Abstract
Mammals harbor a complex gut microbiome, comprised of bacteria that confer immunologic, metabolic and neurologic benefits1. Despite advances in sequence-based microbial profiling and myriad studies defining microbiome composition during health and disease, little is known about the molecular processes employed by symbiotic bacteria to stably colonize the gastrointestinal (GI) tract. We sought to define how mammals assemble and maintain the Bacteroides, one of the most numerically prominent genera of the human microbiome. While the gut normally contains hundreds of bacterial species2,3, we surprisingly find that germ-free mice mono-associated with a single Bacteroides are resistant to colonization by the same, but not different, species. To identify bacterial mechanisms for species-specific saturable colonization, we devised an in vivo genetic screen and discovered a unique class of Polysaccharide Utilization Loci (PUL) that are conserved among intestinal Bacteroides. We named this genetic locus the commensal colonization factors (ccf). Deletion of the ccf genes in the model symbiont, Bacteroides fragilis, results in colonization defects in mice and reduced horizontal transmission. The ccf genes of B. fragilis are up-regulated during gut colonization, preferentially at the colonic surface. When we visualize microbial biogeography within the colon, B. fragilis penetrates the colonic mucus and resides deep within crypt channels, while ccf mutants are defective in crypt association. Remarkably, the CCF system is required for B. fragilis colonization following microbiome disruption with Citrobacter rodentium infection or antibiotic treatment, suggesting the niche within colonic crypts represents a reservoir for bacteria to maintain long-term colonization. These findings reveal that intestinal Bacteroides have evolved species-specific physical interactions with the host that mediate stable and resilient gut colonization, and the CCF system represents a novel molecular mechanism for symbiosis.
International microbiome sequencing initiatives are revealing detailed inventories of diverse bacterial communities across various body sites, diets and human populations2-4. Complex ecosystems have been forged by co-adaptation over millennia between animals and microbes to create stable and specific microbiomes5,6, suggesting the evolution of molecular mechanisms that establish and maintain symbiotic microbial colonization. Bacteroidetes is one of the most numerically abundant Gram-negative phyla in the mammalian GI tract7. Studies in the genus Bacteroides have revealed species that induce glycosylation of the intestinal epithelium8, produce glycoside hydrolases that digest carbohydrates for host nutrient utilization9, direct host immune maturation10 and protect animals from inflammation in experimental models of IBD and multiple sclerosis11-13. To explore the dynamics of microbiome assembly, we sequentially introduced Bacteroides species to germ-free mice and monitored colonization via colony-forming units (CFU) in feces. Animals are readily colonized with Bacteroides fragilis followed by Bacteroides thetaiotaomicron (Fig. 1a) or Bacteroides vulgatus (Fig. 1b), and altering the sequence of microbial exposure does not affect results (Supplementary Fig. 1a). Remarkably however, animals colonized with B. fragilis and then exposed to the same species (marked by an antibiotic resistance gene) are resistant to super-colonization and clear the challenging strain (Fig. 1c). This novel observation of ‘colonization resistance’ by the same species is conserved in three other Bacteroides (Supplementary Fig. 1b-d), regardless of the antibiotic resistance markers used (Supplementary Fig. 1e), but not in Escherichia coli (Supplementary Fig. 1f). As conventional mice typically harbor 1011-1012 CFU per gram of cecal content14 (100-fold greater than Bacteroides in mono-association), there appears to be no shortage of space or nutrients under these conditions, using a nutrient rich standard diet. We thus hypothesized that individual Bacteroides species colonize the gut by saturating a limited and unique niche. Indeed, treatment of B. fragilis mono-associated mice with erythromycin to displace the existing strain permits colonization by an erythromycin-resistant challenge strain (Fig. 1d). These data suggest that Bacteroides colonize the gut in a species-specific and saturable manner.
Figure 1. Bacteroides species occupy species-specific niches in the gut via an evolutionarily conserved genetic locus.
a-c, Germ-free mice were mono-associated with B. fragilis and challenged orally with (a) B. thetaiotaomicron; (b) B. vulgatus; or (c) B. fragilis. d, Mice were mono-associated with erythromycin sensitive B. fragilis, and subsequently challenged with erythromycin resistant B. fragilis. Erythromycin was administered where indicated. e, Genomic organization of the ccf locus. f, Mice were mono-associated with either WT B. fragilis, mutant strains deleted in ccfC, ccfD, ccfE and ccfC-E (BFΔCCF), or complemented strain (BFΔCCF∷CCF) and challenged with WT B. fragilis. CFUs were determined after 30 days. g, Mice were mono-associated with WT B. vulgatus or a mutant strain deleted in ccfC-E genes (BVΔCCF), and challenged with WTB. vulgatus. CFUs were determined after 30 days. In all sequential colonization studies, results are representative of at least 2 independent trials (n=3-4 animals/group). h, Cross-colonization between WT B. fragilis and BFΔCCF mono-associated mice at 7 days after encounter measured by CFUs of the initially colonizing and the horizontally transmitted (challenge) strains. (n=2 animals/encounter, 5 independent trials). All graphs: Dashed line indicates the limit of detection at 100 CFU/g feces, and error bars indicate standard deviation (SD).
We developed a functional in vivo screen to identify genetic factor(s) from B. fragilis that are sufficient to mediate species-specific colonization. Mice were mono-associated with B. vulgatus, then challenged with a library of B. vulgatus clones that each contained a fragment of B. fragilis genomic DNA (schematic in Supplementary Fig. 2a). We reasoned that only those clones containing genes that conferred stable gut colonization by B. fragilis would persist, with the remainder being cleared. We screened 2,100 clones each containing 9-10 kilobases of DNA, providing a 3.8-fold coverage of the B. fragilis genome and 98% probability that a given DNA sequence is present in the library (Supplementary Eq. 1). Remarkably, 30 days after orally gavaging the library into animals, only two clones sustained colonization. The inserts from both clones mapped to the same locus on the B. fragilis genome, BF3579-BF3583 (Supplementary Fig. 2b).
Based on predicted protein sequences, BF3583 and BF3582 is a sigma (σ) factor/anti-σ factor gene pair. BF3581 is a member of the SusC family of outer membrane proteins. BF3580 is a homolog of SusD, a lipoprotein often paired with SusC. These Sus-like systems have been shown to bind and import a range of oligosaccharide molecules15-18. BF3579 encodes a putative chitobiase, suggesting a possible polysaccharide substrate for this system19 (Fig. 1e). Comparative genomic analysis using the Integrated Microbial Genomes database (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) reveals conservation of similar clusters of genes among sequenced intestinal Bacteroides species (Supplementary Fig. 3). Sus-like systems are numerous in Bacteroides within polysaccharide utilization loci (PULs), which are gene cassettes used to harvest dietary sugars and/or forage host glycans during nutrient deprivation15,20,21. As PULs have previously not been implicated in saturable niche colonization, the locus we have identified encodes a unique pathway in Bacteroides for species-specific gut association; we named the genes ccfA-E, for commensal colonization factors (Fig. 1e). Furthermore, as deletion of the most closely related genes from B. fragilis (BFΔ0227-0229; Supplementary Fig. 3) do not impact colonization dynamics (Supplementary Fig. 4), we suggest that the CCF system represents a functionally unique subset of PULs that evolved to promote long-term symbiosis.
To test if the putative structural genes (ccfC-E) are required for gut colonization, we generated in-frame deletion mutants of B. fragilis; ΔccfC, ΔccfD and ΔccfE. All strains exhibit normal morphology on solid agar medium and unimpaired growth in laboratory culture (data not shown). As previously shown, animals mono-colonized with WT B. fragilis completely clear the WT challenge strain after 30 days (Fig. 1f; 1st bars). However, animals mono-associated with ΔccfC or ΔccfD are permissive to colonization by WT bacteria (Fig. 1f; 2nd and 3rd bars), unlike the ccfE mutant (Fig. 1f; 4th bars). A deletion mutant in all three genes (B. fragilisΔCCF) also allows WT B. fragilis to colonize (Fig. 1f; 5th bars). Trans-complementation of the B. fragilisΔCCF strain with ccfA-E restores colonization resistance (Fig. 1f; 6th bars). Similarly, a mutant in the B. vulgatus ccfC-E orthologs (ΔBVU0946-BVU0948) also permits WT B. vulgatus to colonize (Fig. 1g), demonstrating conservation in Bacteroides species. When we tested horizontal transmission of bacteria between WT B. fragilis and B. fragilisΔCCF mono-associated mice, only WT bacteria cross-colonized (Fig. 1h). Thus, the CCF system is involved in colonization resistance by Bacteroides.
Building on the previous discovery that a population of B. fragilis associates with mucosal tissues22, we show that ccfB-E are preferentially expressed by bacteria in contact with the colon, with lower levels in cecal content and feces (Fig. 2a). There is virtually no expression in laboratory culture. Thus, in vivo expression of ccf in gut tissue may be critical for colonization. Indeed, in contrast to laboratory grown bacteria (see Fig. 1c), sustained colonization is conferred to bacteria recovered directly from animals (Supplementary Fig. 5). To examine regulation of gene expression, we deleted the σ factor ccfA, which led to highly reduced expression of all five genes during animal colonization (Supplementary Fig. 6a). Accordingly, germ-free mice mono-colonized with B. fragilisΔccfA mutant are permissive of super-colonization by WT B. fragilis, demonstrating a functional defect in the saturable niche occupancy (Supplementary Fig. 6b). Based on the tissue-associated expression pattern, we tested if the CCF system promotes bacterial localization to mucosal tissue. Mice were mono-associated with either the WT or the ccf deletion strain, and both groups were subsequently challenged with WT B. fragilis. 24 hours after challenge, we observe the same numbers for challenge strains in feces of both groups (Fig. 2b; 1st and 2nd bars, and Supplementary Fig. 7a, b). In contrast, ccf mutant-associated animals show higher levels of challenge strain in colon tissue, suggesting a colonization defect by the mutant strain specifically at the mucosal surface (Fig. 2b; 3rd and 4th bars, and Supplementary Fig. 7a, b). Therefore, CCF-mediated colonization fitness appears to involve physical association with the gut.
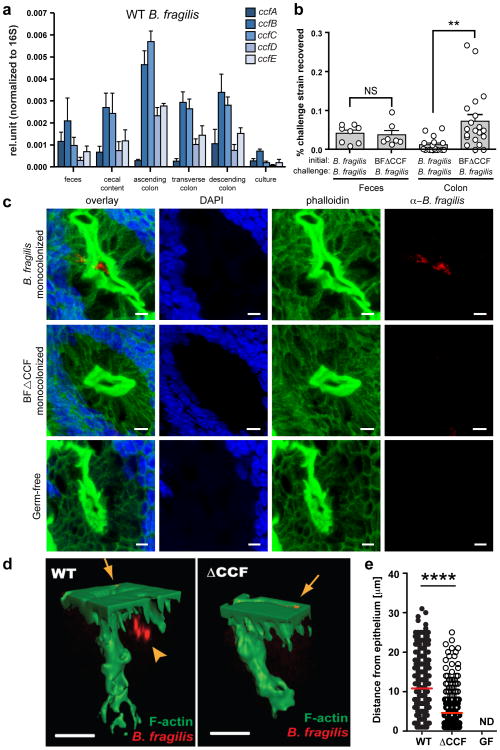
Figure 2. B. fragilis colonization of the colonic crypts is mediated by the CCF system.
a, qRT-PCR of ccf gene expression levels normalized to 16S rRNA (n=3 animals, 2 trials). b, Mice were mono-associated with either WT B. fragilis or B. fragilisΔCCF, and challenged with WT B. fragilis. The percentage of challenge strain was determined in the lumen (feces) and colon after 1 day (n=8 animals/group). c, Confocal micrographs of germ-free, WT B. fragilis or B. fragilisΔCCF mono-associated mice colon whole-mount. Crypts are visualized by DAPI (nuclei, blue) and phalloidin (F-actin, green). Bacteria (red) are stained with IgY polyclonal antibody raised against B. fragilis. Images are representative of 7 different sites analyzed from at least 2 different colons. Scale bar: 5 μm. d, 3D reconstructions of colon crypts from WT B. fragilis or B. fragilisΔCCF mono-associated mice. Bacteria are detected on the apical surface of the epithelium (arrows) and in the crypt space (arrowhead). Scale bar: 10 μm. e, Quantification of bacterial penetration, measured as distance from the epithelial surface per crypt. Error bars indicate standard error of the mean (SEM). NS: not significant. ND: not detected. **p<0.01.****p<0.0001.
Recent studies have revealed microbial communities that colonize intestinal crypts of conventional mice in the absence of disease23 and we have shown that B. fragilis occupies the colonic crypts of mono-associated mice22. Discovering a role for ccf genes near mucosal tissue led us to explore the intriguing hypothesis that the CCF system mediates crypt occupancy. We mono-colonized mice with B. fragilis and visualized bacterial localization in colon tissue by whole-mount confocal microscopy. Indeed, WT B. fragilis co-localize with crypts from the ascending colon, appearing to be located in the center of crypt opening. Remarkably, B. fragilisΔCCFmono-associated mice display virtually no crypt occupancy (Fig. 2c and Supplementary Fig. 8). Colon cross-section imaging also reveals that only WT bacteria are crypt associated (Supplementary Fig. 9). Two-photon imaging of colon explants clearly demonstrates presence of WT B. fragilis on the surface of the epithelium and inside the crypt. While both WT and mutant strains of B. fragilis associate with the surface of the epithelium, only WT bacteria are able to penetrate deep into the colonic crypts of mice (Fig. 2d and Supplementary video 1). Measuring the distance from the surface of the epithelium to bacterial signals in a survey of crypts reveals significantly greater tissue penetration by WT bacteria (Fig. 2e). Collectively, these data reveal that the CCF system allows B. fragilis to reside in a specific niche within crypts during steady-state colonization.
We next investigated the effects of the CCF system in the context of a complex microbiota. Wild-type Bacteroides species do not readily colonize most strains of specific pathogen-free (SPF) mice, namely BALB/c, Swiss Webster and C57BL/6, despite oral administration of high inoculums (Supplementary Fig. 10a, c and data not shown). Furthermore, transfer of an SPF microbiota to mono-colonized mice leads to clearance of WT B. fragilis (Supplementary Fig. 10b, d). To overcome this obstacle, we tested various additional genetic backgrounds and empirically determined that C57BL/6 Rag-/- mice (which lack an adaptive immune system) and non-obese diabetic (NOD) mice can be stably colonized by B. fragilis with a single oral gavage. We introduced either WT or ccf mutant B. fragilis at equal inoculums into separate groups of animals, and measured colonization. Only WT B. fragilis stably colonizes SPF Rag-/- mice, whereas B. fragilisΔCCF establishes a significantly lower colonization in the gut (Fig. 3a). Co-inoculation of equal numbers of WT and ccf mutant bacteria into Rag-/- mice also results in rapid clearance of the mutant strain from the gut (Fig. 3b), demonstrating a cell intrinsic defect that could not be complemented in trans by WT bacteria. NOD animals are also preferentially colonized by WT B. fragilis compared to ccf mutants in separate groups of animals (Fig. 3c) or in equal co-inoculation (Fig. 3d). These data show that deletion of the ccf genes compromises B. fragilis colonization of hosts with a complex microbiota.
Figure 3. B. fragilis requires the ccf genes for stable and resilient colonization of mice.
a, Groups of SPF Rag-/- mice were gavaged with either WT B. fragilis or B. fragilisΔCCF. b, SPF Rag-/- mice were given a 1:1 co-inoculum of WT B. fragilis and B. fragilisΔCCF by single gavage. c, SPF NOD mice were gavaged with either WT B. fragilis or B. fragilisΔCCF. d, SPF NOD mice were given a 1:1 co-inoculum of WT B. fragilis and B. fragilisΔCCF by single gavage. e, SPF mice were co-associated with WT B. fragilis and B. fragilisΔCCF, and infected with Citrobacter rodentium. f, SPF mice were co-associated with WT B. fragilis and B. fragilisΔCCF, and given ciprofloxacin in drinking water for the time period shown. For all analyses, bacterial colonization levels were assessed by real-time qRT-PCR from stool DNA (n=4 animals/group). Results are representative of at least 2 independent trials per experiments. Error bars indicate SEM.*p<0.05. ***p<0.001.
During symbiosis with mammals, the microbiota may be confronted by rapid environmental changes with potentially adverse consequences to bacteria, such as enteric infections or antibiotic exposure. Gastroenteritis is commonly experienced by humans and is known to perturb the microbiota. To test if resilience of B. fragilis colonization is CCF-dependent, we used Citrobacter rodentium infection of mice to mimic human GI infection24. Employing an antibiotic treatment protocol that does not sterilize the gut but promotes colonization of SPF mice by Bacteroides25, we were able to simultaneously colonize mice with equivalent levels of WT and ccf mutant bacteria. Mice were subsequently infected orally with C. rodentium, and colonization of B. fragilis was monitored. WT bacteria decline in number at first, but return to maximal levels 3-4 weeks post infection (Fig. 3e). Importantly, the B. fragilisΔCCF strain is completely cleared from the mouse gut following gastroenteritis (Fig. 3e), but not when animals are left uninfected (Supplementary Fig. 11a). Next, we challenged mice that were co-colonized with WT and B. fragilisΔCCF with oral antibiotics and observe a colonization defect only in ccf mutant bacteria (Fig. 3f and Supplementary Fig. 11b). These results reveal that the CCF system establishes resilient colonization by gut Bacteroides following disruption of the microbiome. Finally, when SPF mice colonized with WT B. fragilis were given an antibiotic treatment that cleared fecal bacteria, crypt-associated microbial populations persisted (Supplementary Fig. 12), suggesting that symbiotic bacteria occupy a protected niche that creates a reservoir for stable gut colonization.
Co-evolution has bound microbes and man in an inextricable partnership, resulting in remarkable specificity and stability of the human microbiome2,3. Our findings reveal a novel pathway required for persistent colonization of the mammalian gut by the Bacteroides. Homology to the Sus family of proteins suggests a role for CCF in uptake and utilization of glycans. Although certain Sus-containing PULs in B. thetaiotaomicron mediate foraging of host mucus15, their contributions to microbial colonization have been previously described only during nutrient deprivation conditions20. Our discovery of CCF-dependent colonization in mice fed a nutrient rich diet suggests a new role whereby Bacteroides evolved specific Sus-like systems to utilize non-dietary glycans during homeostasis. Based on the findings that ccf genes are preferentially expressed in proximity to mucosal tissues and B. fragilis associates with colonic crypts, we find it likely that host factors may promote expression of the CCF system. In support of this notion, N-Acetyl-D-lactosamine (LacNAc)—a component of host mucus—induces the ccf genes and its homologs in B. fragilis and B. thetaiotaomicron20 (Supplementary Fig. 13). But since a closely related PUL (BF0227-31) responds to LacNAc but does not mediate saturable niche colonization (Supplementary Fig. 4), LacNAc alone may be an inducer but is not the substrate utilized by CCF systems. We propose that specific glycan structures within colonic crypts serve as nutrient sources for individual Bacteroides species, and that CCF systems provide a molecular mechanism for a hypothesis proposed decades ago, “that populations of most indigenous intestinal bacteria are controlled by substrate competition, i.e., that each species is more efficient than the rest in utilizing one or a few particular substrates and that the population level of that species is controlled by the concentration of these few limiting substrates”26. Future work will aim to identify the precise glycan(s) for CCF systems from various Bacteroides. Finally, our data suggests that the ccf genes encode for a specific subset of PULs that evolved the novel activity of promoting stable and resilient colonization, and crypt-associated bacterial reservoirs may represent ‘founder’ cells that repopulate the gut following disruption of the microbiome by enteric infections or antibiotic exposure. Discovery of a molecular mechanism for colonization fitness by gut bacteria provides a glimpse into the evolutionary forces that have shaped the assembly and dynamics of the human microbiome.
Methods Summary
All germ-free mice were bred and housed in flexible film isolators until 8 weeks of age, then transferred to microisolator cages and maintained with autoclaved food, bedding and water supplemented with gentamicin and erythromycin. Mice were mono-associated with gentamicin and erythromycin resistant Bacteroides strains by single oral gavage. Colonization level was determined over time by stool serial dilution plating on selective agar media. For qRT-PCR, total RNA was extracted from laboratory bacterial culture, fecal and cecal content (ZR Soil/Fecal RNA MicroPrep™, Zymo Research) and colon tissues (Trizol, Invitrogen) from mono-associated animals, converted to first strand cDNA and analyzed by qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems). For colon whole-mount imaging, tissues were harvested from germ-free or single strain mono-associated animals, fixed with 4% PFA and stained with a B. fragilis specific antibody, DAPI and phalloidin. The colon crypts were visualized by confocal microscopy and two-photon microscopy. SPF mice were colonized with B. fragilis and/or B. fragilisΔCCF by single oral gavage and the colonization level was determined over time by stool DNA extraction (ZR Fecal DNA MiniPrep™, Zymo Research) and qPCR using strain specific primers.
Methods
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids are described in Supplementary Table 1. Bacteroides strains were grown anaerobically at 37°C for two days in brain heart infusion broth supplemented with 5 μg/ml hemin and 0.5 μg/ml Vitamin K (BHIS), with gentamicin (200 μg/ml), erythromycin (5 μg/ml), chloramphenicol (10 μg/ml) and tetracycline (2 μg/ml) added where appropriate. Escherichia coli JM109 containing recombinant plasmids were grown in LB with ampicillin (100 μg/ml) or kanamycin (30 μg/ml). Citrobacter rodentium DBS100 strain was grown in LB at 37°C for 24 hours. For the induction of susC/D homologues, B. fragilis and B. thetaiotaomicron were grown in minimal medium (MM) with either glucose or N-acetyllactosamine as the sole carbon source as described previously20.
Mice
8-10 week old male and female germ-free Swiss Webster mice were purchased from Taconic Farms (Germantown, NY) and bred in flexible film isolators. For gnotobiotic colonization experiments, germ-free mice were transferred to freshly autoclaved microisolator cages, fed ad libitum with a standard autoclaved chow diet and given autoclaved water supplemented with 10 μg/ml of erythromycin and 100 μg/ml of gentamicin. Male SPF (Specific Pathogen-Free) C57BL/6 mice and Swiss Webster mice were purchased from Taconic Farms. Male SPF NOD/ShiLtJ mice and Rag-/- C57BL/6 mice were purchased from the Jackson Laboratory. No randomization or blinding was used to allocate experimental groups. Sample size and standard deviation were based on empirical data from pilot experiments. All procedures were performed in accordance with the approved protocols using IACUC guidelines of the California Institute of Technology.
Construction of chromosomal library and screen
Genomic DNA was isolated from overnight culture of B. fragilis using a commercial kit (Wizard® Genomic DNA Purification Kit, Promega). 20 μg of genomic DNA was incubated with 4U of Sau3AI for 5, 10, 15, or 20 minutes at 37°C in 50 μl volume and the partially digested genomic DNA was separated by electrophoresis on 0.7% agarose gel. 9-10 kb fragment DNA was excised and recovered from the agarose gel (Zymoclean™ Gel DNA Recovery Kit, Zymo Research). Insert DNA was ligated to BglII site of plasmid vector (pFD340-catBII, Supplementary Table 1), transformed into E. coli and amplified on LB-ampicillin plate. Individual clones from the plasmid library were mobilized from E. coli to B. vulgatus by conjugal helper plasmid RK231 generating a library of B. vulgatus hosting B. fragilis chromosomal DNA fragments consisting of approximately ∼2100 clones. To screen the library in vivo, pools of 96 clones (106 CFU of each clone) were gavaged into 22 germ-free Swiss Webster mice (108 CFU per animal) pre-colonized with B. vulgatus pFD340 for 1-2 weeks. Two weeks after gavage, fresh fecal samples were plated on BHIS agar plate containing chloramphenicol to select for clones with colonization phenotype.
Generation of ccfA, ccfC, ccfD, ccfE, ccfC-E (ΔCCF) and ΔBF0227-0229 deletion mutants
∼2 kb DNA segments flanking the region to be deleted were PCR amplified using primers listed in Supplementary Table 2. Reverse primer of the left flanking DNA and forward primer of the right flanking DNA were designed to be partially complementary at their 5′ ends by 18-21 bp. Fusion PCR was performed using the left and right flanking DNA (∼300 ng each after gel purification) as DNA template and forward primer of the left flanking DNA and reverse primer of the right flanking DNA27. The fused PCR product was cloned into BamHI or SalI site of the Bacteroides conjugal suicide vector pNJR6 and mobilized into B. fragilis. Colonies selected for erythromycin resistance (Emr), indicating integration of the suicide vector into the host chromosome were passaged for five days and then plated on nonselective medium (BHIS). The resulting colonies were replica plated to BHIS containing Em, and Ems (erythromycin sensitive) colonies were screened by PCR to distinguish wild-type revertants from strains with the desired mutation. The same strategy was employed to generate ΔccfC-E deletion mutant from B. vulgatus.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from mid-log phase bacterial culture using ZR Fungal/Bacterial RNA MiniPrep™ (Zymo Research), feces and cecal content from mice using ZR Soil/Fecal RNA MicroPrep™ (Zymo Research), and mouse colon tissues after removing luminal content by gently scraping the mucosal surface and PBS rinse using Trizol (Invitrogen). cDNA was made using an iSCRIPT cDNA synthesis kit per manufacturer's instructions (Bio-Rad). All qRT-PCR reactions were performed in ABI PRISM 7900HT Fast Real-Time PCR System (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems). Gene-specific primers are described in Supplementary Table 2.
Immunofluorescent staining of colon whole-mounts and frozen sections
For whole-mount staining, colons were fixed in buffered 4% paraformaldehyde, washed with PBS and subjected to indirect immunofluorescence. Tissues were made permeable by incubation with 0.5% (wt/vol) saponin, 2% (vol/vol) FBS, and 0.09% (wt/vol) azide in PBS for at least 18 hours. The same buffer was used for subsequent incubations with antibodies. Colon fragments were incubated with a primary polyclonal chicken IgY anti-B. fragilis antibodies for 12-16 hours at room temperature (RT) followed by 1-2 hour incubation at 37°C. Following PBS washes, samples were reacted with goat anti-chicken IgY secondary antibodies (Alexa Fluor 488 or Alexa Fluor 633, 2 μg/ml, Molecular Probes), fluorescently labeled phalloidin (fluorescein or AF568, 2 U/ml, Molecular Probes) and DAPI (2 μg/ml, Molecular Probes) for 1 hour at RT. Tissues were mounted in Prolong Gold (Invitrogen) and allowed to cure for at least 48 hours prior to imaging. In some experiments, anti-B. fragilis antibodies were pre-absorbed on tissue fragments derived from either germ-free mice (up to 18 hours at RT) or SPF mice (1 hour at RT).
For frozen sections, colon tissues were embedded in O.C.T. Compound (Sakura Finetek), frozen on dry ice, and stored at -80°C. Frozen blocks were cut with a thickness of 10 μm using a Microm HM505E cryostat, and sections were collected on positively charged slides (Fisher Scientific) for staining. Slides were fixed with 4% buffered paraformaldehyde for 10 minutes and washed 2×10 minutes with PBS. Tissue sections were blocked with 10% normal goat serum and 0.5% bovine serum albumin in PBS for 1 hour at RT. Sections were incubated with anti-B. fragilis antibodies for at least 8 hours at 4°C, washed 2 times for 10 minutes with PBS, reacted with secondary reagents and mounted as described above. In some experiments, anti-B. fragilis antibodies were pre-absorbed for 1 hour at RT on tissue sections derived from germ-free mice.
Fluorescence microscopy
An SP5 resonant laser-scanning confocal and two-photon microscope (both scanning heads mounted on the same DM 6000 upright microscope, Leica Microsystems) with a 40× oil objective (numerical aperture 1.4) or 63× oil objective (numerical aperture 1.4) were used for fluorescence microscopy. Images used for 3D reconstructions were acquired using dual confocal – two-photon mode. For confocal imaging, 488-nm and 543-nm excitation wavelengths were used for Alexa Fluor 488-labeled bacteria and Alexa Fluor 568-labeled phalliodin, and signals were detected with internal photomultiplier tubes. 2-photon imaging was performed with 4 nondescanned detectors (Leica Microsystems) and a Chameleon Ultra Ti: Sapphire laser (Coherent) tuned at 700–800 nm for acquisition. Emitted fluorescence was split with 3 dichroic mirrors (496 nm, 560 nm and 593 nm) and passed through an emission filter (Semrock) at 585/40 nm. Images (512×512) acquired with a 0.5 μm Z step were smoothed by median filtering at kernel size 3 × 3 pixels. 3D reconstructions of crypts and bacteria were performed using Imaris software (version 7.5.1 ×64; Bitplane AG). Crypt structures were visualized by DAPI and phalloidin signals. Images used for quantification were acquired with FluoView FV10i confocal microscope (Olympus) using 60× (numerical aperture 1.35) oil objective.
Image analysis
For bacterial localization with respect to the epithelial layer, frames of 512×512 pixels were acquired with 1 μm Z steps in the crypt length axis. Images were processed using ImageJ software (NIH). Background was subtracted (rolling ball method), images were smoothed by median filtering (3×3 pixels), segmented by threshold and position of the signal in the Z stack was recorded. Data did not follow normal distribution and were analyzed by non-parametric two-sided Mann-Whitney U test.
For quantification of crypt associated bacterial signals from antibiotic treated animals, stacks of 512×512 pixels by 8 frames (1 μm per frame) were flattened by maximum intensity projection and filtered by median (3×3 kernel size). Images were segmented by thresholding. Number of positive spots/1000 μm2 and area occupied by individual spots were analyzed. Data were not normally distributed and were analyzed by Mann-Whitney or Kruskal-Wallis followed by Dunn's multiple comparisons test where appropriate. 11-13 stacks/group were examined. Total area that was analyzed within the group of stacks was between 0.08-0.2 mm2.
Gnotobiotic animal colonization experiments
8-12 week old germ-free Swiss Webster mice were gavaged once with a 100 μl of bacterial suspension for mono-association (∼108 CFU of each bacterial strain harvested from a log-phase culture and resuspended in PBS with 1.5% NaHCO3). For sequential colonization, germ-free mice were mono-associated with an initial strain for 6-7 days and subsequently gavaged with a 100 μl suspension of a challenge strain. All Bacteroides strains used to colonize germ-free animals were resistant to gentamicin inherently, and to erythromycin by plasmid. Unless otherwise indicated, the initial strains carried pFD340-cat (chloramphenicol resistant; Cmr) and the challenge strains, pFD340-tetQ (tetracycline resistant; Tetr). For horizontal transfer by encounter experiment, two single-housed mice that were mono-associated with either WT B. fragilis pFD340-tetQ or B. fragilisΔCCF pFD340-cat for at least 3 weeks were co-housed in a fresh sterile cage for 4 hours and then separated. At each time point, fresh fecal samples were collected, weighed, homogenized and serially diluted in PBS (or BHI broth) for plating on selective media to determine bacterial CFU per g of feces.
SPF animal colonization experiments
7-8 week old male SPF mice (C57BL/6, Swiss Webster, NOD, and Rag-/-) were given a single inoculum of 1×108 CFU of either WT B. fragilis, B. fragilisΔCCF, or 1:1 mixture of the two strains by oral gavage. At each time point, bacterial genomic DNA from fecal samples were isolated using a commercial kit (ZR Fecal DNA MiniPrep™, Zymo Research) following the manufacturer's instructions and the relative densities of bacteria were determined by qPCR using strain-specific primers (Supplementary Table 2).
Citrobacter rodentium infection
8 week old female SPF Swiss Webster mice were treated with metronidazole (100 mg/kg) by oral gavage every 24 hours and ciprofloxacin dissolved in drinking water (0.625 mg/ml; Hikma Pharmaceuticals) for seven days; mice were transferred to a fresh sterile cage every two days. Two days after the cessation of antibiotic treatment, mice were orally gavaged with a single inoculum of 1:1 mixture of WT B. fragilis and B. fragilisΔCCF (∼5×108 CFU total per animal). 6-7 days after B. fragilis gavage, mice were either infected orally with ∼5×108 CFU of overnight culture C. rodentium or PBS-gavaged as control. The relative densities of bacteria were determined by fecal bacterial DNA extraction and qPCR.
Antibiotic treatment
8 week old female SPF Swiss Webster mice were treated with metronidazole (100 mg/kg) by oral gavage every 24 hours and ciprofloxacin dissolved in drinking water (0.625 mg/ml) for seven days; mice were transferred to a fresh sterile cage every two days. Two days after the cessation of antibiotic treatment, mice were orally gavaged with a single inoculum of 1:1 mixture of WT B. fragilis and B. fragilisΔCCF (∼5×108 CFU total per animal). 6-7 days after B. fragilis gavage, one group of mice were treated with ciprofloxacin for 4 days dissolved in drinking water (1 mg/ml) and another group were left untreated. The relative densities of bacteria were determined by fecal bacterial DNA extraction and qPCR.
Antibiotic treatment for colon whole-mount imaging
8 week old female SPF Swiss Webster mice were treated with metronidazole (100 mg/kg) by oral gavage every 24 hours and ciprofloxacin dissolved in drinking water (0.625 mg/ml) for seven days; mice were transferred to a fresh sterile cage every two days. Two days after the cessation of antibiotic treatment, mice were orally gavaged with a 100 μl inoculum of B. fragilis (∼108 CFU) or PBS. 7 days after bacterial gavage (day 16), PBS or B. fragilis inoculated mice were treated with ciprofloxacin in drinking water (1 mg/ml) for 7 days and one group of B. fragilis inoculated mice were left untreated. At the end of the ciprofloxacin treatment (day 23), feces were collected for stool DNA extraction and colon tissues were harvested and fixed with 4% PFA for whole-mount imaging.
Supplementary Material
Acknowledgments
We thank Taren Thron and Sara McBride (Caltech) for the maintenance of germ-free animals, Jane Selicha (Caltech) for assisting with the experimental procedures, and Grzegorz Chodaczek (LIAI) for help with confocal and two-photon microscopy. We are grateful to Dr. Eric C. Martens (Univ. of Michigan) and members of the Mazmanian laboratory for critical review of the manuscript. S.M.L. and G.P.D. were supported by a pre-doctoral training grant (GM007616). This work was supported by grants from the NIH (GM099535 and DK078938) and the Crohn's and Colitis Foundation of America to S.K.M.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Author Contributions. S.M.L. and S.K.M. conceived the project. S.M.L. performed most of the experiments; G.P.D., Z.M. and S.B. contributed data. S.M.L., G.P.D., Z.M., K.L. and S.K.M. interpreted the data. K.L. and S.K.M secured funding. S.M.L. and S.K.M. wrote the manuscript, G.P.D., Z.M. and K.L. edited the manuscript. G.P.D. and Z.M. contributed equally to the work.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. nature08821 [pii] 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. nature07008 [pii] 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 12.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. 0909122107 [pii] 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochoa-Reparaz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. jimmunol.1001443 [pii] 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 14.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. nrmicro2746 [pii] 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. R109.022848 [pii] 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. S0968-0004(08)00101-1 [pii] 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Kawada M, et al. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Invest. 2008;88:883–895. doi: 10.1038/labinvest.2008.47. labinvest200847 [pii] 10.1038/labinvest.2008.47. [DOI] [PubMed] [Google Scholar]
- 20.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. S1931-3128(08)00303-X [pii] 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 22.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedron T, et al. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3 doi: 10.1128/mBio.00116-12. mBio.00116-12 [pii] 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cellular microbiology. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 25.Bloom SM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. S1931-3128(11)00131-4 [pii] 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infection and immunity. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol Tech. 1998;12:653–657. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.