Abstract
Quantitative real-time PCR (qRT-PCR) is a reliable and reproducible technique for measuring and evaluating changes in gene expression. The most common method for analyzing qRT-PCR data is to normalize mRNA levels of target genes to internal reference genes. Evaluating and selecting stable reference genes on a case-by-case basis is critical. The present study aimed to facilitate gene expression studies by identifying the most suitable reference genes for normalization of mRNA expression in qRT-PCR analysis of the beet armyworm Spodoptera exigua (Lepidoptera: Noctuidae). For this purpose, three software tools (geNorm, NormFinder and BestKeeper) were used to investigate 10 candidate reference genes in nine developmental stages and five different tissues (epidermis, head, midgut, fat body and hemolymph) in three larval physiological stages (molting, feeding and wandering stages) of, S. exigua. With the exception of 18S ribosomal RNA (18S), all other candidate genes evaluated, β-actin1(ACT1), β-actin2 (ACT2), elongation factor1(EF1), elongation factor 2 (EF2), Glyceralde hyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L10 (L10), ribosomal protein L17A (L17A), superoxide dismutase (SOD), α-tubulin (TUB),proved to be acceptable reference genes. However, their suitability partly differed between physiological stages and different tissues. L10, EF2 and L17A ranked highest in all tissue sample sets. SOD, ACT2, GAPDH, EF1 and ACT1 were stably expressed in all developmental stage sample sets; ACT2, ACT1 and L10 for larvae sample sets; GAPDH, ACT1 and ACT2 for pupae and adults; SOD and L17A for males; and EF2 and SOD for females. The expression stability of genes varied in different conditions. The findings provided here demonstrated, with a few exceptions, the suitability of most of the 10 reference genes tested in tissues and life developmental stages. Overall, this study emphasizes the importance of validating reference genes for qRT-PCR analysis in S. exigua.
Introduction
Quantification of gene expression levels is fundamentally important for identifying genes relevant to biological processes [1] and provides insights into complex regulatory networks. Quantitative real-time PCR (qRT-PCR) [2], [3] is one of the most reliable and reproducible techniques available to measure and evaluate changes in gene expression[4], which is often used to confirm or refute interpretations of relative gene expression profiles derived from high-throughput systems [4], [5]. The qRT-PCR technique is sensitive enough to detect subtle alterations in gene expression, even for those with fairly low transcript levels [6], [7]. Although this powerful technique is often described as the gold standard, results are inevitably affected by different experimental conditions, such as different amounts of starting material, quality and integrity of template RNA samples, reverse transcription efficiency, recovery and integrity of mRNA, primer design and transcription efficiency [8]. Additionally, random pipetting errors can add technical variability to the data [9], [10]. As these factors can potentially render the quantification of gene transcripts unreliable, having a robust system for normalization of qRT-PCR data is essential to avoid non-specific variations or errors [6], [11]. The most common method for normalizing gene expression levels is to compare mRNA levels of the genes of interest to those of endogenous control genes, which are often called housekeeping or reference genes.
Ideal reference genes should not be regulated or influenced by the experimental procedure or co-regulated with the target gene. They should also be expressed in abundance and have minimal innate variability [12]. However, the indiscriminate use of some internal reference genes is questionable, since their expression levels are regulated according to cellular conditions [13]–[15]. Several studies have shown that this approach can introduce large errors when the expression of such “housekeeping genes” varies under different treatments and in different tissues [16], [17].
The beet armyworm, Spodoptera exigua (Lepidoptera: Noctuidae), is a widespread and polyphagous lepidopteran pest that causes severe economic damage in both dicotyledon (e.g., sugar beet, alfalfa, cotton, chrysanthemum) and monocotyledon (e.g., rice) crops and flower species. Molecular studies have been widely conducted previously in S. exigua [18]–[20], including investigations of insecticide resistance [21]–[23] and the role of important genes in physiological processes of the insect [24]. Understanding the function of important regulatory genes at the molecular level is essential for pest control. The molting, feeding and wandering stages are three larval physiological stages of the Lepidoptera larvae, which are regulated by different specific hormone levels. Tissues and genes in these three larval physiological stages have shown significant differences [25]–[27]. Therefore, molecular studies directed towards the three larval physiological stages are at the center of Lepidoptera physiological research. The fat body is a major tissue found to play an important role in the metabolism and detoxification of xenobiotics in insects [28], [29]. Receptors involved in insecticide resistance have been found in the midgut [30], [31], and several antiviral proteins and genes of potential value in clinical medicine were found in the epidermis and hemolymph [32]. Exploring gene expression profiles in these tissues will help our understanding of the regulation of the three larval physiological stages and facilitate application of useful resources to control the pest. Several genes have been demonstrated to be differentially expressed in some tissues based on sex [33], and some genes related to insecticide resistance, such as P450, are regulated by female mating [34]. Some studies have also reported differences in expression of the sex pheromones of S. exigua [35].
To date, studies have been published on evaluating the stability of reference genes in some insects [36], [37], [38]. Teng et al. [39] chose 4 candidates and screened the relatively stable reference genes in 4 lepidopteran insect species including S. exigua. Several studies have shown that each candidate reference gene should be evaluated under specific experimental conditions for gene expression profiling to ensure that expression occurs at a constant level [40]. The evaluation and selection from just four candidate genes seemed to be insufficient. Therefore, ten commonly used reference genes β-actin1(ACT1), β-actin2 (ACT2), elongation factor1(EF1), elongation factor 2 (EF2), Glyceralde hyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L10 (L10), ribosomal protein L17A (L17A), superoxide dismutase (SOD), α-tubulin (TUB), 18S ribosomal RNA (18S) from S. exigua were tested and their effectiveness for the normalisation of expression studies were further validated by quantitative analysis of a well-studied target diapause-specific peptide (DSP) gene. Three available and commonly used tools (geNorm, NormFinder and BestKeeper) were used to determine a set of the most stably expressed genes in different developmental stages (egg, 1st larvae, 2nd larvae, 3rd larvae, 4th larvae, 5th larvae, prepupae, pupae and adult), in both sexes of pupae and adults, as well as in five different tissues (epidermis, head, midgut, fat body and hemolymph) and three larval physiological stages (molting stage, feeding stage and wandering stage) of S. exigua. The objectives of this investigation were (i) to provide appropriate reference genes to develop an accurate and comprehensive qRT-PCR method for use in S. exigua studies, and (ii) to assess the importance of variations in relative quantification among normalization strategies in different developmental stages, sexes, larval physiological stages and tissues, with a focus on the merits of using multiple versus single reference genes in different studies.
Materials and Methods
Insects
S. exigua were reared on an artificial diet [41] at 27±1°C (14L: 10D). Pupae were selected and sexed on the third day. Adult males and females were allowed to emerge in transparent containers and fed with a 5% honey solution.
Sample collection
The stability of candidate genes was tested in different S. exigua samples of (i) five different tissues in three larval physiological stages, (ii) different developmental stages and (iii) two sexes. Only the tissue samples in three larval physiological stages had been dissected individually and all other samples were whole body. For each of the different sample groups, three replicate cages were used.
Sampling of different tissues in three larval physiological stages
For this study, the beet armyworms were synchronized in the 4th larval molting stage, 5th larval feeding stage (48 h post-molting) or 5th larval wandering stage (96 h post-molting), and then larvae in the three larval physiological stages were dissected individually using a dissection needle in physiological saline. The epidermis, head, midgut, fat body and hemolymph were collected separately. The collected tissues were quickly frozen and homogenized immediately after dissection with liquid nitrogen in a mortar and used for RNA extraction.
Samples of different developmental stages
The beet armyworms in different developmental life stages were collected separately and pooled as follows: eggs (50–80 per pool), 1st larvae (50–80 per pool), 2nd larvae (50–80 per pool), 3rd larvae (10 per pool), 4th larvae (10 per pool), 5th larvae (10 per pool), prepupae (10 per pool), pupae (10 per pool) and adults (10 per pool).
Samples of different sexes
Male pupae (10 per pool), female pupae (10 per pool), male adults (10 per pool) and female adults (10 per pool), were collected separately and placed in 1.5 ml centrifuge tubes.
Selection of gene sequences and primer design
PCR primer sequences used for quantification of the 10 candidate genes are shown in Table 1. The secondary structure of the DNA template was analyzed with UNAFold[42] using the mfold web server (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form) [43] with the following settings: melting temperature, 60°C; DNA sequence, linear, Na+ concentration, 50 mM; Mg++ concentration, 3 mM. Other parameters were set by default. The primers were designed using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome), with the settings: primer melting temperature, 60°C; primer GC content, 40–60%; and PCR product size, 80–200 base pairs. The excluded regions were based on results of analysis by mfold, and other parameters were set by default.
Table 1. Description, primer sequence and amplicon characteristics for the 10 candidate reference genes and a target gene used in this study.
| Gene symbol | Gene name | (putative)Function | Gene ID | Primer sequences [5′→3′] | L(bp)a | E(%)b | r2 c | slope | y intercept |
| ACT1 | β-actin1 | Involved in cell motility, structure and integrity | AEJ38214.1 | For 5′ AAGCCTTCGATGCCACCGGGTA 3′ Rev 5′ TTCGGGCGTGTTTAGTGGAGGC 3′ | 170 | 107.3 | 0.997 | −3.15857 | 52.363 |
| ACT2 | β-actin2 | Involved in cell motility, structure and integrity | AEJ38216.1 | For 5′ GGCTGCCGACATAGACATGCG 3′ Rev 5′ GGGTCCTCCACGCGGATCTT 3′ | 180 | 107 | 0.996 | −3.16485 | 51.900 |
| EF1 | elongation factor1 | Catalysation of GTP-dependent binding of amynoayl-total RNA to the ribosome | AEJ38219.1 | For 5′ TGCAGAGAAGCAAGTATTTGAGCGA 3′ Rev 5′ CCACGAGCTTTCTCTTCCGGAGC 3′ | 180 | 101.5 | 0.990 | −3.2865 | 57.719 |
| EF2 | elongation factor 2 | Catalysation of GTP-dependent binding of amynoayl-total RNA to the ribosome | AAL83698.1 | For 5′ CTGACCGCGCAACCCAGACT 3′ Rev 5′ CACGAACATGGGGGTACCAGCG 3′ | 150 | 99.4 | 0.998 | −3.33639 | 51.986 |
| GAPDH | Glyceralde hyde-3-phosphate dehydrogen ase | Glycolytic enzyme | AEJ38217.1 | For 5′ CTGAGGAACAGGTCGTGTCATCCGA 3′ Rev 5′ GATCGATAACGCGGTTGGAGTAGCC 3′ | 150 | 98.5 | 0.996 | −3.3584 | 50.509 |
| L10 | ribosomal protein L10 | Structural constituent of ribosome | ABX54738.1 | For 5′ GGCTACGGTCGACGACTTCCC 3′ Rev 5′ GCAGCCTCATGCGGATGTGGAAC 3′ | 155 | 102.6 | 0.997 | −3.26116 | 52.455 |
| L17A | ribosomal protein L7A | Structural constituent of ribosome | ABX55885.1 | For 5′ TGAGCTTGTCCTCTTCCTGCCC 3′ Rev 5′ GCTGCACGGTCGCCAGACTC 3′ | 150 | 101 | 0.996 | −3.2982 | 51.733 |
| SOD | Superoxide dismutase | Highly specific superoxide dismutation activity | ABX11259.1 | For 5′ GCCGTGTGTGTTCTCAAGGGCG 3′ Rev 5′ GCGCCAGCTGACGTGCATCC 3′ | 170 | 101.3 | 0.993 | −3.29117 | 53.057 |
| TUB | α-tubulin | Cytoskeleton structural protein | ADL38966.1 | For 5′ CGTGACGACGTGTCTGCGGT 3′ Rev 5′ GCGTGAGCTCGGGTACGGTG 3′ | 167 | 100.2 | 0.998 | −3.31714 | 55.528 |
| 18S | 18S ribosomal RNA | Cytosolic small ribosomal subunit | JN863293.1 | For 5′ GGTCCATCACGATGCGGTGGG 3′ Rev 5′ TACCCAATCGCAACCGAGCAACG 3′ | 150 | 111.4 | 0.991 | −3.07593 | 55.544 |
| DSP | diapause-specific peptide | An endogenous diapause -specific peptide; antifungal activity | HQ128581.1 | For 5′ ATGGCCGCTCTCAAGACCAC 3′ Rev 5′ TCATCAGTAACAGTCCATCCTACCG 3′ | 195 | 108.3 | 0.995 | −3.13785 | 58.553 |
a Amplicon length;
b Real-time qPCR efficiency (calculated by the standard curve method);
c Regression coefficient calculated from the regression line of the standard curve.
Total RNA isolation and cDNA synthesis
All collected samples were preserved in microcentrifuge tubes (1.5 ml) and stored at −80°C after being frozen in liquid nitrogen. Subsequently, three total RNA samples were prepared for each sample set using the SV Total RNA Isolation System (Promega, USA). According to the protocol of the kit, total RNA was incubated for 15 minutes at 20–25°C after adding 5 µl DNase I enzyme (Promega, USA). The purified RNA was stored at −80°C before further processing. The quality and quantity of RNA were assessed with a UV-1800 spectrophotometer (Shimadzu, Japan). cDNA was produced using the PrimeScript 1st Strand cDNA Synthesis Kit (TAKARA, Japan) in a total volume of 20 µl, with 4 µl 5×PrimeScript Buffer,1 µg of total RNA, 1 µl oligo dT primer, 1 µl PrimeScript RTase (200 U/µl), and 0.5 µl RNase Inhibitor (40 U/µl). Following the manufacturer's protocol, the 20 ul mixture was incubated for 60 minutes at 42°C. No-template and no-reverse transcription (no-RT) controls were run for each reverse transcription run for the control treatment. cDNA was stored at−20°C until used.
qRT-PCR
Triplicate first strand DNA aliquots for each sample served as templates for qRT-PCR using SoFast™ EvaGreen® Supermix (Bio-Rad, USA) on an iQ2 Optical System (Bio-Rad). Each amplification reaction was performed in a 20 µl total volume with 1 µl of cDNA and 100 nM of each primer in an iQ™ 96-well PCR plate (Bio-Rad), which was covered with Microseal “B” adhesive seals (Bio-Rad). Thermal cycling conditions included initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 10 s. After all reactions, a melting curve analysis from 65 to 95°C was applied to ensure consistency and specificity of the amplified product. A 10-fold dilution series of cDNA from the whole body of adults was employed as a standard curve, and the qRT-PCR efficiency was determined for each gene and each treatment with the slope of a linear regression model [44]. The corresponding qRT-PCR efficiencies (E) were calculated according to the equation: E = (10[−1/slope] −1)×100 [45].
Stability of gene expression
The stability of candidate genes was evaluated by three commonly used software tools, BestKeeper [46], [47], geNorm (http://medgen.ugent.be/~jvdesomp/genorm/)[48] and NormFinder (http://www.mdl.dk/publicationsnormfinder.htm) [49]. The Excel based tool Bestkeeper, is able to compare expression levels of up to ten housekeeping genes together with ten target genes, each up to hundred biological samples. The raw data of cycle threshold (Ct) values(CP values) and PCR efficiency (E) of the candidate genes were used to determine the best-suited standards by BestKeeper. The underlying principle for identification of stably expressed reference genes by Bestkeeper is that the expression levels of suitable reference genes should be highly correlated. Therefore, the correlation between each candidate and the index is calculated, describing the relation between the index and the contributing candidate reference gene by the coefficient of determination and the P value [46].Ct values converted to linear values (the lowest relative quantity for each gene was set to 1) were used as input data for subsequent analyses with geNorm and NormFinder. Similar with Bestkeeper, the key principle of geNorm is that the expression ratio of two suitable reference genes should be constant across samples. geNorm algorithm first calculates an expression stability value (M) for each gene and then compares the pairwise variation (V) of this gene with the others. Using microarray data as a training set for the algorithm, the value of Vn/Vn+1 indicates the pairwise variation between two sequential normalization factors and determines the optimal number of reference genes required for accurate normalization. A value below 0.15 indicates that an additional reference gene will not significantly improve normalization. Reference genes are ranked according to their expression stability by a repeated process of stepwise exclusion of the least stably expressed genes [48]. NormFinder provides a stability value for each gene which is a direct measure for the estimated expression variation enabling the user to evaluate the systematic error introduced when using the gene for normalizsation [49]. Every gene was ranked by the three software tools and assigned an appropriate weight separately. The final ranking was established after calculating the geometric mean of their weights.
Evaluation of target gene expression
DSP of S. exigua was used as a target gene to evaluate the candidate reference genes. Normalized with different reference genes, relative quantification of DSP in different samples were conducted according to threshold cycle (Ct) value based on 2−△△Ct method.
Results
Amplification efficiencies
The initial screening of 10 candidate reference genes and one target gene by PCR showed that all of the genes were expressed in all S. exigua sample sets, as indicated by the presence of a single amplicon of the expected size on a 2% agarose gel. In order to determine the amplification efficiency of all 11 genes in the study, 5-point standard curves with known concentrations of transcribed reference RNA were made. All amplification efficiencies in the qRT-PCR analysis for the 10 candidate genes and one target gene ranged between 98.5∼111.4% compared with the templates from which the primers were designed. Linear regression coefficients (r2) for all 11 genes were ≥0.990 (Table 1).
Expression levels of 10 candidate reference genes
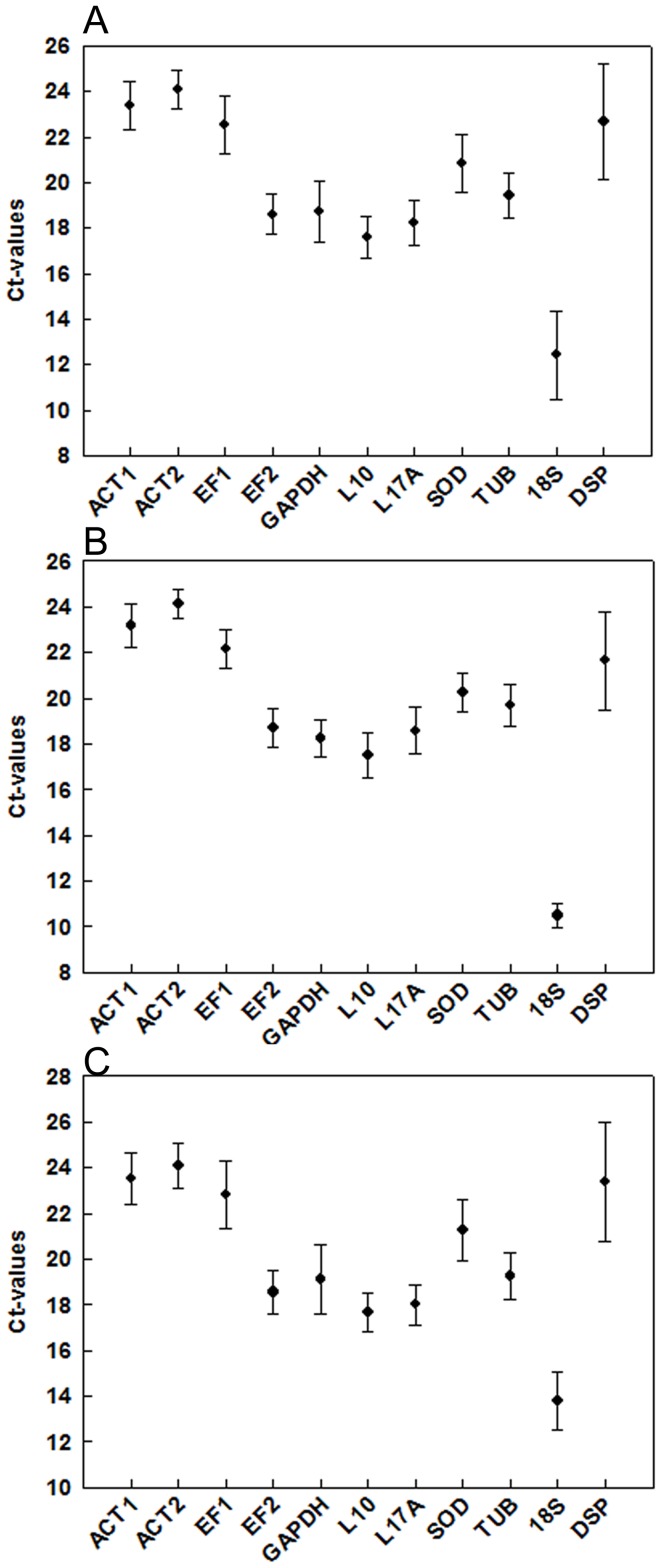
Relative Ct values are widely used as a simple way to identify stably expressed genes by qRT-PCR. Gene expression analyses of the 10 candidate genes exhibited a narrow mean Ct value range across all the experimental samples (Figure 1). The Ct values of the candidate reference genes under the same threshold value for fluorescence ranged from 9.18 for 18S to 26.00 for ACT2, which were the most and least abundant transcripts, respectively. There were no much differences among the average Ct values for each gene, and the range of values was consistently narrower in individuals than in tissue samples, when the two sample sets consisting of the developmental stage samples and the tissue ones in three larval physiological stages were compared (Figure 1B and 1C). The amplification of 18S, which was generally highly expressed, produced much lower Ct values (mean Ct = 12.41) than did other genes overall. The other candidate reference genes were expressed at moderate levels, with mean Ct (n = 26 samples) values of 23.37, 24.11, 22.53, 18.56, 18.75, 17.62, 18.25, 20.85 and 19.45 for ACT1, ACT2, EF1, EF2, GAPDH, L10, L17A, SOD and TUB, respectively (Figure 1A). The Ct values obtained for the target gene DSP varied in different samples, ranging from 18.23 (female pupae) to 27.51 (midgut of 5th feeding larval stage). Therefore, the standard errors of Ct values obtained for DSP were larger than those of all of the candidate reference genes studied (Figure 1).
Figure 1. Range of Ct values in different developmental stages and tissues of S.exigua.
The above plots show expression levels of 10 candidate reference genes and a target gene in (A) all S. exigua samples (n = 26), (B) different developmental stage samples (n = 11) and (C) all tissues samples in three larval physiological stages (n = 15). Values are given as Ct values from the mean of duplicate samples. Bars indicate standard error of the mean.
Expression stability of candidate reference genes
Ct values of the 10 candidate reference genes were obtained in each sample, and variations in their expression were assessed by Bestkeeper. Ct values converted to linear values were used as input data for subsequent analyses with geNorm and NormFinder.
BestKeeper analysis
The high correlation of expression levels is the key principle for identification of stable reference genes, which ideally should display similar expression patterns across samples. The program BestKeeper was used to determine variations in expression and standard deviations (SD) of 10 candidate reference genes and a target gene. Examination of the standard deviations (SD) (Table 2) revealed that the candidate reference genes were not all stable across different samples, because some showed SD values higher than 1.0. The variations were diverse in different sample groups.
Table 2. Descriptive statistic analysis with BestKeeper.
| ACT1 | ACT2 | EF1 | EF2 | GAPDH | L10 | L17A | SOD | TUB | 18S | DSP | ||
| Specific Larval Physiological Stages | ||||||||||||
| Molting | SD (±CPa)b | 0.982 | 0.9 | 1.132 | 0.678 | 0.996 | 0.681 | 0.689 | 0.978 | 0.462 | 1.09 | 2.451 |
| Stage | BK Corr [r]c | 0.688 | 0.771 | 0.942 | 0.962 | 0.9 | 0.971 | 0.968 | 0.937 | 0.667 | 0.77 | −0.025 |
| Feeding | SD (±CP) | 0.744 | 0.757 | 1.128 | 0.931 | 1.513 | 0.835 | 0.75 | 1.194 | 0.579 | 1.111 | 2.734 |
| Stage | BK Corr [r] | 0.556 | 0.675 | 0.937 | 0.959 | 0.967 | 0.983 | 0.97 | 0.952 | 0.846 | 0.907 | 0.469 |
| Wandering | SD (±CP) | 0.864 | 0.643 | 0.987 | 0.497 | 0.885 | 0.528 | 0.692 | 0.608 | 1.009 | 0.665 | 1.136 |
| Stage | BK Corr [r] | 0.773 | 0.746 | 0.836 | 0.821 | 0.81 | 0.902 | 0.902 | 0.479 | 0.862 | −0.083 | 0.257 |
| Five tissues in different stages | ||||||||||||
| Total d | SD (±CP) | 0.882 | 0.757 | 1.224 | 0.672 | 1.322 | 0.677 | 0.73 | 1.133 | 0.763 | 0.94 | 2.267 |
| BK Corr [r] | 0.618 | 0.712 | 0.824 | 0.923 | 0.84 | 0.965 | 0.937 | 0.781 | 0.601 | 0.685 | 0.231 | |
| Epidermis | SD (±CP) | 0.486 | 0.228 | 0.668 | 0.25 | 0.697 | 0.149 | 0.14 | 0.87 | 0.624 | 0.534 | 1.675 |
| BK Corr [r] | 0.411 | 0.647 | 0.552 | 0.101 | 0.746 | 0.343 | 0.402 | 0.831 | −0.023 | −0.186 | 0.111 | |
| Fat body | SD (±CP) | 0.531 | 0.507 | 0.855 | 0.358 | 0.938 | 0.312 | 0.412 | 1.186 | 0.528 | 0.466 | 1.534 |
| BK Corr [r] | 0.4 | 0.452 | 0.663 | 0.84 | 0.742 | 0.909 | 0.795 | 0.797 | −0.016 | −0.05 | 0.302 | |
| Head | SD (±CP) | 0.685 | 0.556 | 0.816 | 0.292 | 1.112 | 0.228 | 0.226 | 1.01 | 0.823 | 0.475 | 0.918 |
| BK Corr [r] | 0.788 | 0.909 | 0.042 | 0.872 | 0.763 | 0.73 | 0.657 | 0.801 | 0.331 | 0.467 | 0.308 | |
| Hemolymph | SD (±CP) | 0.858 | 0.779 | 0.886 | 0.838 | 0.688 | 0.686 | 0.571 | 1.386 | 0.864 | 0.888 | 1.584 |
| BK Corr [r] | 0.946 | 0.953 | 0.908 | 0.982 | 0.942 | 0.967 | 0.955 | 0.901 | 0.903 | 0.953 | 0.874 | |
| Midgut | SD (±CP) | 1.177 | 1.165 | 1.771 | 0.604 | 1.11 | 0.745 | 0.939 | 0.835 | 0.865 | 0.486 | 1.905 |
| BK Corr [r] | 0.979 | 0.994 | 0.994 | 0.908 | 0.838 | 0.983 | 0.976 | 0.839 | 0.972 | 0.461 | −0.798 | |
| Developmental life stages | ||||||||||||
| Developmental | SD (±CP) | 0.688 | 0.481 | 0.654 | 0.661 | 0.645 | 0.778 | 0.81 | 0.714 | 0.743 | 0.471 | 1.572 |
| life stages e | BK Corr [r] | 0.639 | 0.647 | 0.743 | 0.538 | 0.689 | 0.683 | 0.728 | 0.865 | 0.633 | 0.223 | −0.172 |
| Larvae | SD (±CP) | 0.287 | 0.244 | 0.298 | 0.601 | 0.593 | 0.436 | 0.538 | 0.422 | 0.531 | 0.473 | 1.157 |
| BK Corr [r] | 0.573 | 0.423 | 0.761 | 0.849 | 0.809 | 0.919 | 0.808 | 0.463 | 0.714 | −0.335 | 0.276 | |
| Pupae | SD (±CP) | 0.432 | 0.348 | 0.442 | 0.251 | 0.2 | 0.26 | 0.331 | 0.468 | 0.551 | 0.576 | 1.092 |
| BK Corr [r] | 0.949 | 0.95 | 0.731 | 0.772 | 0.892 | 0.851 | 0.782 | 0.953 | 0.933 | 0.955 | −0.599 | |
| Adult | SD (±CP) | 0.859 | 0.615 | 0.917 | 0.329 | 0.71 | 0.447 | 0.335 | 0.77 | 0.779 | 0.404 | 0.552 |
| BK Corr [r] | 0.968 | 0.978 | 0.838 | 0.917 | 0.942 | 0.861 | 0.906 | 0.966 | 0.954 | 0.023 | −0.163 | |
| Sex | ||||||||||||
| Male | SD (±CP) | 0.369 | 0.369 | 0.751 | 0.378 | 0.847 | 0.931 | 0.539 | 0.46 | 0.515 | 0.432 | 0.553 |
| BK Corr [r] | 0.186 | 0.3 | 0.576 | −0.238 | 0.758 | 0.723 | 0.88 | 0.789 | 0.822 | 0.728 | −0.161 | |
| Female | SD (±CP) | 1.496 | 1.159 | 0.564 | 0.853 | 0.173 | 0.297 | 0.221 | 0.958 | 1.048 | 0.466 | 1.08 |
| BK Corr [r] | 0.942 | 0.942 | 0.828 | 0.91 | 0.175 | −0.55 | 0.087 | 0.971 | 0.951 | 0.574 | −0.634 | |
a CP: Crossing point;
b SD (±CP): the standard deviation of the CP;
c BK CorrC [r]: Pearson correlation coefficient, correlation between the BestKeeper index and the contributing gene;
d Total, all the tissues samples in three specific larval physiological stages;
e Developmental Stages, all the developmental life stages samples.
Due to high variability as presented with SD (± CP) >1.0, the following genes in the indicated samples were excluded: EF1, GAPDH, and SOD in all tissues samples; EF1 and 18S in molting stage samples; EF1, GAPDH, SOD, and 18S in feeding stage samples; TUB in wandering stage samples; SOD in fat body and hemolymph samples; GAPDH and SOD in head samples; ACT1, ACT2, EF1, and GAPDH in midgut samples; and ACT1, ACT2, and TUB in female samples. The target gene DSP showed the highest variations with SD values of nearly 1.0. Other genes in each experimental condition were ranked based on Pearson's correlation coefficient (a higher coefficient indicates greater stability of expression) (Table 2). Interestingly, despite displaying acceptable stability levels (i.e., far below the default limit of SD 1.0), SOD had the highest standard deviation, indicating that it was the least stable of the candidate reference genes. Since the expression of 18S was exceptionally high and variable across the different treatments (Figure 1 and Table 2), it was excluded from further analyses.
geNorm analysis
Next, the geNorm software was used to determine the expression stability of the selected candidate genes in different samples. The expression ratio of two suitable reference genes should be constant across different samples, which is the underlying principle followed by the geNorm program. Two parameters defined by the program were used to assess the stability of reference genes: M (average expression stability) and V (pairwise variation). In each group of samples, the M stability value for each gene, which is inversely related to expression stability, was obtained as the average pair-wise variation in the transcript levels of one gene with respect to all other reference genes tested. V values were determined with all other control genes as the SD of the logarithmically transformed expression ratios.
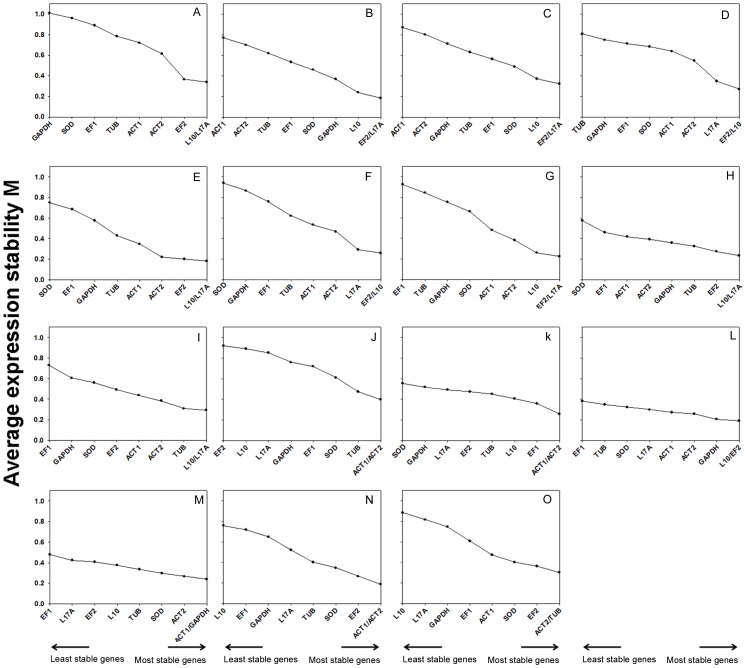
The gene with the highest M value was considered to have the least stable expression. Thus, the tested reference genes were ranked according to the stability of their expression by stepwise exclusion of the gene with the highest M value (Figure 2). Starting from the two most stable genes on the right, the genes are ranked according to reducing expression stability, ending with the least stable gene on the left. From all of the expression data of the tissue sample groups examined, EF2, L10, and L17A were the three most stable genes, suggesting that they play housekeeping roles and may be widely used for multiple conditions (Figure 2). While the ACT1 and ACT2 genes with an M value of 0.3990 were most stably expressed throughout the developmental stages (Figure 2J), EF2 and L10 showed the higher stable expression in pupae (Figure 2L). The highest ranked genes were ACT1 and ACT2 for larvae and male groups (Figure 2K, 2N); ACT1 and GAPDH for adults (Figure 2M); and ACT2 and TUB for female groups (Figure 2O). All tested reference genes reached high expression stability with M values below 1.1, far below the default limit of 1.5 for defining stably expressed genes.
Figure 2. Average expression stability values (M) of the candidate reference genes for tissue samples.
Average expression stability values (M) of the reference genes were measured during stepwise exclusion of the least stable reference genes. A lower M value indicates more stable expression, as analyzed by the geNorm software in S.exigua samples at five tissue samples in molting stage(B), five tissue samples in feeding stage(C), five tissue samples in wandering stage(D), epidermis samples in three specific larval physiological stages (E), fat body samples in three specific larval physiological stages (F), head samples in different stages (G), hemocytes samples in different stages (H), midgut samples in different stages (I), larvae samples (K), pupae samples (L), adult samples (M), male samples (N),female samples (O).The M values calculated for all the samples examined in all specific larval physiological stages(A) and all body samples examined in all developmental stages (J)are also given.
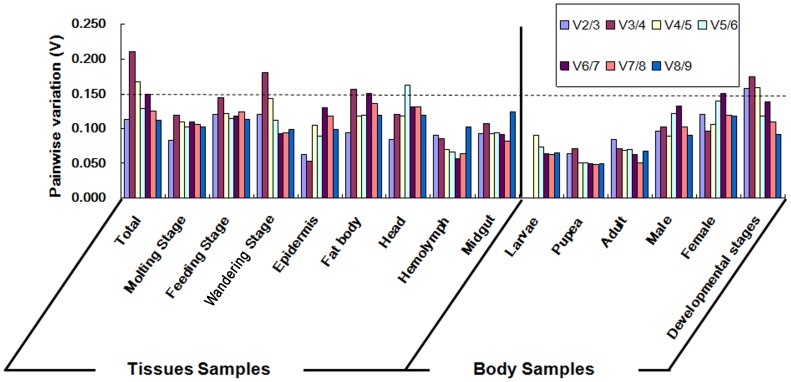
It has been reported that more than one reference gene is required for accurate normalization [16]. When the use of additional genes was equally informative, the pairwise variation (Vn/Vn+1) between the sequential normalization factors (NF) (NFn and NFn+1) was calculated by geNorm to determine the optimal number of reference genes for each experimental condition (Figure 3). The cut-off value of pairwise variation of 0.15 was proposed to indicate that inclusion of an additional reference gene would be unnecessary.
Figure 3. variation (V) analysis of the candidate reference genes.
The pairwise variation (Vn/Vn+1) was analyzed between the normalization factors NFn and NFn+1 by the geNorm software to determine the optimal number of reference genes required for qRT-PCR data normalization.
Analysis of the pairwise variation in all developmental stages samples revealed a significant decrease with the inclusion of a fifth gene (Figure 3). Normalization factors should preferably consist of at least five reference genes, because the pairwise variation of the V2/3, V3/4, and V4/5 values were 0.158, 0.175, and 0.159, respectively, all of which exceeded the threshold of 0.15, while the pairwise variation of the V5/6 value was 0.1118. Based on this analysis, EF1, SOD, TUB, ACT1 and ACT2 should be ideal reference genes for normalizing gene expression data in all developmental stages samples. Analysis of the pairwise variation in other samples revealed that two reference genes may be sufficient to normalize expression values of target genes. Therefore, the two most stably expressed genes, mentioned above for each type of samples were selected as reference genes.
NormFinder analysis
Finally, the NormFinder software tool was also employed to investigate each type of sample. This algorithm is used for identifying the optimal normalization gene among a set of candidate genes. When analyzing expression data using the qRT-PCR method, the software provides a stability value for each gene, which is the estimated expression variation if such gene is used for normalization. The candidate normalization genes were ranked according to the stability of their expression patterns between subgroups of the sample set under a given experimental condition. The lower average expression stability values represented more stable gene expression within the gene set examined.
Similarly to geNorm, the top-ranked candidates in different sample groups were analyzed by NormFinder (Table 3). Among all tissues, L10 ranked one of the three most stable genes, while EF2 and L17A ranked between the top four genes (except in hemolymph and midgut samples), in agreement with results of the other two programs. The ranking in the two sex sample groups showed significant differences compared with other body samples. For example, the L10 gene ranked among the four most stable genes for pupae, adult and all larvae sample groups, but it ranked last in the sex sample groups. The SOD gene also ranked better in male and female samples than in others samples. All ranking results are summarized in Table S1.
Table 3. S.exigua reference genes for normalization and their expression stability values calculated by the NormFinder software.
| Rank | Different tissues | Developmental life stages b | ||||||||||||
| Total a | Epidermis | Fat body | Head | Hemolymph | Midgut | |||||||||
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | L10 | 0.223 | ACT2 | 0.026 | EF2 | 0.089 | EF2 | 0.09 | EF2 | 0.141 | L17A | 0.114 | SOD | 0.327 |
| 2 | L17A | 0.314 | L17A | 0.149 | L10 | 0.089 | ACT2 | 0.163 | ACT2 | 0.184 | ACT2 | 0.137 | ACT2 | 0.339 |
| 3 | EF2 | 0.457 | L10 | 0.163 | L17A | 0.13 | L10 | 0.209 | L10 | 0.189 | L10 | 0.2 | GAPDH | 0.393 |
| 4 | ACT2 | 0.672 | EF2 | 0.23 | ACT2 | 0.438 | L17A | 0.244 | ACT1 | 0.207 | TUB | 0.213 | EF1 | 0.414 |
| 5 | ACT1 | 0.88 | ACT1 | 0.429 | ACT1 | 0.489 | ACT1 | 0.399 | GAPDH | 0.222 | ACT1 | 0.265 | TUB | 0.471 |
| 6 | SOD | 0.889 | GAPDH | 0.456 | EF1 | 0.639 | SOD | 0.582 | L17A | 0.227 | SOD | 0.421 | ACT1 | 0.478 |
| 7 | TUB | 0.91 | TUB | 0.538 | GAPDH | 0.643 | GAPDH | 0.661 | EF1 | 0.306 | EF2 | 0.454 | EF2 | 0.552 |
| 8 | EF1 | 0.917 | EF1 | 0.581 | TUB | 0.652 | EF1 | 0.731 | TUB | 0.318 | GAPDH | 0.458 | L17A | 0.558 |
| 9 | GAPDH | 0.981 | SOD | 0.601 | SOD | 0.732 | TUB | 0.764 | SOD | 0.639 | EF1 | 0.763 | L10 | 0.586 |
a Total, all the tissue samples in three specific physiological stages;
b Developmental Stages, all the developmental life stages samples.
Consensus list of reference genes
Because of the different algorithms used and the different sensitivities toward co-regulated reference gene candidates, the three software tools offered different ranks in each sample group. Although rankings of the most suitable reference genes were not identical, the three best reference genes identified by the different methods were similar, and they only varied in their relative rank positions (Table S1). Finally, the highest ranking reference genes were identified (Table 4). Interestingly, L10, EF2 and L17A ranked highest in different tissue groups, and ACT2 ranked as the fourth most stable (except in molting stage samples), indicating that these four genes could be selected as the best reference genes for tissue research in S. exigua.
Table 4. The best-ranking reference genes across different experimental conditions in S. exigua according to software analysis.
| Experimental conditions | The best-ranking reference genes | |||
| Molting Stage | EF2 | L10 | L17A | |
| Specific Larval Physiological Stages | Feeding Stage | L17A | L10 | EF2 |
| Wandering Stage | L17A | L10 | ||
| Total a | L10 | L17A | EF2 | |
| Epidermis | ACT2 | L17A | L10 | |
| Fat body | EF2 | L10 | L17A | |
| Different Tissues | Head | EF2 | L17A | L10 |
| Hemolymph | EF2 | L10 | L17A | |
| Midgut | L17A | L10 | ||
| Developmental Stages b | SOD | ACT2 | GAPDH | |
| Larvae | ACT1 | ACT2 | L10 | |
| Developmental life stages | Pupae | GAPDH | ACT2 | ACT1 |
| Adult | GAPDH | ACT2 | ACT1 | |
| Sex | Male | SOD | L17A | |
| Female | EF2 | SOD | ||
a Total, all the tissues samples in three Specific Larval Physiological Stages;
b Developmental Stages samples, all the developmental life stages samples.
In different whole body samples, the last ranking reference genes showed significant differences. The best reference genes were ACT2, ACT1 and L10 for larvae groups; GAPDH, ACT1 and ACT2 for pupae and adults; SOD and L17A for males; and EF2 and SOD for females (Table 4). Thus, for all developmental stage samples, using the five genes SOD, ACT2, GAPDH, EF1 and ACT1 together should provide reliable results in expression studies of S. exigua.
Target gene expression
In order to demonstrate the effect of reference genes on target gene expression data, the relative expression of the target gene DSP was investigated under different experimental conditions. Target expression analyses further showed that differences in quantification were detected when normalizing with arbitrary internal controls relative to the best reference genes. The best or the most unstable reference genes were selected based on their rank order of expression stability among the 10 candidates evaluated in this study.
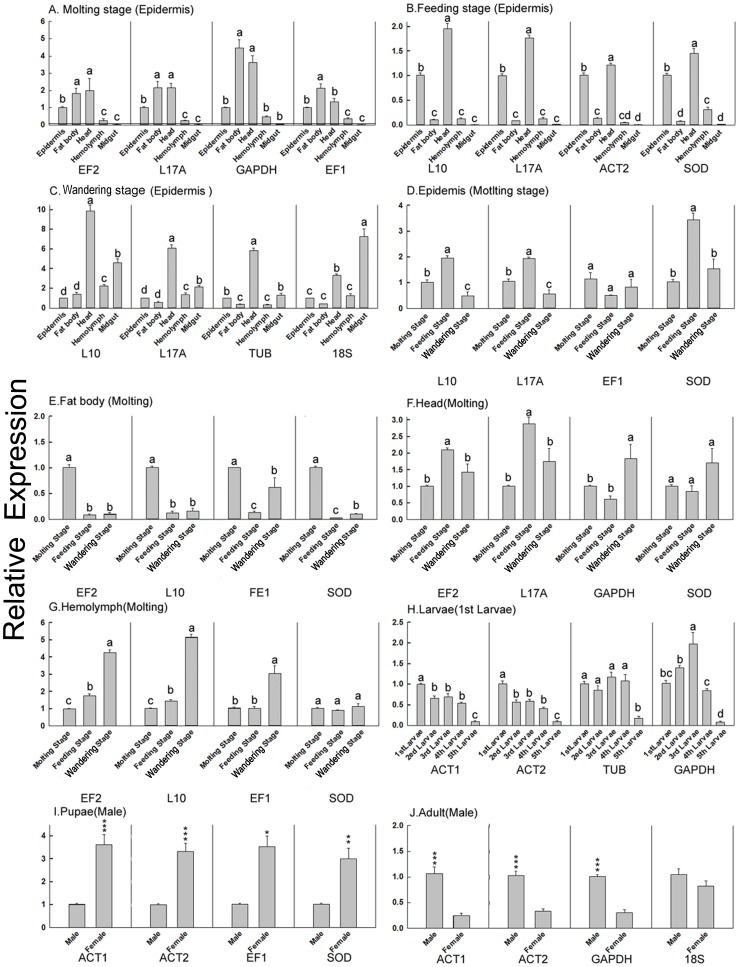
Arbitrary selection of reference genes may thus decrease the accuracy of calculating target gene expression, since such a normalization strategy can over-estimate or under-estimate differences in expression level among different samples. For example, the relative expression level of DSP showed no significant differences between adult male and female samples when calculated using 18S as the reference gene; however, its expression was significantly different when normalized by other reference genes (such as ACT1, ACT2) (Figure 4J). Similar changes also occurred in calculating relative expression levels of DSP after normalization by other unstable reference genes, such as GAPDH in the molting stage, TUB in the molting stage, SOD and 18S in the epidermis, SOD in the head, EF1 and SOD in hemolymph, TUB in all larvae groups and EF1 and SOD in pupae (Figure 4A, 2C, 2D, 2F, 2G, 2H and 2I).
Figure 4. Analysis of expression of the target gene DSP using different reference genes.
The relative expression of the target gene DSP among different samples normalized with different reference genes was investigated. Control groups used in each sample set were: A. molting stage (epidermis): epidermis samples in molting stage; B. feeding stage (epidermis): epidermis samples in feeding stage; C. wandering stage (epidermis): epidermis samples in wandering stage; D. epidermis (molting stage): epidermis samples in molting stage; E. fat body (molting stage): fat body samples in molting stage; F. head (molting stage): head samples in molting stage; G. hemolymph (molting stage): hemolymph samples in molting stage; H. larvae (1st larvae): 1st larvae samples; I. pupae (male): the male pupae samples; J. adult (male): male adult samples. Data are means ± SEM. The comparisons among more than two reference genes were analyzed using one-way ANOVA (from A to H). Those between two reference genes were compared using Student's t-test (I & J). *P<0.05; **P<0.01; ***P<0.001.
Relative expression levels of the target gene in different samples were even more divergent if calculated using arbitrary reference genes. For example, when using EF1 or SOD as reference gene, the fat body DSP expression in the feeding stage was lower than that in the wandering stage; however, after using other reference genes, the conclusion was modified, and they were determined to be at the same level (Figure 4E). Similar errors could be produced when using EF1 in the molting stage, and ACT2 or SOD in the feeding stage (Figure 4A, B). Thus, determination of the optimal reference genes is important for accurate normalization of qRT-PCR data, especially when differences in expression levels are subtle.
Inaccurate conclusions were made when certain reference genes were used for normalization. While the larvae DSP expression level was higher in 3rd larvae when using GAPDH as the reference gene, it exhibited a significant age-dependent decrease in larvae when normalizing with ACT1 or ACT2 (Figure 4H). Similar results were found when using 18S in the molting stage and GAPDH in the head tissue (Figure4C, 4F). Taken together, results of this study showed that the selection of reference genes for qRT-PCR data normalization varied on a case-by-case basis. Thus, in order to obtain accurate expression data, any given sample set must be assessed using the panel of selected candidate reference genes.
Discussion
qRT-PCR quantification requires robust normalization by reference genes to offset confounding variations in experimental data. However, improper selection of reference genes can conceal or magnify real biological changes due to changes in the reference gene expression [50], [51]. Furthermore, using a single endogenous control can also profoundly influence the statistical outcome and may lead to inaccurate data interpretation [52]. Each candidate reference gene should be evaluated under specific experimental conditions for gene profiling to ensure a constant level of expression [40]. In this study, we examined 10 candidate reference genes from S. exigua and analyzed the stability of these genes across various sample sets using three analytical software programs. These genes involved in ubiquitous cellular processes represent those commonly used as single normalizer in S. exigua gene expression studies. The studied sample subgroups included detailed life developmental stages (prepupae and every instar of the larvae), both sexes and five different tissues in three larval physiological stages. Although different ranks were offered by the three analytical tools, the combined results ultimately provided recommendations for the optimal reference genes. The different rankings of the reference genes in different sample sets in this study illustrated the need for evaluating their use under various experimental conditions. Compared with a previous report [39], our work provides a more complete set of information for the selection of reference genes in S. exigua.
A major conclusion arising from our results is that none of the candidate reference genes could serve as a “universal” normalizer that would maintain a constant expression level across all experimental conditions. Although most of these candidates are considered to be classical housekeeping genes and are widely used for data normalization, they exhibited considerable variations in expression stability among the different samples. The results of this study emphasized that the stability of reference gene expression must be verified under all experimental conditions to be investigated. Given that all internal reference genes are regulated to some extent and if none are constitutively expressed for each experimental treatment, then a combination of reference genes that would best fulfill the universality criteria should be selected (i.e., L10, EF2, and L17A across different tissue subgroups in our study).
In contrast to the findings of Teng et al [39], who selected relatively stable reference genes just from four candidate housekeeping genes in certain tissues from final instar larvae and developmental stages, we investigated the influence of many more variables (e.g. five different tissues in three larval physiological stages and life developmental stages) on the expression stability of the studied genes. The authors of the study above chose GAPDH as one of the most stable reference genes, while GAPDH ranked last among the reference genes across most sample sets in the current study (Table S1). Assessing a low number of initial candidate genes would lead to such a misleading result. Although the GAPDH gene ranked in the first three in all developmental stages sample subgroup, normalizing the expression of a target gene just by one reference gene is not ideal. Since the GAPDH gene ranked highly in some sample subgroups, it may be used as the reference gene in certain experimental conditions but not as a sole universal normalizer.
While stability of reference genes still must be determined on a case-by-case basis in S. exigua studies, certain genes may be preferred for normalization in experiments involving different treatments. The lowest ranking reference genes also showed significant differences across different whole body sample subgroups. This observation indicated that candidate genes in body subgroups showed more variations than in tissue subgroups.
Results of this study suggest that more complex sample sets will exhibit higher variability in the reference genes. To determine the optimal number of reference genes, the pairwise variation (Vn/Vn+1) between the sequential NF (NFn and NFn+1) was calculated by geNorm. After the analysis, two reference genes were found to be sufficient for normalizing expression values of target genes in most of the samples, but five reference genes were needed in all of the life developmental stages samples (Figure 3), indicating that larger sample sizes require a higher number of reference genes for accurate normalization. The same results were obtained in a Drosophila reference gene selection study [53]. Along the same way, in an aging-related study, nine reference genes were sufficient for three samples, whereas 13 reference genes were needed when nine samples were tested. Finally in a neurodegeneration-related study, one reference gene was feasible for three samples; however, the number of the reference genes needed to be increased to six for accurate normalization in nine samples. Perhaps additional reference genes are required when adding more samples into a study, because it would be harder to reach the minimum value of Vn/n+1, due to the introduction of more unstable factors.
Compared to the other reference genes tested, 18S ranked low in most experimental conditions and displayed an excessively high expression level, excluding it as a potential reference gene. However, the differences in rRNA and mRNA fractions between samples limit the use of 18S as a normalizer in qRT-PCR analyses [1], [40], [54]. In other words, rRNA cannot be used for correcting sample-to-sample variation in the quantity of mRNA, as it has been shown on occasion to fail to be representative of mRNA levels [55]. This shortcoming may explain the high coefficient of variation of 18S in our study. The low Ct values of 18S observed in our study (Figure 1) reflect the abundance of these transcripts. Thus, in order to use 18S as a reference gene, the samples would need to be diluted sufficiently to keep the rRNA within the range of detection. However, the target gene would also be diluted further, potentially leading to an over-estimation or under-estimation in differences of target gene expression among different conditions (Figure 4). Therefore, we suggest that18S should be excluded as a reference gene in qRT-PCR, since the expression of other candidates proved to be more stable.
SOD was never listed in the top three ranked reference genes across tissue sample sets and some of the whole body sample sets in our study (Table 4 & Table S1). Similarly, the glycolytic enzyme GAPDH was selected as a suitable reference gene for only three out of fifteen samples (i.e., pupae and adult), even though it has been reported as a good normalizer in previous gene expression studies of S. exigua and other insect species [38]. In contrast, L10 and L17A, were found to be stably expressed across the different samples. Though they all encode the structural constituents of the ribosome, it was reported that L17A was related to the pupal diapause regulation in the insect [56] and L10 was a component of an antiviral signaling [57]. These observations indicate that the two ribosomal protein genes are not co-regulated and can be regarded as independent reference genes. Additionally, EF2 ranked at the top as a reference gene in most samples in this study, but it has rarely been previously used as a normalizer. Interestingly, EF1 ranked last in some sample sets in our study. The elongation factors (i.e., EF1 & EF2) play an important role in translation by catalyzing GTP-dependent binding of aminoacyl tRNA to the acceptor site of the ribosome. Recently, a number of studies have reported that it is a suitable reference gene in different species, including salmon [58], [59], humans [60] and Orthoptera [61], [62].
Actin, as the major component of the protein scaffold which supports the cell and determines its shape, was also selected as a good reference gene under some conditions. It was expressed at moderately abundant levels in most samples. Even though two actin genes were selected as candidate reference genes in our study, they have been reported to be unsuitable for normalizing qRT-PCR data due to large measurement errors [4]. On the other hand, actin has ranked at the top as a reference gene in expression studies in the desert locust [61], European honey bee [38], two species of Collembola [63] and the salmon louse [58].
Taken together, the simultaneous measurement of a panel of candidate reference genes is essential for quantification by qRT-PCR. As empirically-determined or pre-validated reference genes may yield inaccurate results, data normalization needs to be optimized for each particular assay.
Conclusion
In our study, several reference genes suitable for normalizing qRT-PCR data in S. exigua were identified. Although most of the selected candidates exhibited stable expression patterns acceptable for reference genes, some showed the highest stability in different experimental conditions. While the expression levels of L10, EF2, and L17A were most stable in different tissue sample sets, the best reference genes selected were ACT2, ACT1, and L10 for larvae samples; GAPDH, ACT1, and ACT2 for pupae and adults; SOD and L17A for males; and EF2 and SOD for females. Overall, five genes, SOD, ACT2, GAPDH, EF1, and ACT1, were determined to be most reliable when used together to analyze all developmental stage sample groups in S. exigua.
Supporting Information
Ranking of candidate reference genes according to their stability value using BestKeeper, geNorm, and NormFinder analyses. Candidates are listed from top to bottom in order of decreasing expression stability.
(DOC)
Acknowledgments
Special thanks go to Dr. Mariana del Vas (Instituto de Biotecnología, CICVyA, Instituto Nacional de Tecnología AgropecuariaI (IB-INTA), Las Cabañas y Los Reseros s/n, Buenos Aires, Argentina), Prof. David Majerowicz, PhD (Universidade Federal do Rio de Janeiro), Prof. Zhou Xuguo (University of Kentucky) for comments on an earlier draft.
Funding Statement
This research was supported by the National Natural Science Foundation of China (No. 31201544), Hubei Province Science & Technology Department of China (No. 2011BFA018), Beijing Natural Science Foundation of China (6102021), and Independent Innovation Foundation of Huazhong Agricultural University (No. 2011PY123). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gibson UE, Heid CA, Williams PM (1996) A novel method for real time quantitative RT-PCR. Genome Res 6: 995–1001. [DOI] [PubMed] [Google Scholar]
- 3. Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6: 986–994. [DOI] [PubMed] [Google Scholar]
- 4. Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization: strategies and considerations. Genes Immun 6(4): 279–284. [DOI] [PubMed] [Google Scholar]
- 5. Provenzano M, Mocellin S (2007) Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv Exp Med Biol 593: 66–73. [DOI] [PubMed] [Google Scholar]
- 6. Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nat Protoc 1: 1559–1582. [DOI] [PubMed] [Google Scholar]
- 7. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR – a perspective. J Mol Endocrinol I34: 597–601. [DOI] [PubMed] [Google Scholar]
- 8.Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription–PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: , 5245–5250. [DOI] [PubMed] [Google Scholar]
- 9. Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse transcription polymerase chain reaction. J Biomol Tech 15: 155–566. [PMC free article] [PubMed] [Google Scholar]
- 10. Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27: 126–139. [DOI] [PubMed] [Google Scholar]
- 11. Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization; strategies and considerations. Genes Immun 6: 279–284. [DOI] [PubMed] [Google Scholar]
- 12. Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856–862. [DOI] [PubMed] [Google Scholar]
- 13. Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, et al. (2001) Beta-actin an unsuitable internal control for RT-PCR. Mol Cell Probes 15: 307–311. [DOI] [PubMed] [Google Scholar]
- 14. Glare EM, Divjak M, Bailey MJ, Walters EH (2002) Beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalizing mRNA levels. Thorax 57: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem 295: 17–21. [DOI] [PubMed] [Google Scholar]
- 16. Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Bioph Meth 46: 69–81. [DOI] [PubMed] [Google Scholar]
- 17. Ruan WJ, Lai MD (2007) Actin, a reliable marker of internal control? Clinica Chimica Acta 385(1-2): 1–5. [DOI] [PubMed] [Google Scholar]
- 18. Pascual L, Jakubowska AK, Blanca JM, Cañizares J, Ferré J, et al. (2012) The transcriptome of Spodoptera exigua larvae exposed to different types of microbes. Insect Biochem Mol Biol 42(8): 557–70. [DOI] [PubMed] [Google Scholar]
- 19. Choi JY, Roh JY, Wang Y, Zhen Z, Tao XY, et al. (2012) Analysis of genes expression of Spodoptera exigua larvae upon AcMNPV infection. PLoS One 7(7): e42462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang D, Chen J, Yao Q, Pan Z, Chen J, et al. (2012) Functional analysis of two chitinase genes during the pupation and eclosion stages of the beet armyworm Spodoptera exigua by RNA interference. Arch Insect Biochem Physiol 79(4-5): 220–34. [DOI] [PubMed] [Google Scholar]
- 21. Swevers L, Soin T, Mosallanejad H, Iatrou K, Smagghe G (2008) Ecdysteroid signaling in ecdysteroid-resistant cell lines from the polyphagous noctuid pest Spodoptera exigua . Insect Biochem Mol Biol 38(9): 825–33. [DOI] [PubMed] [Google Scholar]
- 22. Mosallanejad H, Soin T, Smagghe G (2008) Selection for resistance to methoxyfenozide and 20-hydroxyecdysone in cells of the beet armyworm. Arch Insect Biochem Physiol 67(1): 36–49. [DOI] [PubMed] [Google Scholar]
- 23. Smagghe G, Pineda S, Carton B, Del EP, Budia F, et al. (2003) Toxicity and kinetics of methoxyfenozide in greenhouse-selected Spodoptera exigua (Lepidoptera: Noctuidae). Pest Manag Sci 59(11): 1203–9. [DOI] [PubMed] [Google Scholar]
- 24. Seeon L, Sony S, Surakasi VP, Yonggyun K (2011) Role of a small G protein Ras in cellular immune response of the beet armyworm, Spodoptera exigua . J Insect Physiol 57(3): 356–362. [DOI] [PubMed] [Google Scholar]
- 25. Fernandez AP, Gibbons J, Okkema PG (2004) C. elegans peb-1 mutants exhibit pleiotropic defects in molting, feeding, and morphology. Dev Biol 276(2): 352–366. [DOI] [PubMed] [Google Scholar]
- 26. Riddiford LM, Hiruma K, Zhou XF, Nelson CA (2003) Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster . Insect Biochem Molec 33(12): 1327–1338. [DOI] [PubMed] [Google Scholar]
- 27. Zhang C, Zhou DH, Zheng SC, Liu L, Tao S, et al. (2010) A chymotrypsin-like serine protease cDNA involved in food protein digestion in the common cutworm, Spodoptera litura: Cloning, characterization, developmental and induced expression patterns, and localization. J Insect Physiol 56(7): 788–799. [DOI] [PubMed] [Google Scholar]
- 28. Chung H, Sztal T, Pasricha S, Sridhar M, Batterham P, et al. (2009) Characterization of Drosophila melanogaster cytochrome P450 genes. Proc Natl Acad Sci 106(14): 5731–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J, McCart C, Woods DJ, Terhzaz S, Greenwood KG, et al. (2007) A Drosophila systems approach to xenobiotic metabolism. Physiol Genomics 30(3): 223–231. [DOI] [PubMed] [Google Scholar]
- 30. Hernández CS, Ferré J (2005) Common receptor for Bacillus thuringiensis toxins Cry1Ac, Cry1Fa, and Cry1Ja in Helicoverpa armigera, Helicoverpa zea, and Spodoptera exigua . Appl Environ Microbiol 71(9): 5627–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA (2005) Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptidase N genes. BMC Genomics 6: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carmo AC, Giovanni DN, Corrêa TP, Martins LM, Stocco RC, et al. (2012) Mendonça Expression of an antiviral protein from Lonomia obliqua hemolymph in baculovirus/insect cell system. Antiviral Res 94(2): 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS (2004) A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131(9): 2007–2021. [DOI] [PubMed] [Google Scholar]
- 34. McGraw LA, Gibson G, Clark AG, Wolfner MF (2004) Genes Regulated by Mating, Sperm, or Seminal Proteins in Mated Female Drosophila melanogaster . Curr Biol 14(16): 1509–1514. [DOI] [PubMed] [Google Scholar]
- 35. Deng JY (2004) Enhancement of attraction to sex pheromones of Spodoptera exigua by volatile compounds produced by host plants. J. Chem. Ecol 30(10): 2037–2045. [DOI] [PubMed] [Google Scholar]
- 36. Majerowicz D, Alves-Bezerra M, Logullo R, Fonseca-de-Souza AL, Meyer-Fernandes JR, et al. (2011) Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae). Insect Mol Biol 20(6): 713–722. [DOI] [PubMed] [Google Scholar]
- 37. Spanier KI, Leese F, Mayer C, Colbourne JK, Gilbert D, et al. (2010) Predator-induced defences in Daphnia pulex: Selection and evaluation of internal reference genes for gene expression studies with real-time PCR. BMC Mol Biol 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maroniche GA, Sagadín M, Mongelli VC, Truol GA, del Vas M (2011) Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. J Virol 8: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teng XL, Zhang Z, He GL, Yang LW, Li F (2012) Validation of Reference Genes for Quantitative Expression Analysis by Real-Time RT-PCR in Four Lepidopteran Insects. J Insect Sci 12(60): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, et al. (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75(2-3): 291–295. [DOI] [PubMed] [Google Scholar]
- 41. Jiang XF, Luo LZ, Hu Y (1999) Influence of larval diets on development, fecundity and flight capacity of the beet armyworm, Spodoptera exigua . Acta Entomol Sin 42: 270–272. [Google Scholar]
- 42.Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybriziation. In: Keith, J.M. (Ed.), Bioinformatics, Volume II. Structure, Function and Applications, number 453 in Methods in Molecular Biology. Humana Press, Totowa, Chapter 1, pp. 3–31. [DOI] [PubMed]
- 43.Markham NR, Zuker M (2005) DNAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33: , W577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl M (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Radonic A, Thulke S, Mackay I, Landt O, Siegert W, et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Bioph Res Co 313: 856–862. [DOI] [PubMed] [Google Scholar]
- 46.Pfaffl MW (2004) Quantification Strategies in Real-time PCR. Pages 87–120 in SA B, ed. A-Z of Quantitative PCR. IUL Biotechnology Series, International University Line, La Jolla, CA.
- 47. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 48. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 research0034-research0034: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 50. VanGuilder HD, Vrana KE, Freeman WM (2008) Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44: 619–626. [DOI] [PubMed] [Google Scholar]
- 51. Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15: 155–166. [PMC free article] [PubMed] [Google Scholar]
- 52. Ferguson BS, Nam H, Hopkins RG, Morrison RF (2010) Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE 5: e15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ling D, Salvaterra PM (2011) Robust RT-qRT-PCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS ONE 6: e17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bustin S (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29: 23–39. [DOI] [PubMed] [Google Scholar]
- 55. Solanas M, Moral R, Escrich E (2001) Unsuitability of using ribosomal RNA as loading control for northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal Biochem 288: 99–102. [DOI] [PubMed] [Google Scholar]
- 56. Li A, Michaud MR, Denlinger DL (2009) Rapid elevation of Inos and decreases in abundance of other proteins at pupal diapause termination in the flesh fly Sarcophaga crassipalpis . Biochimica et Biophysica Acta 1794: 663–668. [DOI] [PubMed] [Google Scholar]
- 57. Rocha CS, Santos AA, Machado JPB, Fontes EPB (2008) The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology 380: 165–169. [DOI] [PubMed] [Google Scholar]
- 58. Frost P, Nilsen F (2003) Validation of reference genes for transcription profiling in the salmon louse Lepeophtheirus salmonis, by quantitative real-time PCR. Vet Parasitol 118: 169–174. [DOI] [PubMed] [Google Scholar]
- 59. Olsvik P, Lie K, Jordal A, Nilsen T, Hordvik I (2005) Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon . BMC Mol Biol 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hamalainen H, Tubman J, Vikman S, Kyrola T, Ylikoski E, et al. (2001) Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem 299: 63–70. [DOI] [PubMed] [Google Scholar]
- 61. Van Hiel MB, Wielendaele PV, Temmerman L, Soest SV, Vuerinckx K, et al. (2009) Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol Biol 10: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chapuis MP, Tohidi-Estahani D, Dodgson T, Blondin L, Ponton F, et al. (2011) Assessment and validation of a suite of reverse transcription-quantitative PCR reference genes for analyses of density-dependent behavioural plasticity in the Australian plague locust. BMC Mol Biol 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Boer M, de Boer T, Marien J, Timmermans M, Nota B, et al. (2009) Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta Collembola). BMC Mol Biol 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ranking of candidate reference genes according to their stability value using BestKeeper, geNorm, and NormFinder analyses. Candidates are listed from top to bottom in order of decreasing expression stability.
(DOC)