Abstract
The extraordinary biological diversity of tropical forests harbors a rich chemical diversity with enormous potential as a source of novel bioactive compounds. Of particular interest are new environments for microbial discovery. Sloths – arboreal mammals commonly found in the lowland forests of Panama – carry a wide variety of micro- and macro-organisms on their coarse outer hair. Here we report for the first time the isolation of diverse and bioactive strains of fungi from sloth hair, and their taxonomic placement. Eighty-four isolates of fungi were obtained in culture from the surface of hair that was collected from living three-toed sloths (Bradypus variegatus, Bradypodidae) in Soberanía National Park, Republic of Panama. Phylogenetic analyses revealed a diverse group of Ascomycota belonging to 28 distinct operational taxonomic units (OTUs), several of which are divergent from previously known taxa. Seventy-four isolates were cultivated in liquid broth and crude extracts were tested for bioactivity in vitro. We found a broad range of activities against strains of the parasites that cause malaria (Plasmodium falciparum) and Chagas disease (Trypanosoma cruzi), and against the human breast cancer cell line MCF-7. Fifty fungal extracts were tested for antibacterial activity in a new antibiotic profile screen called BioMAP; of these, 20 were active against at least one bacterial strain, and one had an unusual pattern of bioactivity against Gram-negative bacteria that suggests a potentially new mode of action. Together our results reveal the importance of exploring novel environments for bioactive fungi, and demonstrate for the first time the taxonomic composition and bioactivity of fungi from sloth hair.
Introduction
Despite vast increases in spending on international healthcare over the last 20 years, communicable diseases continue to represent an enormous burden to global health [1]. Chronic infectious illnesses such as malaria and diverse neglected tropical diseases (NTDs) affect millions of people each year, mainly women and children in developing countries [2]. The rapid spread of antibiotic resistance has decreased the arsenal available to treat many infectious diseases, with recent emergence of pandrug resistance yielding illnesses that are recalcitrant to treatment with any known drug [3]. Concurrently non-communicable diseases are rising in incidence compared to communicable diseases: in 2008 cancer was responsible for about 7.8 million deaths (about 13% of all deaths worldwide; [4]).
Natural products represent one of the most significant sources of new drugs today. Approximately 50% of all medicines introduced between 1981 and 2006 were of natural product origin [5], with an additional 34 natural product-based drugs launched between 1997 and 2007 [6]. Since the discovery of penicillin over 80 years ago, fungi have contributed enormously to natural product drug discovery, providing a host of invaluable medicines including the β-lactam antibiotics, griseofulvin, cyclosporine, fusidic acid, and lovastatin [7]. However, in recent years the hit-rate for bioactivity from fungi has slowed, possibly pointing to the impending exhaustion of ‘low hanging fruit’ – the easily and frequently accessed fungi such as soil microfungi – as sources of new bioactive metabolites.
Conservative estimates suggest that the total number of fungal species in existence exceeds 5 million, yet fewer than 100,000 fungal species have been described [8] [9]. This suggests that exploring new environments that may be home to previously undescribed fungi could be extremely productive. For example, recent examination of tropical fungal endophytes has described tropical leaves as a ‘biodiversity hotspot’ [10] [11] [12] that is proving fruitful as a source of new bioactive compounds (see [13] for a review).
Gloer [14] proposed that production of secondary metabolites by fungi may be influenced by the selective pressures imparted by other organisms. This suggests that environments where many different species exist in close proximity may have particular potential as sources of bioactive metabolites. One such unexplored environment is the fungal microbiome associated with the coarse, sponge-like hair of the three-toed sloth (Bradypus variegatus), an arboreal mammal commonly found in the lowland tropical forests of Central America.
The coat of three-toed sloths consists of two distinct layers: an inner layer of fine, soft hair close to the skin, and an outer layer of coarse hairs that are oval in cross section and are approximately 0.4 mm wide [15]. These outer hairs carry transverse cracks that are home to an apparently ubiquitous green alga, most commonly of the genus Trichophilus [16]. The alga is popularly thought to provide camouflage for sloths against the mottled background of their habitat, but this has not been confirmed with data. Suutari et al. [16] suggested that exopolymeric substances produced by the alga might encourage the growth of beneficial bacteria. Sloth hair is commonly home to cyanobacteria and diatoms, as well as a variety of macro-organisms (e.g., cockroaches, roundworms, and moth larvae; [16]). However, little is known about communities of fungi on sloth hair.
In an effort to uncover new sources of drugs for treating vector-borne diseases, cancer, and bacterial infections, we used a culture-based approach to examine fungal communities associated with the coarse, outer hair of Bradypus variegatus in Panama. Here we report the isolation of fungi with bioactivity against Trypanosoma cruzi, the causal agent of Chagas disease; Plasmodium falciparum, the causal agent of malaria; the human breast cancer cell line MCF-7; and a range of Gram-negative and Gram-positive human pathogenic bacteria. Phylogenetic analyses were used to identify these fungi at a finer and more robust level than is possible using typical database-matching methods alone. Our results suggest that sloth hair is an interesting new source of important bioactive fungi with much scope for exploration of the five other extant sloth species found across the neotropics.
Materials and Methods
Approval for sampling of sloth hair was obtained from the Institutional Animal Care and Use Committee (IACUC) of the Smithsonian Tropical Research Institute and collection permits were obtained from Panama's Autoridad Nacional del Ambiente (ANAM). A single sample of coarse outer hair was collected from around the lower back of each of nine living three-toed sloths encountered in February 2011 along Pipeline Road in Soberanía National Park, Republic of Panamá (N 9°9′, W 79°44′; [17]). Hair samples were placed in sterile Falcon tubes half filled with silica gel and stored at ambient temperature until processed.
Isolation and cultivation of fungi
Each hair was rinsed with sterile water to remove debris and loosely associated microorganisms. Under aseptic conditions hairs were cut into pieces approximately 2.5 cm long and placed on the surface of potato dextrose agar (PDA) or 2% malt extract agar (MEA) in sterile Petri plates. Plates were sealed with Parafilm, incubated at room temperature, and checked for new fungal growth daily for 2 weeks. Hyphae were cut aseptically from each plate and transferred to axenic culture on the same medium. Isolates were stored as living vouchers at room temperature (i.e., as agar plugs with mycelium in sterile distilled water) and have been archived in the collection of the International Cooperative Biodiversity Group (ICBG) at the Smithsonian Tropical Research Institute in Panama (accessions available on request).
Fungal identification
Total genomic DNA was extracted from fresh cultures of fungi according to Arnold & Lutzoni [11]. The nuclear ribosomal internal transcribed spacers and 5.8S gene (ITSrDNA) and an adjacent portion of the nuclear ribosomal large subunit (LSUrDNA) were amplified as a single fragment using primers ITS1F or ITS5 and LR3 following Hoffman et al. [18]. PCR products were visualized using SYBR green following electrophoresis on a 1% agarose gel and submitted to the University of Arizona Genetics Core for cleanup, normalization, and bidirectional Sanger sequencing. Sequences were assembled automatically and bases called using phred and phrap [19] [20] with orchestration by Mesquite [21], followed by manual editing in Sequencher 4.5 (GeneCodes Corp.). Sequences were compared against the NCBI non-redundant database using BLASTn [22] to estimate taxonomic placement and were submitted to GenBank under accession nos. KF746076-KF746159 (Table S1).
Because identification based only on BLAST matches must be treated with caution (e.g., [23]) we used phylogenetic analyses to provide stronger inference regarding taxonomic affiliation. Sequence data for 84 fungal isolates were loaded as one group into Mesquite [21] and aligned by MUSCLE [24]. The resulting alignment, which identified clusters of easily alignable sequences, was inappropriate for a single phylogenetic analysis due to the prevalence of ambiguous and unalignable regions. Sequences that aligned coherently to one another were partitioned into groups of apparently closely related strains. Resulting groups were then analyzed as separate data sets as follows: All sequences in each group were compared against the NCBI database using BLASTn. The top 50 hits for each sequence were downloaded and filtered to remove (1) redundant sequences, and (2) sequences from potentially mis-identified strains and unvouchered specimens (see [23]). At least one sequence from a reliable culture collection was included in each data set, and one or two outgroups were selected from species closely related to named sequences in the dataset (chosen by literature review).
Each resulting data set was aligned separately in MUSCLE. Sequences in each alignment were trimmed to consistent starting and ending points, and alignment quality was verified by visual inspection in Mesquite. All alignments were submitted to TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S14945). Models of evolution were inferred using jModeltest [25] [26]. Phylogenetic trees were inferred by maximum likelihood in GARLI [27], with support determined by 100 bootstrap replicates implemented in the CIPRES web portal [28], and using Bayesian methods in MrBayes (accessed via CIPRES [28]). For the latter, analyses consisted of 5 million generations, with four chains, random starting trees, and sampling every 1000th tree. Completeness was assessed based on asymptotes of the –ln li values and evaluation of standard deviations of split frequencies. Posterior probabilities provided inferentially independent values to support the topological relationships and associated support values from maximum likelihood analyses. Results of each phylogenetic analysis were compared against the existing literature for each taxonomic group to ensure that relationships of known taxa were appropriate, thus providing confidence with regard to our placement of sloth hair fungi.
Preparation of fungal extracts
A single agar plug of each strain selected for further analysis was transferred under sterile conditions to a fresh plate of 2% MEA and incubated on the benchtop at room temperature until approximately 50% of the plate was covered with mycelial growth. Fifteen agar plugs (each 5 mm in diameter) were cut with a sterile cork borer under sterile conditions and transferred to flasks containing 37 ml of 2% malt extract broth (MEB). Flasks were incubated on an orbital shaker (28°C, 125 rpm) for 2 weeks.
Liquid cultures were mixed with an equal volume of ethyl acetate and blended for 2 minutes at 9000 rpm with a Polytron (Lauda-Brinkmann, Delran, NJ, USA). Fungal biomass was removed by vacuum filtration through Whatman filter paper (#1) and the filtrate was extracted twice with a 1∶1 volume of ethyl acetate. The aqueous layer was discarded and the organic layer was dried and stored at −80°C.
Bioassays
Crude organic extracts of fungal cultures were used in bioassays against focal strains of the causal agents of malaria (Plasmodium falciparum) and Chagas disease (Trypanosoma cruzi), and against breast cancer cell line MCF-7, as described by Higginbotham et al. [29]. Bioactivity of extracts, which were diluted in DMSO (10 µg/ml), was measured as percent inhibition of growth (% IG) compared to the negative control (DMSO with no extract; 0% IG).
BioMAP analyses
An antibiotic activity profile screen, BioMAP (Antibiotic mode of action profile; [30]), was used to test activity of 50 crude organic extracts against 15 human pathogenic bacteria. Extracts with potent bioactivity were serially diluted (16 2-fold dilutions) and re-screened against the same panel of bacteria. Growth curves were plotted by recording OD600 values each hour for 24 hours and minimum inhibitory concentrations (MIC) were subsequently calculated for each extract. To establish concentration-independent bioactivity profiles for each extract, raw MIC values were normalized resulting in a range of values from 0 (inactive) to 1 (most bioactive). These activity profiles were then compared to profiles of known antibiotics belonging to all major structural classes. This method is effective in predicting the structural class of unknown antibiotic compounds from complex or crude mixtures [30].
Results and Discussion
A total of 84 fungal isolates was examined from the coarse outer hair of nine living individuals of the three-toed sloth (Bradypus variegatus) encountered along Pipeline Road in Soberanía National Park, Republic of Panama. Hair samples were transported to the lab in sterile Falcon tubes which had been half filled with silica gel, a desiccant that is effective for storing fungal tissue [31] [32]. Every piece of sloth hair placed on agar yielded multiple fungal isolates representing a variety of morphotypes. Phylogenetic analyses of axenic strains revealed a diverse group of fungi, some of which appear to be novel relative to previously observed or sequenced taxa (Table 1; Figure 1). Many of these isolates display bioactivity in vitro against parasites that cause malaria and Chagas disease, breast cancer cells, and both Gram-positive and Gram-negative human pathogenic bacteria (Table 2; Table 3).
Table 1. Eighty-four fungal isolates from the coarse outer hair of nine individuals sloths (B. varieagatus), their top BLAST matches, and maximum identity value from BLAST searches; group ID and tree (Fig. 1) revealing phylogenetic placement; and taxonomic placement based on phylogenetic analyses at the family (order) and genus levels (Fig. 1).
| Fungus ID | Sloth hair Sample # | Top BLAST Hit, (Accession Number) | Max ID (%) | Group ID | Tree | Family (Order) | Genus | OTU |
| F4847 | S006 | Fungal sp., (HQ832955.1)¥ | 99 | 18 | B | Apiosporaceae (Incertae sedis) | Arthrinium | S* |
| F4821 | S007 | Bionectria cf. ochroleuca, (EU552110.1)¥ | 99 | 3 | P | Bionectriaceae (Hypocreales) | Bionectria | I |
| F4889 | S008 | Bionectria cf. ochroleuca, (EU552110.1)¥ | 99 | 3 | P | Bionectriaceae (Hypocreales) | Bionectria | I |
| F4913 | S009 | Bionectria cf. ochroleuca, (EU552110.1)¥ | 99 | 3 | P | Bionectriaceae (Hypocreales) | Bionectria | I |
| F4874 | S005 | Bionectria pityrodes, (JQ411387.1)¥ | 99 | 3 | P | Bionectriaceae (Hypocreales) | Bionectria | W* |
| F4806 | S008 | Colletotrichum sp., (DQ300348.1)¥ | 99 | 7 | F | Glomerellaceae (Glomerellales) | Colletotrichum | R* |
| F4839 | S002 | Ascomycota sp., (HQ608154.1) [58] | 99 | 10 | K | Valsaceae (Diaporthales) | Cytospora | D |
| F4818 | S002 | Sordariomycetes sp., (JX174122.1)¥ | 99 | 10 | K | Valsaceae (Diaporthales) | Cytospora | D |
| F4801 | S003 | Endomelanconiopsis endophytica, (GQ469968.1)¥ | 100 | 5 | E | Botryosphaeriaceae (Botryosphaeriales) | Endomelanconiopsis | U* |
| F5071 | S003 | Fungal sp., (FJ612981.1) [23] | 99 | 15 | H | Xylariaceae (Xylariales) | Entonaema | N* |
| F4876 | S006 | Fungal sp., (EU563515.1) [59] | 99 | 13 | N | Nectriaceae (Hypocreales) | Fusarium | X |
| F4828 | S005 | Fungal sp., (FJ612934.1) [23] | 99 | 12 | M | Nectriaceae (Hypocreales) | Fusarium | O* |
| F4898 | S003 | Fungal sp., (FJ613085.1) [23] | 100 | 12 | M | Nectriaceae (Hypocreales) | Fusarium | Y* |
| F4881 | S009 | Fusarium concolor, (GQ505763.1) [60] | 99 | 13 | N | Nectriaceae (Hypocreales) | Fusarium | X |
| F4897 | S003 | Uncultured soil fungus clone, (DQ420777.1) [35] | 98 | 11 | L | Nectriaceae (Hypocreales) | Fusarium | AA* |
| F4877 | S003 | Uncultured soil fungus clone, (DQ420802.1) [35] | 99 | 11 | L | Nectriaceae (Hypocreales) | Fusarium | K |
| F4890 | S008 | Uncultured soil fungus clone, (DQ420802.1) [35] | 98 | 11 | L | Nectriaceae (Hypocreales) | Fusarium | K |
| F4900 | S004 | Fungal sp., (FJ613088.1) [23] | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | E |
| F4812 | S001 | Hypocrea jecorina, (AF510497.1)¥ | 99 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4813 | S001 | Hypocrea jecorina, (EU280094.1) [61] | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4815 | S001 | Hypocrea jecorina, (EU280094.1) [61] | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4814 | S001 | Hypocrea jecorina, (JQ070073.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4852 | S001 | Hypocrea jecorina, (JQ070073.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4853 | S001 | Hypocrea jecorina, (JQ070073.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4855 | S001 | Hypocrea jecorina, (JQ070073.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4856 | S001 | Hypocrea jecorina, (JQ070073.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4854 | S001 | Hypocrea jecorina, (JQ411369.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4882 | S009 | Hypocrea jecorina, (JQ411369.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4901 | S004 | Hypocrea lixii, (AY605743.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | E |
| F4831 | S005 | Hypocrea lixii, (JF923806.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | E |
| F4902 | S005 | Hypocrea lixii, (JF923806.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | E |
| F4873 | S005 | Hypocrea muroiana, (JN943367.1) [62] | 99 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | H |
| F4883 | S006 | Hypocrea nigricans, (JN943369.1) [62] | 99 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | E |
| F4816 | S001 | Trichoderma reesei, (JQ979435.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | A |
| F4830 | S005 | Uncultured Trichoderma clone, (JX317342.1)¥ | 100 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | H |
| F4845 | S005 | Uncultured Trichoderma clone, (JX317342.1)¥ | 99 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | H |
| F4860 | S005 | Uncultured Trichoderma clone, (JX317342.1)¥ | 99 | 1 | I | Hypocreaceae (Hypocreales) | Hypocrea | H |
| F4825 | S004 | Lasiodiplodia parva, (GQ469961.1)¥ | 100 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F4844 | S004 | Lasiodiplodia parva, (GQ469961.1)¥ | 100 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F4802 | S004 | Lasiodiplodia parva, (GQ469964.1)¥ | 100 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F5068 | S009 | Lasiodiplodia theobromae, (FJ478103.1)¥ | 99 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F4807 | S008 | Lasiodiplodia theobromae, (GQ469934.1)¥ | 100 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F4906 | S007 | Lasiodiplodia theobromae, (GQ469934.1)¥ | 100 | 4 | Q | Botryosphaeriaceae (Botryosphaeriales) | Lasiodiplodia | C |
| F4904 | S006 | Fungal sp., (GU566256.1) [63] | 99 | 14 | S | Leptosphaeriaceae (Pleosporales) | Leptosphaeria | J |
| F4905 | S006 | Fungal sp., (GU566256.1) [63] | 100 | 14 | S | Leptosphaeriaceae (Pleosporales) | Leptosphaeria | J |
| F4819 | S006 | Fungal endophyte sp., (EU977287.1) [55] | 99 | 20 | D | Lophiostomataceae (Pleosporales) | Unknown | P* |
| F4831a | S002 | Fungal endophyte sp., (EU561602.1)¥ | 99 | 19 | C | Montagnulaceae (Pleosporales) | Paraconiothyrium † | A |
| F4884 | S008 | Penicillium herquei, (EU833220.1)¥ | 99 | 6 | O | Trichocomaceae (Eurotiales) | Penicillium | V* |
| F4863 | S009 | Penicillium sp., (JF288548.1) [64] | 99 | 6 | O | Trichocomaceae (Eurotiales) | Penicillium | T* |
| F4803 | S004 | Fungal endophyte sp., (EU561622.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4846 | S006 | Fungal endophyte sp., (EU561622.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4878 | S008 | Fungal endophyte sp., (EU561622.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4896 | S002 | Fungal endophyte sp., (EU561622.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4842 | S003 | Pestalotiopsis microspora, (FJ478120.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4848 | S006 | Pestalotiopsis microspora, (FJ478120.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4872 | S004 | Pestalotiopsis sp., (EU605882.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4820 | S006 | Pestalotiopsis sp., [EU605882.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4824 | S003 | Pestalotiopsis sp., (EU605882.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4829 | S005 | Pestalotiopsis sp., (EU605882.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4858 | S004 | Pestalotiopsis sp., (EU605882.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4908 | S007 | Pestalotiopsis sp., (EU605882.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4857 | S002 | Pestalotiopsis sp., (EU605882.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | Q* |
| F4817 | S002 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4826 | S004 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4827 | S005 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4837 | S002 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4841 | S003 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4861 | S005 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4862 | S007 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4895 | S002 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4909 | S008 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F5074 | S003 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4823 | S003 | Pestalotiopsis sp., (HQ832816.1)¥ | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4894 | S002 | Pestalotiopsis theae, (HQ832793.1)¥ | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4870 | S002 | Uncultured fungus clone, (JN890176.1) [65] | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | G |
| F4875 | S005 | Uncultured fungus clone, (JN890258.1) [65] | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F4879 | S008 | Uncultured fungus clone, (JN890258.1) [65] | 99 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F5069 | S006 | Uncultured fungus clone, (JN890258.1) [65] | 100 | 2 | J | Amphisphaeriaceae (Xylariales) | Pestalotiopsis | B |
| F5073 | S008 | Sordariomycetes sp., (JQ760525.1) [65] | 99 | 17 | A | Calosphaeriaceae (Calosphaeriales) | Phaeoacremonium | L* |
| F5070 | S006 | Uncultured fungus clone, (GU721790.1)¥ | 99 | 16 | T | Cephalotheceae (Sordariales) | Phialemonium | F |
| F5072 | S003 | Uncultured fungus clone, (GU721790.1)¥ | 99 | 16 | T | Cephalotheceae (Sordariales) | Phialemonium | F |
| F4886 | S004 | Robillarda sessilis, (HQ608017.1) [58] | 99 | 8 | G | Amphisphaeriaceae (Xylariales) | Robillarda † | Z* |
| F4891 | S009 | Phoma sp., (JQ621876.1)¥ | 99 | 9 | R | Pleosporales† | Unknown | BB* |
| F4850 | S009 | Uncultured soil fungus clone, (DQ421180.1) [35] | 96 | 9 | R | Unknown | Unknown | M* |
OTU codes indicate operational taxonomic units based on 95% sequence similarity. Order and family level identifications were confidently assigned to 82 isolates and genus level identifications were confidently assigned to 80 isolates.
Unpublished strains.
*OTUs occurring only once (singletons).
Tentative phylogenetic placement.
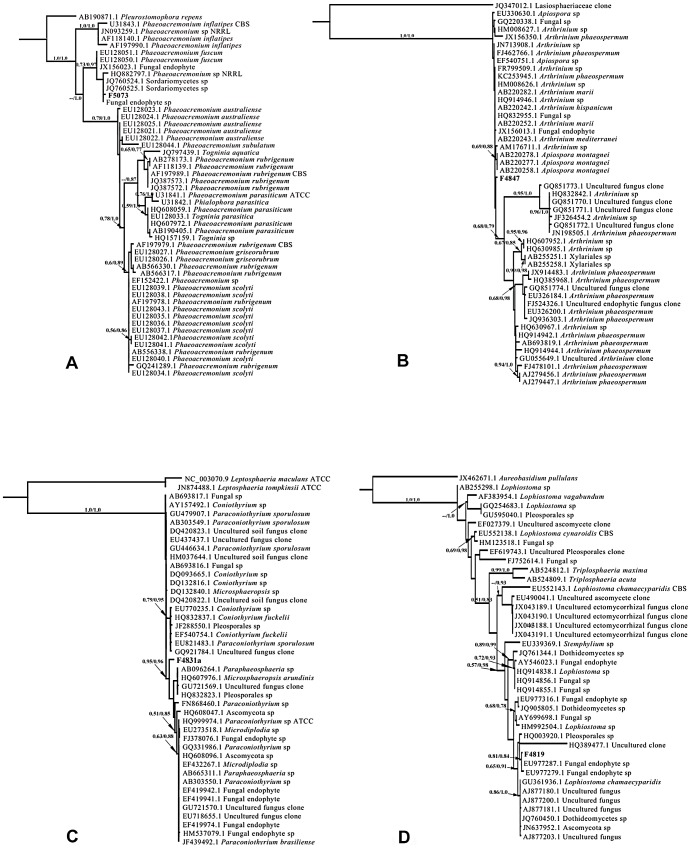
Figure 1. Results of maximum likelihood and Bayesian analyses of ITSrDNA data for fungi isolated in culture from coarse, outer hair of three-toed sloths in Soberanía National Park, Panama.
Topology of each tree reflects ML analysis, and values above branches indicate ML bootstrap values and Bayesian posterior probabilities (>0.50 and >0.75, respectively). Outgroups and taxon sampling for each tree were validated by literature surveys (see methods). Taxonomic conclusions are presented in Table 1. Figure 1(A): Placement of F5073 in group 17; (B) F4847 in group 18; (C) F4831a in group 19; (D) F4819 in group 20; (E) F4801 in group 5; (F) F4806 in group 7; (G) F4886 in group 8; (H) F5071 in group 15; (I) F4812–F4816, F4830, F4831, F4845, F4852–F4856, F4860, F4873, F4882, F4883 and F4900–F4902 in group 1; (J) F4803, F4817, F4820, F4823, F4824, F4826, F4827, F4829, F4837, F4841, F4842, F4846, F4848, F4857, F4858, F4861, F4862, F4870, F4872, F4875, F4878, F4879, F4894–F4896, F4908, F4909, F5069 and F5074 in group 2; (K) F4818 and F4839 in group 10; (L) F4877, F4890 and F4897 in group 11; (M) F4828 and F4898 in group 12; (N) F4876 and F4881 in group 13; (O) F4863 and F4884 in group 6; (P) F4821, F4874, F4889 and F4913 in group 3; (Q) F4802, F4807, F4825, F4844, F4906 and F5068 in group 4; (R) F4850 and F4891 in group 9; (S) F4904 and F4905 in group 14; and (T) F5070 and F5072 in group 16.
Table 2. Bioactivity of sloth hair surface associated fungi isolated on potato dextrose agar (PDA) or 2% malt extract agar (MEA) against causative agents of malaria (P. falciparum) and Chagas disease (T. cruzi), and against the MCF-7 breast cancer cell line.
| Fungus ID | OTU | Isolation Media | Putative Genus | P. falciparum | T. cruzi | MCF-7 |
| F4813 | A | PDA | Hypocrea | A | ||
| F4814 | A | MEA | Hypocrea | A | ||
| F4815 | A | PDA | Hypocrea | A | A | |
| F4816 | A | PDA | Hypocrea | A | A | |
| F4818 | D | PDA | Cytospora | A | A | |
| F4828 | O | PDA | Fusarium | A | ||
| F4853 | A | MEA | Hypocrea | - | A | |
| F4854 | A | MEA | Hypocrea | A | ||
| F4855 | A | PDA | Hypocrea | A | A | |
| F4863 | T | PDA | Penicillium | A | A | A |
| F4874 | W | MEA | Bionectria | A | ||
| F4881 | X | MEA | Fusarium | A | A | A |
| F4882 | A | MEA | Hypocrea | - | A | |
| F4890 | K | MEA | Fusarium | - | A | |
| F4894 | G | MEA | Pestalotiopsis | A | ||
| F4898 | Y | PDA | Fusarium | A | A |
Bioactive fungi (A) are those causing ≥50% inhibition of growth of parasite or cancer cells in in vitro assays.
- = extract not tested in this bioassay; empty cell = not highly active.
Table 3. Bioactivity of fungi from sloth hair isolated on potato dextrose agar (PDA) or malt extract agar (2%; MEA) against a range of Gram-positive and Gram-negative bacteria in the BioMAP assay [30].
| Gram-positive | Gram-negative | |||||||||||||||||
| Fungus ID | OTU | Isolation Media | Putative Genus | B. sub | E. fae | L. iva | S. epi | S. au | MRSA | Y. pse | P. aer | S. typ | V. chol | E. coli | A. baum | E. aero | O. ant | P. alc |
| F4850 | M | PDA | Unknown | A | ||||||||||||||
| F4847 | S | MEA | Arthrinium | A | ||||||||||||||
| F4821 | I | MEA | Bionectria | A | A | A | A | |||||||||||
| F4806* | R | MEA | Colletotrichum | A | A | A | A | A | A | |||||||||
| F4818* | D | PDA | Cytospora | A | A | A | A | A | A | |||||||||
| F4839 | D | PDA | Cytospora | A | A | A | A | A | ||||||||||
| F4904 | J | PDA | Leptosphaeria | A | A | A | A | |||||||||||
| F4905 | J | PDA | Leptosphaeria | A | ||||||||||||||
| F4801 | U | MEA | Endomelanconiopsis | A | ||||||||||||||
| F4828 | O | PDA | Fusarium | A | A | A | A | A | A | |||||||||
| F4898* | Y | PDA | Fusarium | A | A | A | A | A | A | |||||||||
| F4830 | H | MEA | Hypocrea | A | ||||||||||||||
| F4807* | C | PDA | Lasiodiplodia | A | A | A | WA | A | WA | WA | ||||||||
| F4825 | C | PDA | Lasiodiplodia | A | ||||||||||||||
| F4844* | C | MEA | Lasiodiplodia | A | A | A | A | A | A | |||||||||
| F4824 | B | MEA | Pestalotiopsis | A | ||||||||||||||
| F4829 | B | MEA | Pestalotiopsis | A | A | A | A | |||||||||||
| F4842 | B | MEA | Pestalotiopsis | A | A | A | A | A | ||||||||||
| F4846 | G | MEA | Pestalotiopsis | A | ||||||||||||||
| F4848 | B | MEA | Pestalotiopsis | A | ||||||||||||||
Fungi with particularly potent bioactivity were selected for further study and are marked with an asterisk (*). Fungal extracts causing full cell death are marked ‘A’ and those causing partial cell death are marked ‘WA’.
B. sub = Bacillus subtilis 168; E. fae = Enterococcus faecium ATCC 6569; L. iva = Listeria ivanovii BAA-139; S. epi = Staphylococcus epidermis ATCC 14990; S. au = Staphylococcus aureus ATCC 29213; MRSA = Methicillin Resistant Staphylococcus aureus BAA-44; Y. pse = Yersinia pseudotuberculosis IP2666 pIBI; P. aer = Pseudomonas aeruginosa ATCC 27835; S. typ = Salmonella typhimerium LT2; V. chol = Vibrio cholerae O1 (biotype El Tor A1552); E. coli = Escherichia coli K12 (BW 25113); A. baum = Acinetobacteria baumanii NCIMB 12457; E. aero = Enterobacter aerogenes ATCC 35029; O. ant = Ochrobactrum anthropi ATCC 49687; P. alc = Providencia alcallifaciens ATCC 9886.
Phylogenetic analyses
The 84 fungal isolates represented 28 operational taxonomic units (OTUs) based on 95% sequence similarity (Table 1). The two most abundant OTUs represented 34 (40.5%) of the isolates. The remaining 50 isolates were represented by 26 distinct OTUs.
BLAST comparisons with GenBank provided preliminary estimations of taxonomic placement and similarity to previously sequenced fungi (Table 1). All isolates were Ascomycota. Fourteen isolates had a top match to uncultured fugal clones and 23 had a top match to cultured but unidentified fungi. The remaining 47 isolates had matches to named strains that tentatively suggested placement in the Sordariomycetes (Xylariales, Glomerellales, Hypocreales), Dothideomycetes (Botryosphaeriales and Pleosporales), and Eurotiomycetes (Eurotiales). To more confidently determine taxonomic placement, sequences were analyzed using maximum likelihood and Bayesian methods.
Sequences that aligned well to one another were partitioned into clusters to create 20 groups of apparently similar species (Table 1; Figure 1). One group contained 29 sequences from sloth-hair fungi, one group contained 20 sequences, and the remaining 18 groups contained between 1 and 6 sequences each. Between 34 and 165 sequences from closely related taxa were compiled for each group to make non-redundant datasets to which one or two appropriate outgroups were added based on literature review.
Following phylogenetic analyses, high level identities (class, order, family) could be confidently assigned to 82 fungi representing 15 families and 10 orders (Table 1; Figure 1). The majority (81.7%) of these were Sordariomycetes. Fungi from this class are well documented sources of bioactive metabolites (e.g., [33] [34]). The remaining isolates were Dothideomycetes (15.9%) and Eurotiomycetes (2.4%).
Eighty isolates could confidently be assigned to 15 genera (Table 1; Figure 1). Two isolates were given more tentative phylogenetic placements. F4886 (Figure 1(G)) was identified as Robillarda sp. or as a member of a closely related genus within the Amphisphaeriaceae. Isolate F4831a (Figure 1(C)) was identified as a member of Paraconiothyrium sp. or a closely related genus within the Montagnulaceae. The two most common genera, Pestalotiopsis sp. and Hypocrea sp., were isolated from 7 of 9 and 5 of 9 sloth hair samples, respectively.
Isolate F4891 (Figure 1(R)) did not appear to be closely related to any named sequences in GenBank and could only be identified as possibly a member of the order Pleosporales (Dothideomycetes). Isolate F4850 (Figure 1(R)) had 96% sequence similarity to its closest match in GenBank, an uncultured soil fungus clone (Table 1; [35]). Further analyses are warranted in order to confirm whether isolates F4891 and F4850 are novel fungal species. In general, identifications assigned by phylogenetic analyses closely matched those of the top BLAST hits for each isolate (Table 1).
Bioactivity of sloth hair isolates
Sloth hair fungi that were cultivated in liquid medium and extracted with ethyl acetate were tested for bioactivity. We considered extracts to be highly bioactive if they caused at least 50% inhibition of the growth (i.e., ≥50% IG) of parasites or cancer cells in vitro.
Overall, two of 70 (2.5%) extracts tested were highly active against P. falciparum, eight of 62 (12.9%) extracts tested were highly active against T. cruzi, and 15 of 73 (20.6%) extracts tested were highly active against the MCF-7 breast cancer cell line (Table 2). Most extracts with bioactivity were active in only one assay (Table 2). Bioactive strains were isolated with equal frequency on MEA and PDA (Table 2). Notably, closely related isolates often differed in bioactivity: for example, three of 20 isolates identified as Hypocrea sp. (Table 1) had bioactivity only against MCF-7 breast cancer cells; three were active against MCF-7 breast cancer cells and T. cruzi (Table 2); and one was active only against methicillin-resistant S. aureus (Table 3).
Of the 16 fungi that had high % IG in at least one in vitro assay, 13 belonged to the Hypocreales (Hypocreaceae, Bionectriaceae and Nectriaceae). The remaining three fungi belonged to the Valsaceae (Diaporthales), Amphisphaeriaceae (Xylariales) and Trichocomaceae (Eurotiales). Anti-malarial and anti-cancer activities have been previously reported in the literature from these relatively well-studied fungal lineages (e.g., [36] [37] [38] [39] [40] [41] [42] [43]). However our results represent, to the best of our knowledge, the first reports of anti-trypanosomal activity in Cytospora (Valsaceae) and Bionectria (Bionectriaceae).
The number of isolates highly bioactive against T. cruzi (Table 2) is of particular interest as we so rarely encounter microbes with bioactivity in this assay: only 104 of 2698 fungal endophytes (3.9%) from our overall culture collection are highly bioactive against that parasite [29]. Currently the only treatments available for this illness are nitrofurane and benznidazole, both of which are associated with such toxic side effects that treatment is often abandoned [44].
Of 50 fungal extracts screened by BioMAP, 20 were bioactive against at least one test organism (Table 3). The majority of isolates (16 of 20) active against bacteria in the BioMAP assay belonged to the families Lasiosphaeriaceae (Sordariales), Glomerellaceae (Glomerellales), Valsaceae (Diaporthales), Botryosphaeriaceae (Botryosphaeriales), and Amphisphaeriaceae (Xylariales). The remaining four fungi belonged to the Nectriaceae, Bionectriaceae and Hypocreaceae (Hypocreales). Activity against Gram-positive bacteria was detected far more frequently than against Gram-negative bacteria (Table 3).
Five isolates (Colletotrichum sp. F4806 (Figure 1(F)); Cytospora sp. 1, strain F4818 (Figure 1(K)); Fusarium sp. 1, strain F4898 (Figure 1(M)); Lasiodiplodia sp. 1, strain F4807 (Figure 1(Q)); and Lasiodiplodia sp. 1, strain F4844 (Figure 1(Q)) had particularly potent activity in the BioMAP assay and were selected for further testing. Serially diluted extracts were re-tested for activity against the panel of 15 bacteria and bioactivity profiles were constructed using normalized MIC values (Table 4).
Table 4. Minimum inhibitory concentrations (MIC) of 5 sloth hair associated fungal extracts in the BioMAP antibiotic profile screen [30].
| Minimum Inhibitory Concentration (Normalized 0–1) | |||||
| Fungus ID | F4806 | F4807 | F4818 | F4844 | F4898 |
| Bacillus subtilis | 1 | 0.85 | 1 | 1 | 1 |
| Staphylococcus epidermis | 0.46 | 0 | 0.88 | 0.87 | 0.69 |
| Enterococcus faecium | 0.6 | 0 | 0.52 | 0.6 | 0.53 |
| Listeria ivanovii | 0.6 | 0 | 0.4 | 0.46 | 0.53 |
| Staphylococcus aureus | 0.87 | 0 | 1 | 0.6 | 0.85 |
| MRSA | - | - | - | - | - |
| Yersinia pseudotuberculosis | 0 | 0.69 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 0 | 0 | 0 | 0 | 0 |
| Salmonella typhimerium | 0 | 0.53 | 0 | 0 | 0 |
| Vibrio cholerae | 0 | 0 | 0 | 0 | 0 |
| Escherichia coli | 0 | 0.69 | 0 | 0 | 0 |
| Acinetobacter baumanii | 0 | 1 | 0 | 0 | 0 |
| Enterobacter aerogenes | 0 | 0 | 0 | 0 | 0 |
| Ochrobactrum anthropi | 0 | 0.69 | 0 | 0 | 0 |
| Providencia alcalifaciens | 0 | 0.53 | 0 | 0 | 0 |
To generate a concentration- independent bioactivity profile, raw assay results were normalized giving a range of values from 0 (inactive) to 1 (most bioactive). These bioactivity fingerprints were then compared to fingerprints of known antibiotics from all the major structural classes, which had previously been tested in the BioMAP assay. The bioactivity fingerprint of F4807 was of particular interest as it did not match that of any antibiotic previously tested in the BioMAP screen.
- = Test failed, data not available.
The extract from Lasiodiplodia sp. 1 (strain F4807) was of particular interest as it had potent and specific activity against Gram-negative bacteria (Table 4). The bioactivity profile of the isolate did not match that of any of the known antibiotic classes previously tested in the BioMAP assay, suggesting a potentially novel mode of action (see [30]). Infections caused by emerging multidrug-resistant (MDR) Gram-negative bacteria are becoming a major clinical concern worldwide [45]. These infections most commonly include MDR Pseudomonas aeruginosa, extended-spectrum β-lactamase producing Enterobacteriaceae, and MDR Acinetobacter baumannii [46]. Recent data suggest that infections traced to Staphylococcus aureus bloodstream isolates are in decline, but that infections by Escherichia coli, the most common Gram-negative species responsible for infections in human blood isolates, are increasing in frequency [47]. That report [47] also highlights the paucity of antibacterial agents under development for Gram-negative bacteria, with most effort being focused on Gram-positive MRSA. The increasing prevalence of MDR organisms is mainly due to overuse of broad-spectrum antibiotics and poor antibiotic stewardship [48]. Hence the specificity of the extract from strain F4807 for Gram-negative strains is highly valuable and worth pursuing for further analysis.
Fungi isolated in this study were taxonomically consistent with groups of fungi known to occur in soil and in plants as pathogens, saprotrophs, or endophytes [11] [49]. Sloths may encounter such fungi incidentally from air spora, or via direct contact when they descend from trees in order to defecate and urinate, at which time they dig a hole in the soil that they subsequently cover with leaf litter [50]. Strikingly, comparison with a large collection of endophytes from terrestrial plants in Panama ([29]; 1269 strains) revealed that sequences from 29 isolates from sloth hair were identical to strains obtained from plants (100% similarity over the full sequence length). Thus this study extends the known host range and ecological mode of putative endophytes. Some of these fungi may affiliate directly with the green alga that inhabits coarse hair of sloths, much like endolichenic fungi associate with green algae in lichen thalli and are often taxonomically similar to endophytes in the same environments [51] [52].
Potential roles of these fungi in the health of sloths have not been investigated. Araújo Xavier et al. [50] reported fungal infections in B. variegatus caused by the fungal pathogen Microsporum (which also causes dermatitis in humans; [53]), but this genus was not recovered in the present study or in previous work by Suutari et al. [16]. In contrast to the human microbiome, where fungi comprise <0.01% of microbial communites on external surfaces such as skin [54], Suutari et al. [16] reported that fungi represented 8% of the flora in surveys of sloth-hair microbes. The high abundance and diversity of fungi associated with sloth hair, coupled with their bioactivity, may speak to a biological importance to sloths that is yet unexplored.
The pressing need for new medications continues to represent one of humanity's greatest challenges. It is commonly agreed that the vast majority of bioactive microbes remain to be discovered [9] and newly discovered microbial taxa hold an important promise of novel chemistry. Abundant evidence points to novel environments as promising sources of as yet undescribed microorganisms [55]. Here we have demonstrated that hair of the three-toed sloth (Bradypus variegatus) in Panama is a rich source of diverse bioactive fungi. Strains isolated here included members of some well-studied lineages (e.g., Hypocreales; Pleosporales), but also members of understudied or potentially novel groups (e.g., Endomelanconiopsis sp. F4801; Robillarda sp. F4886; Ascomycota sp. F4891; Ascomycota sp. F4850). We anticipate that additional novel taxa could be found on sloth hair by expanding survey methods to diverse types of media and using high-throughput methods to assess community composition. We also anticipate that different taxa may be isolated from sloths in other regions, much as communities of endophytes in tropical plants differ markedly at a regional scale [56] [57]. Thus our work suggests that fruitful exploration of the sloth microbiota is warranted for potential applications in drug discovery.
Supporting Information
Sloth hair fungus ID and Genbank accession numbers.
(DOCX)
Acknowledgments
We thank Bryson Voirin for collection of sloth hair samples. We thank Phyllis D. Coley, Thomas A. Kursar, Meg Crofoot and Santosh Jagadeeshan for helpful comments and other assistance, and Malkanthi Gunatilaka and Jakob Riddle for sequence analysis. We thank the government of Panama and, in particular, the personnel of Panama's Autoridad Nacional del Ambiente (ANAM), for facilitating this research. We thank the Smithsonian Tropical Research Institute (STRI) for supporting all logistical aspects of this work and the many students and assistants who contributed to this body of research.
Funding Statement
This work was supported by the International Cooperative Biodiversity Groups program (ICBG-Panama http://www.icbg.org/, grant number 2 U01 TW006634-06) and the College of Agriculture and Life Sciences at the University of Arizona. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dye C, Mertens T, Hirnschall G, Mpanju-Shumbusho W, Newman RD, et al. (2013) WHO and the future of disease control programmes. Lancet 381: 413–418. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Neglected tropical diseases [Online]. Available: http://www.who.int/neglected_diseases/en./ Accessed 2013 Apr 30.
- 3. Falagas ME, Bliziotis IA (2007) Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int J Antimicrob Agents 29: 630–636. [DOI] [PubMed] [Google Scholar]
- 4. Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, et al. (2012) Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 380: 1840–1850. [DOI] [PubMed] [Google Scholar]
- 5. Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75: 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler MS (2008) Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 25: 475–516. [DOI] [PubMed] [Google Scholar]
- 7. Aly AH, Debbab A, Proksch P (2011) Fungal endophytes: unique plant inhabitants with great promises. Appl Microbiol Biotechnol 90: 1829–1845. [DOI] [PubMed] [Google Scholar]
- 8. Hawksworth DL, Rossman AY (1997) Where are all the undescribed fungi? Phytopathology 87: 888–891. [DOI] [PubMed] [Google Scholar]
- 9. Blackwell M (2011) The Fungi: 1, 2, 3 … 5. 1 Million Species? Am J Bot 98: 426–438. [DOI] [PubMed] [Google Scholar]
- 10. Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA (2000) Are tropical fungal endophytes hyperdiverse? Ecol Lett 3: 267–274. [Google Scholar]
- 11. Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 88: 541–549. [DOI] [PubMed] [Google Scholar]
- 12. Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gunatilaka AA (2006) Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J Nat Prod 69: 509–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gloer JB (1995) Chemistry of fungal antagonism and defense. Can J Bot 73(S1): 1265–1274. [Google Scholar]
- 15. Gilmore DP, Da Costa CP, Duarte DPF (2001) Sloth biology: an update on their physiological ecology, behavior and role as vectors of arthropods and arboviruses. Braz J Biol Res 34: 9–25. [DOI] [PubMed] [Google Scholar]
- 16. Suutari M, Majaneva M, Fewer DP, Voirin B, Aiello A, et al. (2010) Molecular evidence for a diverse green algal community growing in the hair of sloths and a specific association with Trichophilus welckeri (Chlorophyta, Ulvophyceae). BMC Evol Biol 10: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goyette JL, Howe RW, Wolf AT, Robinson WD (2011) Detecting tropical nocturnal birds using automated audio recordings. J Field Ornithol 82: 279–287. [Google Scholar]
- 18. Hoffman M, Arnold AE (2010) Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76: 4063–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ewing B, Green P (1998) Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 20. Ewing B, Hiller L, Wendl M, Green P (1998) Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 21.Maddison WP, Maddison DR (2011) Mesquite: A modular system for evolutionary analysis. Available: http://mesquiteproject.org. Accessed 2013 Apr 30.
- 22. Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic Local Alignment Search Tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 23. U'Ren JM, Dalling JW, Gallery RE, Maddison DR, Davis EC, et al. (2009) Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: a case study for analysing fungal environmental samples. Mycol Res 113(PT4): 432–449. [DOI] [PubMed] [Google Scholar]
- 24. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guindon S, Gascuel O (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 26. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristic and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin.
- 28.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. pp. 1–8. [Online] Available: http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=5676129. Accessed 2013 Apr 04.
- 29. Higginbotham S, Arnold AE, Ibañez A, Spadafora C, Coley PD, et al. (2013) Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants: evidence from a Panamanian drug discovery project. PLoS ONE 8(9): e73192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong WR, Oliver AG, Linington RG (2012) Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem Biol 19: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pérez-García A, Mingorance E, Rivera ME, Pino D, Romero D, et al. (2006) Long-term Preservation of Podosphaera fusca Using Silica Gel. J Phytopathology 154: 190–192. [Google Scholar]
- 32. Arabi MIE, Jawhar M, Al-Daoude A (2007) Viability of Cochliobolus sativus cultures after storage under different conditions. Journal of Plant Pathology 89: 79–83. [Google Scholar]
- 33. You J, Dai H, Chen Z (2010) Trichoderone, a novel cytotocix cyclopentenone and cholesta-7, 22-diene-3 beta, 5 alpha, 6 beta-triol, with new activities from the marine-derived fungus Trichoderma sp. J Ind Microbiol Biotechnol 37: 245–252. [DOI] [PubMed] [Google Scholar]
- 34. Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Carcache de Blanco EJ, et al. (2012) Peptaibols from two unidentified fungi of the order Hypocreales with cytotoxic, antibiotic, and anthelmintic activities. J Pept Sci 18: 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Waldrop MP, Zak DR, Blackwood CB, Curtis CD, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9: 1127–1135. [DOI] [PubMed] [Google Scholar]
- 36. Figueroa M, Graf TN, Ayers S, Adcock AF, Kroll DJ, et al. (2012) Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J Antibiot 65: 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frisvad JC, Houbraken J, Popma S, Samson RA (2013) Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum producing the anticancer compound asperphenamate. FEMS Microbiol Lett 339: 77–92. [DOI] [PubMed] [Google Scholar]
- 38. Iwatsuki M, Takada S, Mori M, Ishiyama A, Namatame M, et al. (2011) In vitro and in vivo antimalarial activity of puberulic acid and its new analogs, viticolins A–C, produced by Penicillium sp. FKI-4410. J Antibiot 64: 183–188. [DOI] [PubMed] [Google Scholar]
- 39. Lee HB, Kim Y, Jin HZ, Lee JJ, Kim CJ, et al. (2005) A new Hypocrea strain producing harzianum A cytotoxic to tumour cell lines. Lett Appl Microbiol 40: 497–503. [DOI] [PubMed] [Google Scholar]
- 40. Liu L, Li Y, Liu S, Zheng Z, Chen X, et al. (2009) Chloropestolide A, an antitumor metabolite with an unprecedented spiroketal skeleton from Pestalotiopsis fici . Org Letters 11: 2836–2839. [DOI] [PubMed] [Google Scholar]
- 41. Mohamed HF (2012) Molecular analysis and anticancer properties of two identified isolates, Fusarium solani and Emericella nidulans isolated from Wady El-Natron soil in Egypt against Caco-2 (ATCC) cell line. Asian Pac J Trop Biomed 2: 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh SB, Zink DL, Polishook JD, Dombrowski AW, Darkin-Rattray SJ, et al. (1996) Apicidins: Novel cyclic tetrapeptides as coccidiostats and antimalarial agents from Fusarium pallidoroseum . Tetrahedron Lett 37: 8077–8080. [Google Scholar]
- 43. Singh MP, Janso JE, Brady SF (2007) Cytoskyrins and cytosporones produced by Cytospora sp. CR200: taxonomy, fermentation and biological activities. Mar Drugs 5: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castro JA, De Mecca MM, Bartel LC (2006) Toxic side effects of drugs used to treat Chagas disease (American trypanosomiasis). HumExpToxicol 25: 471–479. [DOI] [PubMed] [Google Scholar]
- 45. Peleg AY, Hooper DC (2010) Hospital-acquired infections due to Gram-negative bacteria. N. Engl. J. Med 362(19): 1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kunz AN, Brook I (2010) Emerging resistant Gram-negative aerobic bacilli in hospital-acquired infections. Chemotherapy 56: 492–500. [DOI] [PubMed] [Google Scholar]
- 47.ECDC/EMEA Join Technical Report (2009) The bacterial challenge: time to react [Online]. Available: http://www.ecdc.europa.eu/en/publications/Publications/0909_TER_The_Bacterial_Challenge_Time_to_React.pdf. Accessed 2013 Apr 04.
- 48. Ramsamy Y, Muckart DJJ, Han KSS (2013) Microbiological surveillance and antimicrobial stewardship minimise the need for ultrabroad-spectrum combination therapy for treatment of nosocomial infections in a trauma intensive care unit: An audit of an evidence-based empiric antimicrobial policy. S Afr Med J 103: 371–376. [DOI] [PubMed] [Google Scholar]
- 49. Wang P, Liu Y, Yin Y, Jin H, Wang S, et al. (2011) Diversity of microorganisms isolated from soil sample surround Chroogomphus rutilus in the Beijing region. Int J Biol Sci 7: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Araújo Xavier GA, Galiza da Silva LB, Rubem da Silva D, de Moraes Peixoto R, Camara Lino G, et al. (2008) Dermatophytosis caused by Microsporum canis and Microsporum gypseum in free-living Bradypus variegatus (Schiz, 1825) in the state of Pernambuco, Brazil. Braz J Microbiol 39: 508–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. U'Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE (2012) Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot 99: 898–914. [DOI] [PubMed] [Google Scholar]
- 52. Arnold AE, Miadlikowska KL, Higgins L, Sarvate SD, Gugger A, et al. (2009) A phylogenetic estimation of trophic transition networks for ascomycetous fungi: Are lichens cradles of symbiotrophic fungal diversification? Syst Biol 58: 283–297. [DOI] [PubMed] [Google Scholar]
- 53. Elewski BE (2005) Clinical diagnosis of common scalp disorders. J Investig Dermatol Symp Proc 10: 190–193. [DOI] [PubMed] [Google Scholar]
- 54. Huffnagle GB, Noverr MC (2013) The emerging world of the fungal microbiome. Trends Microbiol 21: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith SA, Tank DC, Boulanger LA, Bascom-Slack CA, Eisenman K, et al. (2008) Bioactive endophytes warrant intensified exploration and conservation. PLoS ONE 3(8): e3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arnold AE, Herre A (2003) Canopy cover and leaf age affect colonization by tropical fungal endophytes: Ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 68: 388–398. [PubMed] [Google Scholar]
- 57. Higgins KL, Coley PD, Kursar TA, Arnold AE (2011) Culturing and direct PCR suggest prevalent host-generalism among fungal endophytes of tropical grasses. Mycologia 103: 247–260. [DOI] [PubMed] [Google Scholar]
- 58. Rodrigues A, Mueller UG, Ishak HD, Bacci M Jr, Pagnocca FC (2011) Ecology of microfungal communities in gardens of fungus-growing ants (Hymenoptera: Formicidae): a year-long survey of three species of attine ants in Central Texas. FEMS Microbiol Ecol 78: 244–255. [DOI] [PubMed] [Google Scholar]
- 59. Kluger CG, Dalling JW, Gallery RE, Sanchez E, Weeks-Galindo C, et al. (2008) Host-generalists dominate fungal communities associated with seeds of four neotropical pioneer species. J Trop Ecol 24: 351–354. [Google Scholar]
- 60. O'Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, et al. (2009) Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum, F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol 47: 3851–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hoyos-Carvajal L, Orduz S, Bissett J (2009) Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol 46: 615–631. [DOI] [PubMed] [Google Scholar]
- 62. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bukovská P, Jelínková M, Hrselová H, Sykorová Z, Gryndler M (2010) Terminal restriction fragment length measurement errors are affected mainly by fragment length, G+C nucleotide content and secondary structure melting point. J Microbiol Methods 82(3): 223–228. [DOI] [PubMed] [Google Scholar]
- 64. Glenn A, Bodri MS (2012) Fungal endophyte diversity in Sarracenia . PLoS ONE 7(3): e32980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McGuire KL, Zak DR, Edwards IP, Blackwood CB, Upchurch R (2010) Slowed decomposition is biotically mediated in an ectomycorrhizal, tropical rain forest. Oecologia 164: 785–795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sloth hair fungus ID and Genbank accession numbers.
(DOCX)