Introduction
Arthrodesis is a critical component of spine surgery for both degenerative and oncologic pathologies, with durable clinical benefits requiring successful bony fusion. The gold standard for bone grafting remains the autograft, optimally from the iliac crest. However, the effectiveness of an autograft varies due to the inconsistent quality of the bone procured as well as risks of donor site morbidity (1, 2). Several technologies exist as alternatives to autograft, either as a graft extender or replacement. These include treatment with bone morphogenetic protein (BMP-2), use of synthetic ceramics, demineralized bone matrix (DBM), and allografts; all with varying strengths and weaknesses in terms safety and/or efficacy (3, 4).
Alternatively, stem cells have become increasingly popular as cell-based therapeutics for musculoskeletal applications. Mesenchymal stem cells (MSCs) have been obtained from adipose tissue, bone marrow, peripheral blood, and synovial fluid, then combined with various osteoconductive scaffolds(5, 6). The rationale for their use is to add an osteogenic component to enhance formation of new bone via differentiation into osteoblasts. However, despite the appeal of this approach, there is a paucity of data supporting the efficacy of using stem cells in a clinical setting for spinal surgery. Furthermore, the best method for incorporating this technology into spinal surgery has not yet been determined.
One approach has been to process an allograft such that endogenous progenitor cells are retained during the processing of freshly procured cadaveric bone. This approach has the advantage that cells potentially benefit from microenvironmental cues derived from maintaining their attachment to the native cancellous bone scaffold. Indeed, signaling in terms of chemical and mechanical cues between the cell and its scaffold is critically important for new bone formation (7-9).
While cellularized allografts are known to harbor endogenous cells, the identity of these cells remains obscure, largely due to the lack of bona fide markers for stem and progenitor cells. In this study, we hypothesized that a cellular allograft bone matrix (Osteocel Plus) contains a population of mesenchymal stem and bone progenitor cells, the former capable of self-renewal and multi-lineage differentiation(10). Currently, no single cell marker can unequivocally distinguish stem cells from progenitor cells. The use of cell surface marker combinations allows for enrichment of the stem cell population but is inadequate for prospective isolation (11-15). A novel use of lineage mapping allowed identification of highly proliferative clones and permitted us to determine whether cells endogenous to a cellular allograft undergo extensive self-renewal-a functional hallmark of stem cells (16).
Further, we used genetic and proteomic profiling as well as functional assays to examine whether these cells in the Osteocel Plus allograft are capable of multipotential differentiation (the second functional hallmark of stem cells). We postulated that the use of these two functional hallmarks could enable us to establish corroborative evidence for the existence of a stem and progenitor cell population in cellular allografts.
Materials and Methods
Allograft cellular bone matrix preparation
The allograft cellular bone matrix material used in this study is commercially available as Osteocel Plus (NuVasive®, Inc.) and is prepared from freshly procured donated cadaver tissue, between the ages of 18 and 44 years old, brought to the processing facility on wet ice, and promptly processed. Detailed donor screening consisting of medical and social history, medical record review and physical examinations, as well as a complete review is performed by a licensed medical director. Safety testing is also performed, including evaluation for viral, bacterial and fungal contamination. The collected cortical bone is separated and processed by acid demineralization into demineralized bone matrix (DBM) and is kept hydrated in physiologic saline. The remaining cellular cancellous bone is processed and undergoes a selective depletion of immunogenic components, through a specific and validated series of washes to remove unwanted cells. These include hematopoietic-lineage cells. Viable bone lining cells remain attached to the cancellous bone matrix. A broad-spectrum antimicrobial treatment is performed on all tissue. Then demineralized cortical bone from the same donor is mixed with the cellular cancellous bone component. A cryopreservation solution is added and the aseptically processed product is cryogenically frozen, permitting its 5-year shelf life. Quality testing is performed on every lot post-cryopreservation to verify sterility and that the number of retained cell’s yield, viability, and osteogenicity meet or exceed established criteria.
Cell isolation and culture
4 samples of the cryopreserved Osteocel Plus cellular allograft samples (1ml each) were thawed and washed 3 times with PBS. For released cell assays, four samples (5mL each) were treated similarly and then serially dissociated in 0.25% trypsin (Invitrogen) for up to 1 hour at 37°C. Dissociated cells were centrifuged at 300×G. Cell pellets were resuspended in MSC media [advanced DMEM (Invitrogen), 200mM Glutamax (Invitrogen), Penicillin-Streptomycin (Invitrogen), 10% fetal bovine serum (Invitrogen)]. Cell viability was assessed by Trypan blue dye exclusion. Viable cells for culture expansion were seeded in T75 pre-coated collagen flasks (Thermo Scientific) at a density of 5×103 cells/cm2. Dissociated cells from these four 5ml samples were used for assessment of tri-lineage differentiation capacity.
Cellular allograft bone explant culture and time-lapse video
Osteocel Plus samples were washed and resuspended in MSC media and cultured in 25mm collagen-coated cell culture dishes. Image acquisition (24 and 72 hours) for time-lapse video was performed with a Carl Zeiss Axio Observer with a fully automated Carl Zeiss Cell Observer System using a Plan Neofluar 20x Ph3 objective, incubator XL, heating insert P and 5% CO2 cover HP for pH and humidity control.
Assessment of differentiation capacity of cells derived from cellular allograft bone
Osteogenic differentiation
To induce osteogenic differentiation, low (P1-4) and high (P10) passage cells enzymatically dissociated from Osteocel Plus were placed in osteogenic differentiation medium (ODM) [Advanced DMEM (Invitrogen), 200mM Glutamax (Invitrogen), Penicillin-Streptomycin (Invitrogen), 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 10 mM glycerol phosphate (Sigma-Aldrich), 50 mM ascorbic acid (Sigma-Aldrich), and 10nM dexamethasone (Sigma-Aldrich)] for 28 days, with medium changes twice weekly. Osteogenesis differentiation capacity was assessed by 2% Alizarin red S-calcium staining (Sigma-Aldrich) according to the manufacturer’s instructions.
Adipogenic differentiation
To induce adipogenic differentiation, low (1-4) and high (10) passage dissociated cells were placed in adipogenic medium [Advanced DMEM (Invitrogen), 200mM Glutamax (Invitrogen), Penicillin-Streptomycin (Invitrogen), 10% fetal bovine serum (Invitrogen), 100uM indomethacin (Sigma-Aldrich Ltd., St. Louis, MO), 50uM linoleic acid (Sigma-Aldrich), 0.5 mM isobutylmethylxanthine (Sigma-Aldrich), and 1uM dexamethasone (Sigma-Aldrich)] for 28 days, with medium changes twice weekly. Adipogenic differentiation was assessed by microscopic detection of intracellular lipid droplets and confirmed by Oil Red O staining (Thermo Scientific).
Chondrogenic differentiation
To induce chondrogenic differentiation, low (1-4) and high (10) passage dissociated cells were placed in chondrogenic medium [advanced DMEM (Invitrogen), 200mM Glutamax (Invitrogen), Penicillin-Streptomycin (Invitrogen), 10% fetal bovine serum (Invitrogen), 1% Insulin/Transferrin/Selenium (Invitrogen), 0.05mM ascorbic acid (Sigma-Aldrich), 0.35mM proline (Sigma-Aldrich), 10ng/ml TGF beta (Millipore)] for 28 days, with medium changes twice weekly. Chondrogenic differentiation was assessed by Alcian blue staining (Sigma-Aldrich), according to the manufacturer’s instructions.
Quantitative Polymerase Chain Reaction (qPCR) arrays
Total RNA was isolated using the RNeasy 96 Kit (Qiagen, Valencia, CA). 500 ng RNA was converted to cDNA with the RT2 First Strand Kit (Qiagen) and combined with RT2 SYBR Green qPCR Mastermix (Qiagen), according to the manufacturer’s instructions. Equal volumes (25 μl) of this mixture were loaded into each well of both the Osteogenesis and the Mesenchymal Stem Cells RT2 Profiler PCR array plates (Qiagen). qPCR was performed on an iQ5 Real-time PCR Detection System (Bio-Rad, Hercules, CA). Relative gene expression levels were calculated using the ΔΔCt method.
LOLIG Plasmid construction
To create a plasmid for lentiviral vector production that includes a 24-bp barcode library and a green fluorescent protein (GFP) identifier, we used an 83-bp oligo containing 24 degenerate nucleotide sequence bases, an EcoRI recognition site at the 5′ end and an NheI site at the 3’end as PCR template. To construct pLV-library-ZsGreen1, the PCR product was cut with the appropriate restriction enzymes and cloned into pLVX-IRES-ZsGreen1 (Clontech, Palo Alto, CA). TOP10 E. coli (Invitrogen, Carlsbad, CA) was transformed with the ligation mix and grown overnight in LB medium in the presence of 100 μg/ml ampicillin. The plasmids were isolated using the Qiagen Plasmid Maxi kit. The final plasmid product termed LOLIG was used for lentiviral vector production.
Vector production
293T cells were co-transfected with 20 μg of LOLIG and 10 μg of pPACK packaging plasmid mix (SBI) using calcium phosphate precipitation. The culture medium was replaced with fresh medium after 6 h, and supernatants were collected 24 and 48 h after transfection. To determine viral titer, 105 HT1080 cells were seeded in a six-well plate and transduced with various dilutions of vector in the presence of 4 μg/ml Polybrene (Sigma-Aldrich). The percentage of GFP-positive cells was determined by flow cytometry after 24 h.
Massively parallel sequencing
Genomic DNA was isolated from 1×106 cells of a 5ml sample at passages 1 and 10 using the QiaAmp DNA mini kit (Qiagen). A 232-bp fragment of genomic DNA or the LOLIG plasmid was amplified in 50 μl reactions containing 175 ng of genomic DNA or 170 ng of the LOLIG plasmid, 20 pmol of each primer, 3% DMSO and 25 μl of Phusion High-Fidelity Mastermix (NEB). The primer sequences, which contained the adaptors required for deep sequencing, were: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTGCTCGTCAAGAAGACAG and 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC T TCCGATCTTGACAGCTACTAGCGTA. The reactions were carried out in an ABI 2720 Thermal Cycler (Applied Biosystems) as follows: 98° C for 1 min (initial denaturation), 45 cycles of 98°C for 10s, 58°C for 30s and 72°C, followed by a final extension step at 72°C for 5 min. Illumina Solexa was used for massively parallel leverage single-read de novo sequencing. The Illumina Solexa GA II sequencing system pipeline along with Smith-Waterman alignment algorithm on a graphics processing unit (GPU) using Compute Unified Device Architecture (CUDA) for sequence alignment was used.
Immunocytochemistry and flow cytometry analysis
Cells were fixed with 4% paraformaldehyde at 4°C. Samples were blocked for 1h in 0.3% Triton X-100 (Sigma) and 10% normal goat serum in PBS. Primary antibodies BMP-2 (Abcam), CD44 (Millipore), CD146 (Millipore), Collagen (Millipore), Integrin (Millipore), Runx-2 (Millipore), SMAD (Millipore), Sto-1 (Millipore), Thy1 (Millipore), Osteocalcin (Millipore), Osterix (Millipore), CD254/RANKL (Millipore) were diluted in carrier solution (1% normal goat serum/0.3% Triton X-100 in PBS) and incubated overnight at 4°C. After washing, IgG or IgM secondary antibodies conjugated to Alexa 488 (green) or Cy3 (red) (Jackson Immuno Research) were incubated for 1hr at room temperature. Primary and secondary antibody information is available upon request. Samples for immunocytochemistry were imaged on a Carl Zeiss Axio Observer fluorescent microscope using Axiovision software (Germany). Samples for flow cytometry analysis were measured on the CyAn ADP (DAKO, Inc.) or MoFlo MLS Flow Cytometer (DAKO, Inc) and data analyzed using FlowJo software (Ashland, OR).
ELISA and multiplex immunoassays
Four sample groups were evaluated: 1) Acellular (dead) cancellous bone alone (1cc), 2) 1×105 bone marrow huMSCs (Lonza Group Ltd.), 3) cells dissociated from Osteocel Plus (1×105 cells), and 4) Osteocel Plus cellular allograft bone explants (1cc) maintained in ODM for 28 days in vitro. Total of four different acute phase proteins indicative of bone metabolism were analyzed for the Multiplex Immunoassay, these included RANKL, Osteoprotegrin, Osteocalcin, and Osteopontin (Millipore). These cytokines where measured on a Luminex 100 analyzer (Millipore). For BMP-2 ELISA, BMP-2 Quantikine Kits (R&D Systems) were used according to the manufacturer’s instructions.
Statistical analyses
Data are represented as mean values ± standard error of the mean. Statistical analyses for all experiments including more than two groups were performed using a two-way ANOVA followed by posthoc comparisons using Dunnett’s posthoc test. For experiments with two groups, Student’s t-test was used.
Study Funding and Potential Conflict of Interest
Funding for this work was provided by NuVasive Inc. The authors disclose no potential conflict of interest.
Results
Quantity, viability, and proliferative capacity of cells isolated from Osteocel Plus
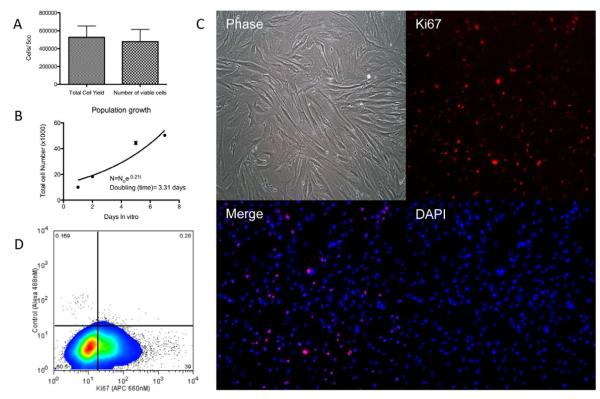
Serial trypsinization of 5ml samples from four different donors yielded 5.25×105 ± 4.6×103 cells/5ml (mean ± s.d.). Of these, 91% were viable, or 4.77×105 ± 4.9×103 cells/5ml as determined by Trypan blue dye exclusion staining(17) (Figure 1A).
Figure 1. Quantity, viability, and proliferative capacity of cells isolated from Osteocel Plus.
(A) Cell yield and viability of 5ml of Osteocel Plus was determined by serial enzymatic digestion and Trypan blue exclusion staining (n=4). (B) Population growth curves used to calculate doubling time of isolated cells. (C) Immunoflourescent staining (10x) reveals proliferative capacity of cells released from Osteocel Plus. Phase contrast, Ki67 (APC, red), nuclear DAPI (blue) and merged images show that a high percentage of cells co-localize. (D) flow cytometry analysis for the proliferative marker Ki67; 97% of cells were gated (left panel) and 39% expressed Ki67 (right panel).
To assess proliferative capacity, released cells were cultured in MSC proliferation media for 7 days in vitro. Under these conditions, an initial population of 10,000 dissociated cells doubled in 3.31 ± 0.05 (mean ± s.d.) days (Figure 1B). Additionally, immunocytochemistry showed that cells dissociated from Osteocel Plus exhibited robust staining for proliferative marker Ki67 (Figure 1C). Flow cytometry analysis revealed that Ki67-positive cells comprised 39% of the total population (Figure 1D). Collectively, these results demonstrate that isolates from the allografts showed consistent numbers of viable cells and proliferation capacity between all samples.
Explant of cellular allograft cultures to investigate cell fate and migration
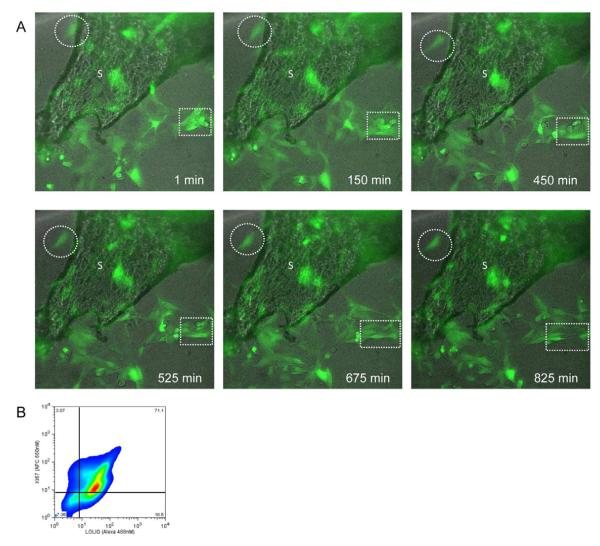
Three-dimensional properties of a culture system can exert substantial influence on cell fate (9). To investigate molecular pathways underlying osteogenesis, explant culture systems have been generated to recapitulate the physiological milieu(18, 19). Accordingly, we evaluated cells endogenous to Osteocel Plus in explant culture to investigate cell migration from the scaffold without removal of the cells by trypsinization. Osteocel Plus in its entirety was transduced with LOLIG in order to visualize the cellular compartment. Time-lapse imaging revealed an active and dynamic cellular environment on the cancellous bone scaffold of the allograft. We observed migration of cells off of the cancellous bone over a 12-hour period (Figure 2A, broken circles). Moreover, we observed the migrating cells divide and proliferate off of the Osteocel Plus (Figure 2A, broken boxes). Flow cytometry analyses of cells taken directly from explant culture show that a greater percentage of cells express Ki67 (74%; Figure 2B) compared to cells released through serial trypsinization (39%; Figure 1D) (20). These results demonstrate that cells attached to the bony scaffold can produce migratory progeny with a robust proliferative capacity in vitro.
Figure 2. Migratory and proliferative capacity of Osteocel Plus cells in 3D culture.
(A) Time-lapse imaging of cancellous bone (phase) and LOLIG-transduced cells (green). Migration of cells (broken circles) and proliferation (broken squares) were observed over 13.75 hours (Images taken at 20x). (B) Flow cytometry analysis for proliferative capacity (Ki67+) of LOLIG-positive cells.
Comparative gene and protein expression profiles of Osteocel Plus cells and MSCs
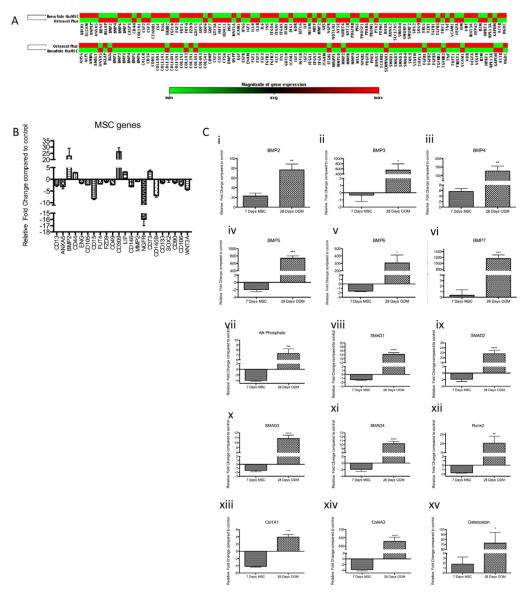
We hypothesized that cells isolated from Osteocel Plus represent a mixed population of MSCs and other progenitor cells. To characterize endogenous cells dissociated from Osteocel Plus, we determined gene and protein expression profiles in comparison to those of a known pure population of huMSCs. Both populations were cultured for 7 days in vitro in MSC media to allow for cellular recovery after enzymatic dissociation. We then evaluated 84 genes associated with MSC pluripotency and self-renewal. Cells from Osteocel Plus showed overall similarities and exhibited less than 2.5-fold difference in gene expression compared to huMSCs for most genes (Figure 3A, top cluster panel). Osteocel Plus cells showed a relative increase in 11 of 84 mRNA transcripts, including BMP-2, VEGFr/CD309 (21), CD44 (22), leukemia inhibitory factor (LIF) (23), and CD73 (24), compared to huMSCs after 7 days of culture in vitro (Figure 3B). In addition, 14 of 84 integral MSC-associated genes showed decreased expression in the Osteocel Plus population, including CD13, Annexin A5(ANAX5), fucosyltransferase 1 (FUT1), alpha 6 integrin (ITGA6), CD146, CD140B, CD106 and WNT3A (Figure 3B). Overall, there was high similarity and little relative change in expression in most (59 of 84) of the MSC-associated genes, including CD105, CD15, matrix metallopeptidase 2 (MMP2), type IV collagenase (MMP2), CD133, and CD90 (25) (Figure 3A and B).
Figure 3. Expression profiling of cells released from Osteocel Plus.
(A) MSC-focused qPCR array results of Osteocel Plus cells versus huMSCs cultured for 7 days in vitro in MSC expansion media (top cluster panel). The Osteogenesis-focused qPCR array results of Osteocel Plus cells versus huMSCs cultured in osteogenic differentiation media for 28 days in vitro (bottom cluster panel). Red= increase in expression relative to huMSCs; Green= decrease in expression relative to huMSCs. (B) Relative expression of selected MSC genes from the MSC PCR array of Osteocel Plus cells compared to huMSCs. Both were cultured in MSC media 7 days prior to analysis. (C) Relative expression of osteogenesis-related genes BMP 2-7 (i-vi), alkaline phosphate (vii), Smad 1-4 (viii-xi), Runx2 (xii), Col1a1 (xiii), col4a3 (xiv), osteocalcin (xv) in MSC- and osteogenesis- focused qPCR arrays. Osteocel Plus cells were cultured in MSC and ODM media for 7 and 28 days in vitro, respectively (n=3, *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001; 95% Confidence interval of difference).
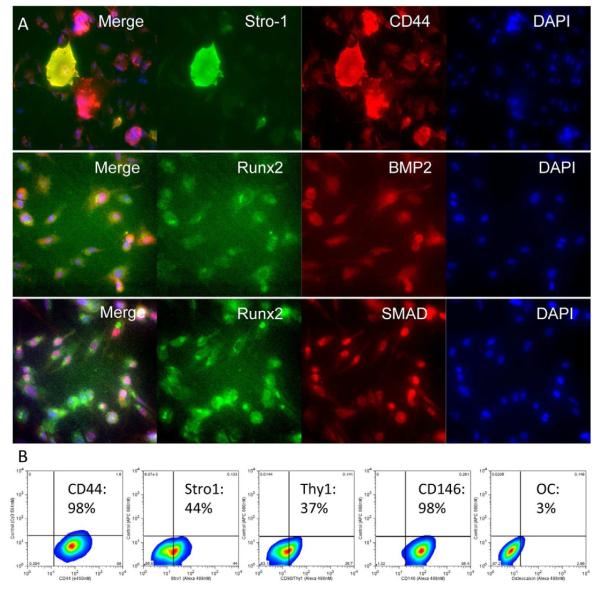
Proteomic analysis revealed that Osteocel Plus cells maintained in MSC media for 7 days in vitro express the MSC markers CD44 and Stro-1. Qualitative analysis revealed 3 subpopulations of Osteocel Plus cells: CD44+/Stro-1−, CD44+/Stro-1+, and CD44−/Stro-1+ (Figure 4A). Flow cytometry analysis revealed a broad range of MSC marker expression for CD44 (99%), Stro-1 (44%), CD90/Thy1 (37%), and CD146 (99%) (Figure 4B). Osteocel Plus dissociated cells also expressed BMP-2 and downstream transcription factors Runx2 and SMADs, which are required for signal transduction and differentiation into osteo- and chondrogenic lineages (Figure 4A).
Figure 4. Proteomic profiling of Osteocel Plus cells cultured for 7 days in MSC media.
(A) Immunofluorescent staining (20x magnification) for early stem and progenitor cell markers and transcription factors: Stro-1 (Alexa 488), CD44 (Cy3), Runx2 (Alexa 488), BMP2 (Cy3), SMAD 1,4 (Cy3), and nuclear DAPI (Blue). (B) Flow cytometry analysis for stem cell markers CD44, Stro-1, CD90/thy1, CD146, and the mature osteoblast marker Osteocalcin.
Genetic and proteomic profiling of the osteogenic potential of Osteocel Plus
We then determined the osteogenic potential of cells released from Osteocel Plus versus huMSCs. Cells were maintained in osteogenic media for 28 days in vitro and then evaluated for expression of 84 osteogenic differentiation genes functioning in cell growth, proliferation, extracellular matrix formation, cell adhesion, skeletal system development, and bone mineral metabolism. Analysis revealed overall upregulation of osteogenic gene expression in cells isolated from Osteocel Plus relative to huMSCs at 28 days in vitro (Figure 3A, bottom cluster panel). In all, 63 of 84 genes were expressed at 2.5 fold higher levels in the Osteocel Plus population than in huMSCs. In addition, 7 of 84 genes were expressed at 2.5 fold lower levels in the Osteocel Plus population relative to huMSCs; 14 of 84 genes were expressed at similar levels. We were particularly interested in expression of the soluble growth factors bone morphogenetic proteins (BMPs) 2-7 (26). We found that BMPs 2-7, which induce osteogenesis, are significantly upregulated in Osteocel Plus cells compared to huMSCs when both populations were grown in osteogenic conditions with ODM (Figure 3C i-vi). Furthermore, the intracellular proteins SMAD1, 2, 3, and 4, which transduce BMP signaling, were also significantly upregulated in Osteocel Plus cells (Figure 3C viii-xi). Runx2, a transcription factor interacting with SMADs and a multifunctional regulator of early bone formation, was also significantly upregulated in Osteocel Plus cells (Figure 3C xii). Other genes functioning in bone and skeletal development, such as Col1a1, Col4a3, osteocalcin, and alkaline phosphate showed significantly higher expression in Osteocel Plus cells relative to huMSCs (Figure 3C, vii, vxiii-xv).
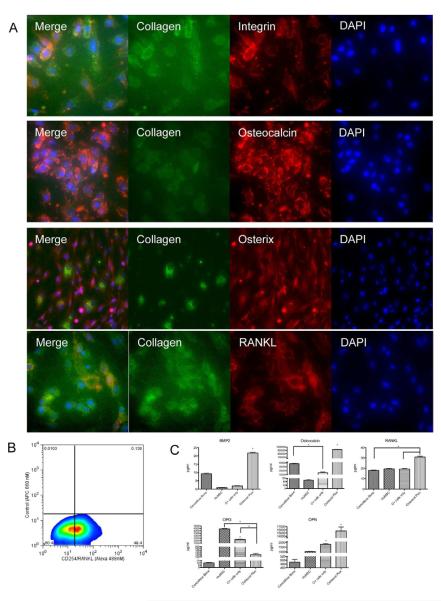
Protein analysis revealed that when maintained in ODM for 28 days in vitro, Osteocel Plus cells express mature osteoblast markers, such as collagen, integrin, osteocalcin, osterix, and RANKL (Figure 5A). Flow cytometry analysis revealed that 49% of released and differentiated Osteocel Plus cells express the mature osteoblast marker CD254/RANKL at 28 days in vitro (Figure 5B). In addition, at 7 days in non-differentiating MSC growth media in vitro, flow cytometry analysis revealed that 3% of the cells express the mature osteoblast marker osteocalcin (Figure 4B), suggesting that Osteocel Plus cells also contain a small subpopulation of mature osteoblasts.
Figure 5. Proteomic profiling of Osteocel Plus cells cultured for 28 days in ODM.
(A) Immunofluorescent staining (20x) for the osteoblast markers Collagen (Alexa 488), Integrin (Cy3), Osteocalcin (Cy3), Osterix (Cy3), RANKL (Cy3), and for nuclear DAPI (blue). (B) Flow cytometry analysis for CD254/RANKL. (C) ELISA and multiplex immunoassay for BMP-2, Osteocalcin, RANKL, Osteoprotegrin (OPG), and Osteopontin (OPN). Comparative analysis was performed on acellular cancellous bone, huMSCs, Osteocel Plus cells only, and native Osteocel Plus cells adherent to the bone. Concentrations are expressed as pg/ml. (n=3, *p<0.05, 95% Confidence interval of difference).
To quantify secretion of ligands critical for osteogenesis, we used ELISA and custom multiplex immunoassays to assess amounts of soluble BMP-2, osteocalcin, RANKL, osteoprotegrin (OPG), and osteopontin (OPN) secreted by Osteocel Plus cells explant cultures, Osteocel Plus released cells alone, acellular cancellous bone, and HuMSCs after 28 days in ODM. Osteocel Plus explant cultures secreted significantly higher levels of BMP-2, OPG, RANKL, and OPN compared to cancellous bone, huMSCs, and Osteocel Plus released cells alone (Figure 5C). Osteocel Plus explants contained significantly lower levels of OPG compared to media from huMSCs or Osteocel Plus released cells only.
Clonal analysis and identification of stem cells based on proliferative capacity from cells released from Osteocel Plus
Genomic and proteomic analysis indicate that cells dissociated from Osteocel Plus consist of a heterogeneous population of progenitors and cells that could differentiate along an osteogenic lineage under the appropriate culture conditions. Whether there is a subpopulation of bona fide stem cells (capable of extensive self-renewal) within the progenitor cell population released from Osteocel Plus remains uncertain due to the lack of markers that allow their prospective isolation and identification. Therefore, to determine whether cells attached to allograft bone are capable of undergoing extensive self renewal, we employed a novel use of a lineage tracing strategy to label individual cells with unique and indelible genetic “tags” passed faithfully to progeny (Figure 6A). This approach uses a lentivirus modified to express the green fluorescent protein ZsGreen1, with each viral particle carrying one of 108 possible unique 24-oligonucleotide sequences. The oligonucleotide tag in a single cell can be sequenced, allowing discrimination of clonal relationships in vitro.
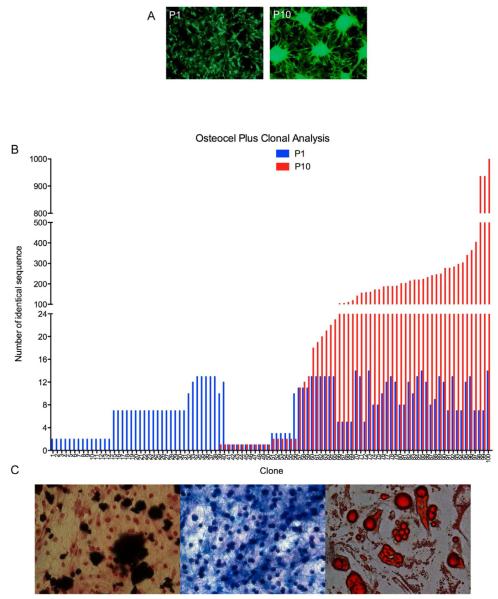
Figure 6. Lineage mapping clonal analysis and identification of stem cells from Osteocel Plus.
(A) Passage 1 (P1) and 10 (P10) cells from Osteocel Plus transduced with LOLIG (Images taken at 10x). (B) Parallel sequencing of cells from P1 (blue) and P10 (red) demonstrating polyclonal population distribution of 100 random clones. (C) Tri-lineage differentiation potential of P10 stem cells into osteoblasts (Alizarin Red with H&E counterstain, left panel), chondrocytes (Alcian Blue, middle panel), and adipocytes (Oil red O, right panel) lineages (Images taken at 20x magnification).
To assess clonal relationships and self-renewal capacity, we analyzed cells from passage 1 (P1) and their passage 10 (P10) progeny that express LOLIG (Figure 6B). Massive parallel sequencing data revealed that at P1 there were 174,041 total individual clones, indicative of a polyclonal population. When clonal populations of cells from P1 were maintained through 10 serial passages, the total number decreased to 118,263 individual clones. Analysis of 100 randomly selected clones revealed a polyclonal population. At P1 all cells showed integration and a detectable LOLIG tag. At this point, cells have gone through approximately 1-4 doublings. Some clones showed no proliferative capacity (clones 1-38), others were relatively quiescent (clones 39-66), and others expanded due to proliferation of the original clone (clones 66-100) (Figure 6C). When we examined cells at P10 we found that clones that failed to divide through P10 were likely differentiated cells (clones 1-38). The limited self-renewal exhibited by the second cluster (39-66) suggested that they were MSCs fated to the progenitor lineage. The third cluster of P10 clones (66-100) generated the most abundant progeny indicative of extensive self-renewal. These clones that show at least one self- renewal tag through 10 serial passages (clones 39-100) represent 68% clones sampled and likely represent a true MSC population.
Differentiation potential of clones exhibiting extensive self-renewal capacity
To confirm that the highly proliferative P10 clones are MSCs, we assessed their capacity to differentiate into osteogenic, chondrogenic, and adipogenic lineages. P10 clones were then switched to appropriate differentiation media for 28 days in vitro. Osteogenic, chondrogenic, and adipogenic differentiation was confirmed using Alizarin Red S staining, Alcian blue staining of proteoglycans, and oil red staining of lipid deposits, respectively (Figure 6D). These results indicated that late passage P10 clones have the capacity for trilineage differentiation potential as well as proliferation, and are thus, by definition, mesenchymal stem cells.
Discussion
Advances in minimally invasive spine surgery along with recent progress in progenitor and stem cell isolation and transplantation present an opportunity to improve treatment and outcomes from spinal fusion. The gold standard for bone grafting remains the autograft (27). However, its benefits must be balanced against donor site morbidity, limited total volume available for grafting, and variable quality of progenitor cells related to patient age and co-morbidities(28). To mimic the autograft—and its fundamental properties of osteoconductivity, osteoinductivity, and osteogenicity—materials, such as cellular allografts in which native progenitor cells are preserved, are available as a bone grafting option. In this study we used complementary methods to quantify and characterize endogenous cells from Osteocel Plus.
Using serial trypsinization to dissociate cells from the bony scaffold, we consistently isolated an average of 5.25×105 ± 4.6×103 cells/5ml sample of allograft. This quantity is comparable to the manufacturer’s claims; however, serial trypsinization may only partially release all the cells from allograft. In addition, this approach may not release the cells embedded in deep portions of the bone matrix, and underestimates the actual total cell count. Importantly, trypan blue staining confirmed the viability of 91% of cells after they were thawed from cryopreservation. Furthermore, 39% of that population stained positively for the proliferative marker Ki67, indicating their potential for growth and self-renewal.
Dissociation processes requiring repeated trypsinization could alter the biology and viability of cells and is not representative of the clinical method of use of the tissue. To circumvent potentially deleterious effects of the enzyme, we directly evaluated cell migration and proliferation in explant cultures of bony scaffold with native Osteocel Plus cells transduced with ZsGreen lentivirus. Explant cultures showed proliferation of cells attached to the scaffold and of their progeny that had migrated away. In fact, flow cytometry analyses of cells taken directly from explant culture showed greater (74%) expression of Ki67 compared to cells enzymatically dissociated from Osteocel Plus through serial trypsinization (39%). These results suggest that microenvironmental cues provided by attachment to the mineralized bony scaffold may be a major determinant of how many cells survive and proliferate. Moreover, cellular allograft may produce a variety of growth factors as opposed to supraphysiologic rhBMP-2 and what is exposed in traditional DBMs alone. Equally, failure of this bone graft to receive adequate mechanical or additional growth and survival signals could lead to significant cell loss during grafting. This concern remains a fundamental challenge for cell therapy (29).
To identify cells dissociated from Osteocel Plus, we compared them to commercially available human bone marrow MSCs. After a recovery period of 7 days in MSC expansion media, a MSC gene-focused expression array revealed that 70% (59/84) of genes from Osteocel Plus were expressed at levels similar to those seen in huMSCs. This result suggests that at least some of the enzymatically released cells are MSCs. On the other hand, the divergence may imply that Osteocel Plus contains a heterogeneous population of MSCs and other cells at various stages of maturation along the osteocyte lineage.
For correlative studies we evaluated expression of the most commonly used MSC surface protein markers in Osteocel Plus cells cultured for 7 days in stem cell media. Using immunocytochemistry and flow cytometry analysis, we found expression of CD44 (99%), Stro-1 (44%), CD90/Thy1 (37%), and CD146 (99%) did not correlate with each other. This expected result highlights the discordancy that occurs using surface markers for stem cell quantification and why this method alone is insufficient to prospectively identify MSCs.
HuMSCs and cells derived from the Osteocel Plus scaffold also show differences in osteogenic capacity. After 28 days in osteogenic differentiation media, 75% (64/84) of genes in an osteogenesis-focused PCR array showed greater than 2.5-fold increases in expression in Osteocel Plus cells relative to huMSCs. Protein profiling (namely, collagen, integrin, osteocalcin, osterix, and RANKL) of cells after 28 days of exposure to osteogenic differentiation media also showed a pattern consistent with increased osteogenesis in Osteocel Plus cells versus huMSCs. Further analysis using flow cytometry revealed that 49% of differentiated Osteocel Plus cells express the mature osteoblast marker CD254/RANKL. Consistent with this data, assessment of soluble factors secreted by Osteocel Plus released cells in osteogenic differentiation conditions produced significantly higher levels of BMP-2, RANKL, and OPN compared to cancellous bone alone and MSC-derived osteoblasts. However, Osteocel Plus cells attached to the scaffold showed significantly lower OPG levels compared to those seen in huMSCs and dissociated Osteocel Plus cells. That difference may be attributable to the fact that OPG specifically acts on bone, increasing bone mineral density and bone volume. Therefore, cancellous bone may provide an environment of optimal mineral density resulting in less feedback for OPG release. Nevertheless, mRNA and protein data overall indicates greater osteogenic potential in Osteocel Plus cells compared to huMSCs. The increased ability of attached cells to produce osteogenesis-related proteins may be a consequence of microenvironmental cues received from attachment to and interaction with the scaffold, as geometry and mechanical forces are known to alter cell fate (30, 31).
Our conclusions on the stemness and osteogenic potential of Osteocel Plus cells should be tempered by recognition of the limitations of genomic and proteomic profiling. One concern is the timing of experiments assessing “stemness”, which were undertaken 7 days after dissociation, and therefore profiles may not be truly reflective of cells’ native state. However, we find this experimental protocol necessary to allow cells to recover from harsh enzymatic dissociation techniques that may cleave or damage cell surface markers. Of equal concern is our use of an arbitrary 2.5-fold difference in relative gene expression as a threshold to analyze qPCR data. Expression levels do not always correlate to protein production and function, nor do they definitively reflect post-transcriptional events.
Quantifying the stem cell population remains challenging with current methodologies. Colony forming unit (CFU) assays can determine proliferative capacity and suggest the presence of stem/progenitor populations, but are unable distinguish between true stem cells and progenitor cells. Surface markers can also be used to estimate the size of a stem cell population. Based on surface markers alone, our data suggests an initial Osteocel Plus population consisting of 37-99% MSC/progenitors and approximately 3% osteoblasts.
Our lineage mapping analysis greatly enhances and improves on analysis compared to using surface markers or colony forming assays. Endogenous cells released from Osteocel Plus were transduced with unique tags and then cultured 10 passages to evaluate which cells exhibited no proliferation (differentiated cells and progenitor cells), limited proliferation (stem cells with osteo-progenitor progeny), and extensive proliferation (stem cells with emergent clones). The cells that demonstrated extensive self-renewal—that is, persisted at 10 passages in conditions unfavorable to progenitors also demonstrated capacity for multipotentiality; Satisfying the two functional hallmarks of true stem cells. Therefore, out of the 174,041 total individual clones at P1, 118,263 (or 68%) show at least one self renewal tag through 10 serial passage exhibiting extensive self -renewal and thus are potentially mesenchymal stem cells.
As of the date of this publication, there have been no well-controlled prospective clinical studies published on the effectiveness of Osteocel Plus. There have been however several case studies in foot and ankle surgery (32, 33) as well as case series in trauma, tibial non-unions (34), oral and maxillofacial surgery (35, 36), and retrospective study in lumbar spinal fusion (37) demonstrating varying success. Additional clinical research has been published on the effectiveness of delivering concentrated MSCs from bone marrow describing a correlation between the number of delivered cells and clinical outcome when treating tibial non-unions (38). That research demonstrated that it required greater than 50,000 osteoprogenitor cells to reproducibly heal the tibial non-unions.
Taken together, these data provide corroborative evidence that the Osteocel Plus cellular allograft contains a heterogeneous cell population with some cells demonstrating the capacity for extensive self-renewal and multipotential differentiation in this in vitro environment. Both of these findings suggest these cells could be stem/progenitor cells in this in vitro investigation. Pre-clinical animal models used to test efficacy and biological mechanisms are limited to immunodeficient animals, and large animal immune-compromised models do not exist. Ultimately, determining whether allografts containing a viable population of stem cells function comparably to an autograft will require further study, and their efficacy in facilitating arthrodesis will depend on well-controlled prospective clinical studies.
Acknowledgement
The authors would like to thank the City of Hope & Beckman Research Institute’s (1) Brian Armstrong, PhD. and Mirako Lee of light microscopy digital core (2) Harry Gao PhD. of DNA sequencing core for help in data acquisition.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bueno EM, Glowacki J. Cell-free and cell-based approaches for bone regeneration. Nat Rev Rheumatol. 2009;5:685–97. doi: 10.1038/nrrheum.2009.228. [DOI] [PubMed] [Google Scholar]
- 2.Delecrin J, Takahashi S, Gouin F, Passuti N. A synthetic porous ceramic as a bone graft substitute in the surgical management of scoliosis: a prospective, randomized study. Spine (Phila Pa 1976) 2000;25:563–9. doi: 10.1097/00007632-200003010-00006. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal R, Williams K, Umscheid CA, Welch WC. Osteoinductive bone graft substitutes for lumbar fusion: a systematic review. J Neurosurg Spine. 2009;11:729–40. doi: 10.3171/2009.6.SPINE08669. [DOI] [PubMed] [Google Scholar]
- 4.Rihn JA, Kirkpatrick K, Albert TJ. Graft options in posterolateral and posterior interbody lumbar fusion. Spine (Phila Pa 1976) 2010;35:1629–39. doi: 10.1097/BRS.0b013e3181d25803. [DOI] [PubMed] [Google Scholar]
- 5.Witkowska-Zimny M, Wrobel E. Perinatal sources of mesenchymal stem cells: Wharton’s jelly, amnion and chorion. Cell Mol Biol Lett. 2011;16:493–514. doi: 10.2478/s11658-011-0019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockmann P, Park J, von Wilmowsky C, Nkenke E, Felszeghy E, Dehner JF, et al. Guided bone regeneration in pig calvarial bone defects using autologous mesenchymal stem/progenitor cells - A comparison of different tissue sources. J Craniomaxillofac Surg. 2011 doi: 10.1016/j.jcms.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Sarraf CE, Otto WR, Eastwood M. In vitro mesenchymal stem cell differentiation after mechanical stimulation. Cell Prolif. 2011;44:99–108. doi: 10.1111/j.1365-2184.2010.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnsdorf EJ, Tummala P, Castillo AB, Zhang F, Jacobs CR. The epigenetic mechanism of mechanically induced osteogenic differentiation. J Biomech. 2010;43:2881–6. doi: 10.1016/j.jbiomech.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MY, Jandial R. Environmental cues for mesenchymal stem cell fate determination. Neurosurgery. 2010;67:N20. doi: 10.1227/01.neu.0000389735.93437.08. [DOI] [PubMed] [Google Scholar]
- 10.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 11.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer research. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 12.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Jones EA, Kinsey SE, English A, Jones RA, Straszynski L, Meredith DM, et al. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheum. 2002;46:3349–60. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 15.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 16.Cepko CL, Ryder E, Austin C, Golden J, Fields-Berry S, Lin J. Lineage analysis with retroviral vectors. Methods Enzymol. 2000;327:118–45. doi: 10.1016/s0076-6879(00)27272-8. [DOI] [PubMed] [Google Scholar]
- 17.Strober W. Trypan blue exclusion test of cell viability. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.ima03bs21. Appendix 3:Appendix 3B. [DOI] [PubMed] [Google Scholar]
- 18.Rauh J, Milan F, Gunther KP, Stiehler M. Bioreactor systems for bone tissue engineering. Tissue Eng Part B Rev. 2011;17:263–80. doi: 10.1089/ten.TEB.2010.0612. [DOI] [PubMed] [Google Scholar]
- 19.Yeatts AB, Fisher JP. Bone tissue engineering bioreactors: dynamic culture and the influence of shear stress. Bone. 2011;48:171–81. doi: 10.1016/j.bone.2010.09.138. [DOI] [PubMed] [Google Scholar]
- 20.Schonk DM, Kuijpers HJ, van Drunen E, van Dalen CH, Geurts van Kessel AH, Verheijen R, et al. Assignment of the gene(s) involved in the expression of the proliferation-related Ki-67 antigen to human chromosome 10. Hum Genet. 1989;83:297–9. doi: 10.1007/BF00285178. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Sun Z, Chen B, Han Q, Liao L, Jia M, et al. Ex vivo expansion and in vivo infusion of bone marrow-derived Flk-1+CD31-CD34- mesenchymal stem cells: feasibility and safety from monkey to human. Stem Cells Dev. 2006;15:349–57. doi: 10.1089/scd.2006.15.349. [DOI] [PubMed] [Google Scholar]
- 22.Martins AA, Paiva A, Morgado JM, Gomes A, Pais ML. Quantification and immunophenotypic characterization of bone marrow and umbilical cord blood mesenchymal stem cells by multicolor flow cytometry. Transplant Proc. 2009;41:943–6. doi: 10.1016/j.transproceed.2009.01.059. [DOI] [PubMed] [Google Scholar]
- 23.Sims NA, Walsh NC. GP130 cytokines and bone remodelling in health and disease. BMB Rep. 2010;43:513–23. doi: 10.5483/bmbrep.2010.43.8.513. [DOI] [PubMed] [Google Scholar]
- 24.Delorme B, Ringe J, Gallay N, Le Vern Y, Kerboeuf D, Jorgensen C, et al. Specific plasma membrane protein phenotype of culture-amplified and native human bone marrow mesenchymal stem cells. Blood. 2008;111:2631–5. doi: 10.1182/blood-2007-07-099622. [DOI] [PubMed] [Google Scholar]
- 25.Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol. 2011;12:126–31. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H, Jiang W, Phillips FM, Haydon RC, Peng Y, Zhou L, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J Bone Joint Surg Am. 2003;85-A:1544–52. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 27.Lad SP, Nathan JK, Boakye M. Trends in the use of bone morphogenetic protein as a substitute to autologous iliac crest bone grafting for spinal fusion procedures in the United States. Spine (Phila Pa 1976) 2011;36:E274–81. doi: 10.1097/BRS.0b013e3182055a6b. [DOI] [PubMed] [Google Scholar]
- 28.Dimitriou R, Mataliotakis GI, Angoules AG, Kanakaris NK, Giannoudis PV. Complications following autologous bone graft harvesting from the iliac crest and using the RIA: A systematic review. Injury. 2011 doi: 10.1016/j.injury.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Tian M, Zhang H. PET molecular imaging in stem cell therapy for neurological diseases. Eur J Nucl Med Mol Imaging. 2011 doi: 10.1007/s00259-011-1860-7. [DOI] [PubMed] [Google Scholar]
- 30.Hwang NS, Varghese S, Li H, Elisseeff J. Regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in PEG-ECM hydrogels. Cell Tissue Res. 2011;344:499–509. doi: 10.1007/s00441-011-1153-2. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Zhou J, Chowdhury F, Cheng J, Wang N, Wang F. Role of mechanical factors in fate decisions of stem cells. Regen Med. 2011;6:229–40. doi: 10.2217/rme.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brosky TA, 2nd, Menke CR, Xenos D. Reconstruction of the first metatarsophalangeal joint following post-cheilectomy avascular necrosis of the first metatarsal head: a case report. J Foot Ankle Surg. 2009;48:61–9. doi: 10.1053/j.jfas.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Clements JR. Use of allograft cellular bone matrix in multistage talectomy with tibiocalcaneal arthrodesis: a case report. J Foot Ankle Surg. 2012;51:83–6. doi: 10.1053/j.jfas.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Hollawell SM. Allograft cellular bone matrix as an alternative to autograft in hindfoot and ankle fusion procedures. J Foot Ankle Surg. 2012;51:222–5. doi: 10.1053/j.jfas.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 35.McAllister BS, Haghighat K, Gonshor A. Histologic evaluation of a stem cell-based sinus-augmentation procedure. J Periodontol. 2009;80:679–86. doi: 10.1902/jop.2009.080345. [DOI] [PubMed] [Google Scholar]
- 36.Gonshor A, McAllister BS, Wallace SS, Prasad H. Histologic and histomorphometric evaluation of an allograft stem cell-based matrix sinus augmentation procedure. Int J Oral Maxillofac Implants. 2011;26:123–31. [PubMed] [Google Scholar]
- 37.Kerr EJ, 3rd, Jawahar A, Wooten T, Kay S, Cavanaugh DA, Nunley PD. The use of osteo-conductive stem-cells allograft in lumbar interbody fusion procedures: an alternative to recombinant human bone morphogenetic protein. J Surg Orthop Adv. 2011;20:193–7. [PubMed] [Google Scholar]
- 38.Hernigou P, Mathieu G, Poignard A, Manicom O, Beaujean F, Rouard H. Percutaneous autologous bone-marrow grafting for nonunions. Surgical technique. J Bone Joint Surg Am. 2006;88(Suppl 1 Pt 2):322–7. doi: 10.2106/JBJS.F.00203. [DOI] [PubMed] [Google Scholar]