189 |
| LC | MGMT | 4255 | 0.188 |
| LC | NQO1 | 1728 | 0.185 |
| LC | RALBP1 | 10928 | 0.183 |
| LC | ING1 | 3621 | 0.183 |
| LC | LGALS3BP | 3959 | 0.182 |
| LC | SEMA3B | 7869 | 0.17 |
| LC | IGF1 | 3479 | 0.169 |
| LC | FAS | 355 | 0.167 |
| LC | IL8 | 3576 | 0.166 |
| LC | MYO18B | 84700 | 0.161 |
| LC | CDKN1B | 1027 | 0.155 |
| LC | GRP | 2922 | 0.154 |
| LC | CTNNB1 | 1499 | 0.154 |
| LC | ASCL1 | 429 | 0.15 |
| LC | SLPI | 6590 | 0.146 |
| LC | NKX2-1 | 7080 | 0.145 |
| LC | AREG | 374 | 0.144 |
| LC | SOCS3 | 9021 | 0.142 |
| LC | MET | 4233 | 0.142 |
| LC | CDH13 | 1012 | 0.142 |
| LC | SFTPB | 6439 | 0.14 |
| LC | ERCC2 | 2068 | 0.14 |
| LC | CXCL12 | 6387 | 0.138 |
| LC | MMP9 | 4318 | 0.137 |
| LC | MAPK1 | 5594 | 0.137 |
| LC | CTAG2 | 30848 | 0.137 |
| LC | PTEN | 5728 | 0.136 |
| LC | CASP8 | 841 | 0.136 |
| LC | SMARCA4 | 6597 | 0.135 |
| LC | RBL2 | 5934 | 0.133 |
| LC | TUBB2A | 7280 | 0.131 |
| LC | PRKCE | 5581 | 0.129 |
| LC | ITGA9 | 3680 | 0.128 |
| LC | RHOA | 387 | 0.127 |
| LC | MAGEC2 | 51438 | 0.124 |
| LC | FEN1 | 2237 | 0.123 |
| LC | COX17 | 10063 | 0.116 |
| LC | ABCG2 | 9429 | 0.115 |
| LC | VEGFC | 7424 | 0.113 |
| LC | RBM6 | 10180 | 0.108 |
| LC | PRKCA | 5578 | 0.108 |
| LC | FGF2 | 2247 | 0.108 |
| LC | CDKN2B | 1030 | 0.106 |
| LC | TYMS | 7298 | 0.105 |
| LC | THPO | 7066 | 0.104 |
| LC | DLC1 | 10395 | 0.103 |
| LC | JUP | 3728 | 0.102 |
| LC | ELAVL4 | 1996 | 0.102 |
| LC | TOP1 | 7150 | 0.101 |
| LC | TSPYL2 | 64061 | 0.1 |
| LC | PLUNC | 51297 | 0.099 |
| LC | CTSB | 1508 | 0.099 |
| LC | CSF2 | 1437 | 0.098 |
| LC | TOP2A | 7153 | 0.097 |
| LC | RARB | 5915 | 0.096 |
| LC | NME1 | 4830 | 0.095 |
| LC | MYC | 4609 | 0.094 |
| LC | SFTPD | 6441 | 0.093 |
| LC | XRCC1 | 7515 | 0.091 |
| LC | CAV1 | 857 | 0.091 |
| LC | IL10 | 3586 | 0.089 |
| LC | UBA7 | 7318 | 0.088 |
| LC | MVP | 9961 | 0.088 |
| LC | AKR1C1 | 1645 | 0.088 |
| LC | TXN | 7295 | 0.086 |
| LC | KIT | 3815 | 0.086 |
| LC | ADH5 | 128 | 0.086 |
| LC | CYR61 | 3491 | 0.085 |
| LC | ALDH3A1 | 218 | 0.085 |
| LC | TERT | 7015 | 0.084 |
| LC | SMAD2 | 4087 | 0.084 |
| LC | ZMYND10 | 51364 | 0.083 |
| LC | RB1 | 5925 | 0.083 |
| LC | CDKN1A | 1026 | 0.083 |
| LC | PRDX1 | 5052 | 0.082 |
| LC | MYCL1 | 4610 | 0.082 |
| LC | RRM1 | 6240 | 0.081 |
| LC | TUSC1 | 286319 | 0.08 |
| LC | TP63 | 8626 | 0.08 |
| LC | EPHX1 | 2052 | 0.08 |
| LC | TNC | 3371 | 0.079 |
| LC | PPARG | 5468 | 0.079 |
| LC | IFRD2 | 7866 | 0.079 |
| LC | GRPR | 2925 | 0.079 |
| LC | LRP1B | 53353 | 0.078 |
| LC | CACNA2D2 | 9254 | 0.078 |
| LC | CYP3A4 | 1576 | 0.077 |
| LC | CASP9 | 842 | 0.077 |
| LC | OPRM1 | 4988 | 0.076 |
| LC | HGF | 3082 | 0.076 |
| LC | MARCKSL1 | 65108 | 0.074 |
| LC | ABCB1 | 5243 | 0.074 |
| LC | CD34 | 947 | 0.073 |
| LC | RAD1 | 5810 | 0.072 |
| LC | HYAL2 | 8692 | 0.072 |
| LC | SEMA3F | 6405 | 0.071 |
| LC | NBN | 4683 | 0.071 |
| LC | APEH | 327 | 0.071 |
| LC | MIF | 4282 | 0.068 |
| LC | IL10RA | 3587 | 0.068 |
| LC | HYAL1 | 3373 | 0.067 |
| LC | AIFM1 | 9131 | 0.067 |
| LC | HIF1A | 3091 | 0.066 |
| LC | DPP4 | 1803 | 0.066 |
| LC | MAX | 4149 | 0.065 |
| LC | EPB41L3 | 23136 | 0.065 |
| LC | CASP5 | 838 | 0.065 |
| LC | CASP3 | 836 | 0.065 |
| LC | TUSC4 | 10641 | 0.064 |
| LC | REST | 5978 | 0.064 |
| LC | PKM2 | 5315 | 0.064 |
| LC | LATS2 | 26524 | 0.064 |
| LC | HYAL3 | 8372 | 0.064 |
| LC | HPSE | 10855 | 0.063 |
| LC | RET | 5979 | 0.062 |
| LC | MUC16 | 94025 | 0.062 |
| LC | CEACAM5 | 1048 | 0.062 |
| LC | PTENP1 | 11191 | 0.061 |
| LC | IGF2 | 3481 | 0.061 |
| LC | TMEM115 | 11070 | 0.06 |
| LC | SLIT2 | 9353 | 0.06 |
| LC | NAT6 | 24142 | 0.06 |
| LC | MALAT1 | 378938 | 0.06 |
| LC | DMP1 | 1758 | 0.06 |
| LC | CYP2C9 | 1559 | 0.06 |
| LC | CYB561D2 | 11068 | 0.06 |
| LC | WEE1 | 7465 | 0.059 |
| LC | TAP1 | 6890 | 0.059 |
| LC | SPARC | 6678 | 0.059 |
| LC | RAPGEF1 | 2889 | 0.059 |
| LC | FASLG | 356 | 0.059 |
| LC | ENO2 | 2026 | 0.059 |
| LC | DMBT1 | 1755 | 0.059 |
| LC | CTSL1 | 1514 | 0.059 |
| LC | CCNB1 | 891 | 0.059 |
| LC | TPX2 | 22974 | 0.058 |
| LC | TGFB1 | 7040 | 0.058 |
| LC | SPON2 | 10417 | 0.058 |
| LC | CD9 | 928 | 0.058 |
| LC | ATF2 | 1386 | 0.058 |
| LC | CCDC34 | 91057 | 0.056 |
| LC | PTGER1 | 5731 | 0.055 |
| LC | CPB2 | 1361 | 0.054 |
| LC | CHFR | 55743 | 0.054 |
| LC | CD44 | 960 | 0.054 |
| LC | ZBTB1 | 22890 | 0.053 |
| LC | TMED8 | 283578 | 0.053 |
| LC | TEX10 | 54881 | 0.053 |
| LC | RSL1D1 | 26156 | 0.053 |
| LC | PDLIM5 | 10611 | 0.053 |
| LC | NOL11 | 25926 | 0.053 |
| LC | NBPF3 | 84224 | 0.053 |
| LC | MED10 | 84246 | 0.053 |
| LC | KIAA0101 | 9768 | 0.053 |
| LC | GAPDH | 2597 | 0.053 |
| LC | FAM60A | 58516 | 0.053 |
| LC | DIABLO | 56616 | 0.053 |
| LC | C18orf10 | 25941 | 0.053 |
| LC | ATAD2 | 29028 | 0.053 |
| LC | TNFSF10 | 8743 | 0.052 |
| LC | PYCARD | 29108 | 0.052 |
| LC | STAT3 | 6774 | 0.051 |
| LC | SCGB3A1 | 92304 | 0.051 |
| LC | MAP3K1 | 4214 | 0.051 |
| LC | AVP | 551 | 0.051 |
| LC | ABCC5 | 10057 | 0.051 |
| LC | DDIT3 | 1649 | 0.05 |
| LC | ADCYAP1 | 116 | 0.05 |
Contributor Information
IVAN P. GORLOV, Department of Genitourinary Medical Oncology, Unit 1374, The University of Texas MD Anderson Cancer Center, 1155 Pressler Street, Houston, Texas 77030-3721, USA.
JINYOUNG BYUN, Department of Genitourinary Medical Oncology, Unit 1374, The University of Texas MD Anderson Cancer Center, 1155 Pressler Street, Houston, Texas 77030-3721, USA jbyun@mdanderson.org.
HONGYA ZHAO, Department of Genitourinary Medical Oncology, Unit 1374, The University of Texas MD Anderson Cancer Center, 1155 Pressler Street, Houston, Texas 77030-3721, USA hongya.zhao@gmail.com.
CHRISTOPHER J. LOGOTHETIS, Department of Genitourinary Medical Oncology, Unit 1374, The University of Texas MD Anderson Cancer Center 1155 Pressler Street, Houston, Texas 77030-3721, USA clogothe@mdanderson.org
OLGA Y. GORLOVA, Department of Epidemiology, Unit 1340 The University of Texas MD Anderson Cancer Center 1155 Pressler Street, Houston, Texas 77030-3721, USA oygorlov@mdanderson.org
References
- 1.Pritchard CC, Nelson PS. Gene expression profiling in the developing prostate. Differentiation. 2008;76:624–640. doi: 10.1111/j.1432-0436.2008.00274.x. [DOI] [PubMed] [Google Scholar]
- 2.Sikaroodi M, Galachiantz Y, Baranova A. Tumor markers: the potential of “omics” approach. Curr Mol Med. 2011;10:249–257. doi: 10.2174/156652410790963277. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annu Rev Pathol. 2006;1:243–271. doi: 10.1146/annurev.pathol.1.110304.100047. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao YH, Chou MC, Fowler C, Mason JT, Man YG. Breast cancer heterogeneity: mechanisms, proofs, and implications. J Cancer. 2010;1:6–13. doi: 10.7150/jca.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol. 2009;220:538–547. doi: 10.1002/jcp.21799. [DOI] [PubMed] [Google Scholar]
- 6.Berry D. Breast cancer heterogeneity may explain peaks in recurrence. Int J Surg. 2005;3:287. doi: 10.1016/j.ijsu.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Symmans WF, Liu J, Knowles DM, Inghirami G. Breast cancer heterogeneity: evaluation of clonality in primary and metastatic lesions. Hum Pathol. 1995;26:210–216. doi: 10.1016/0046-8177(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 8.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran UR, Ma C, Dhir R, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology, The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 13.Gorlov IP, Byun J, Gorlova OY, Aparicio AM, Efstathiou E, Logothetis CJ. Candidate pathways and genes for prostate cancer: a meta-analysis of gene expression data. BMC Med Genomics. 2009;2:48. doi: 10.1186/1755-8794-2-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlov IP, Sircar K, Zhao H, et al. Prioritizing genes associated with prostate cancer development. BMC Cancer. 2010;10:599. doi: 10.1186/1471-2407-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlov IP, Gallick GE, Gorlova OY, Amos C, Logothetis CJ. GWAS meets microarray: are the results of genome-wide association studies and gene-expression profiling consistent? Prostate cancer as an example. PLoS One. 2009;4:e6511. doi: 10.1371/journal.pone.0006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molinié V. [Gleason’s score: update in 2008] Ann Pathol. 2008;28:350–353. doi: 10.1016/j.annpat.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Blasberg JD, Goparaju CM, Pass HI, Donington JS. Lung cancer osteopontin isoforms exhibit angiogenic functional heterogeneity. J Thorac Cardiovasc Surg. 2010;139:1587–1593. doi: 10.1016/j.jtcvs.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai Y, Miyazawa H, Huqun, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–7282. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- 19.Tsubokawa F, Nishisaka T, Takeshima Y, Inai K. Heterogeneity of expression of cytokeratin subtypes in squamous cell carcinoma of the lung: with special reference to CK14 overexpression in cancer of high-proliferative and lymphogenous metastatic potential. Pathol Int. 2002;52:286–293. doi: 10.1046/j.1440-1827.2002.01353.x. [DOI] [PubMed] [Google Scholar]
- 20.Krause FS, Feil G, Bichler KH, Schrott KM, Akcetin ZY, Engehausen DG. Heterogeneity in prostate cancer: prostate specific antigen (PSA) and DNA cytophotometry. Anticancer Res. 2005;25:1783–1785. [PubMed] [Google Scholar]
- 21.Meghani SH, Lee CS, Hanlon AL, Bruner DW. Latent class cluster analysis to understand heterogeneity in prostate cancer treatment utilities. BMC Med Inform Decis Mak. 2009;9:47. doi: 10.1186/1472-6947-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6:454–460. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 23.Brower V. Watchful waiting beats androgen deprivation therapy in early prostate cancer. J Natl Cancer Inst. 2008;100:1494–1496. doi: 10.1093/jnci/djn402. [DOI] [PubMed] [Google Scholar]
- 24.Ganti AK, Huang CH, Klein MA, Keefe S, Kelley MJ. Lung cancer management in 2010. Oncology (Williston Park) 2010;25:64–73. [PubMed] [Google Scholar]