Abstract
Chromosomal translocations in hematologic and mesenchymal tumors form overwhelmingly by nonhomologous end-joining (NHEJ). Canonical NHEJ, essential for the repair of radiation-induced and some programmed double-strand breaks (DSBs), requires the Xrcc4/ligase IV complex. For other DSBs, the requirement for Xrcc4/ligase IV is less stringent, suggesting the existence of alternative end-joining (alt-NHEJ) pathways. To understand the contribution of the canonical and alt-NHEJ pathways, we examined translocation formation in Xrcc4/ligase IV-deficient cells. We find that Xrcc4/ligase IV is not required for, but rather suppresses, translocations. Translocation breakpoint junctions have similar characteristics in wild-type and Xrcc4/ligase IV-deficient cells, including an unchanged bias toward microhomology, unlike what is observed for intrachromosomal DSB repair. Complex insertions in some junctions demonstrate that joining can be iterative, encompassing successive processing steps prior to joining. Our results imply that alt-NHEJ is the primary mediator of translocation formation in mammalian cells.
Keywords: chromosomal translocation, Xrcc4/ligase IV, nonhomologous end-joining, DNA double-strand break, alternative NHEJ
Introduction
NHEJ is a major pathway for the repair of DSBs in mammalian cells1. NHEJ is loosely defined as the joining of DNA ends without the use of extensive homology, and it is important for the correct repair of DSBs generated by ionizing radiation, as well as programmed DSBs generated in developing lymphocytes during V(D)J recombination and class switch recombination (CSR)2–4. Paradoxically, some type of NHEJ also appears to be responsible for generating reciprocal translocations found in human hematologic and mesenchymal tumors, since breakpoint junctions rarely occur within homologous sequences5–7. Rather, breakpoint junctions occur at sites of no homology or just a few bp of homology, termed microhomology. Translocation breakpoint junctions have other features of NHEJ in that they contain DNA end modifications, most often deletions, but also insertions.
A set of NHEJ factors has been identified that is required for the repair of DSBs generated by ionizing radiation and during V(D)J recombination. These canonical NHEJ factors include the DNA ligase complex, Xrcc4/ligase IV, used to seal DNA ends, and the Ku70/80 (Ku) heterodimer, which binds DNA ends and recruits a number of processing factors, including nucleases, polymerases, and the ligase complex itself 8,9. Xrcc4/ligase IV may also participate in DNA end processing through its interaction with a variety of factors10–12. The range of activities of the recruited factors allows the NHEJ pathway to process DSBs with a variety of end structures so that they can be ligated by Xrcc4/ligase IV8,9. End processing is complex, such that a variety of junctional sequences are possible at a single lesion. Biochemical studies support this complexity, and have even shown that the two strands of a DNA end can be independently processed and ligated in an iterative manner8,13. These NHEJ components also suppress the other major DSB repair pathway in mammalian cells, homologous recombination (HR), presumably by protecting DNA ends from the initial resection step necessary for HR14,15.
While critical for the repair of radiation-induced and V(D)J recombination DSBs, the canonical NHEJ factors are not absolutely required for other types of DSB repair. For example, endonuclease-generated DSBs in plasmids and chromosomes can be repaired at reduced levels in Ku and Xrcc4-deficient cells16–19. In the immune system, Xrcc4/ligase IV promotes the efficient repair of DSBs generated during CSR, but is not essential for this process20. These studies point to the existence of other pathways for the nonhomologous repair of DSBs. The poorly defined pathway(s) operating in the absence of the canonical NHEJ pathway has been termed alternative NHEJ, or alt-NHEJ.
Linked to their roles in the immune system, deficiency of several of the canonical NHEJ components in mice on a p53 background leads to pro-B cell lymphomas with Igh/Myc amplification and translocations at nonhomologous sequences21,22. Translocations involving nonhomologous sequences at V(D)J and CSR programmed DSBs have also recently been reported in cells deficient in Xrcc423. In a more generalized system, a translocation reporter was used to demonstrate that translocations are not reduced in Ku70-deficient mouse embryonic stem (ES) cells24, suggesting a role of alt-NHEJ in non-immune system translocations. However, absence of Xrcc4/ligase IV has a more severe phenotype than loss of Ku in several contexts, including telomere fusions25, mouse development26, and the joining of two nearby chromosome DSBs27, suggesting that loss of the terminal activity of NHEJ has a more profound phenotype. Moreover, Ku is implicated in other cellular processes besides NHEJ, such as apoptosis28,29, that may affect the recovery of translocations.
Given that Xrcc4/ligase IV does not have any other known cellular role outside of NHEJ and ligation of DNA ends is essential to NHEJ, we have now examined the role of this canonical NHEJ component in translocation formation outside of the immune system. We find that Xrcc4/ligase IV is not required for NHEJ leading to translocations, but rather suppresses such events. Moreover, translocation breakpoint junctions have similar characteristics in wild-type and Xrcc4/ligase IV-deficient mouse cells, including a similar bias to microhomology use, as well as iterative processing of the ends to generate complex insertions. Our results imply that non-canonical NHEJ (alt-NHEJ) is the primary mediator of translocation formation in mammalian cells.
Results
Chromosomal translocations do not require Xrcc4
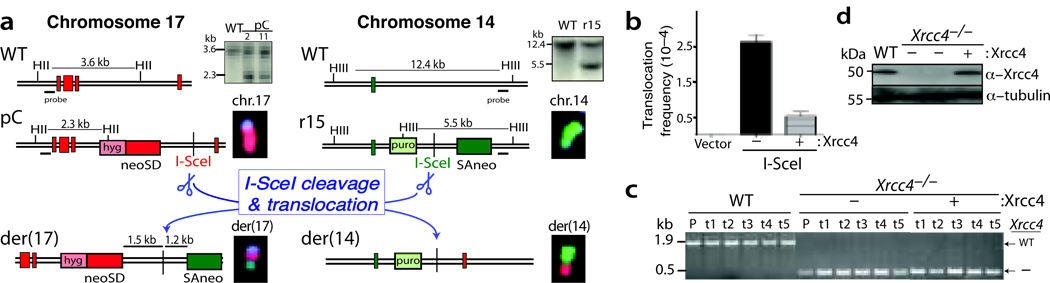
Ligase IV requires Xrcc4 for its stability, such that Xrcc4-deficient cells are effectively ligase IV-deficient20,30. Xrcc4−/− mouse cells were previously constructed by gene targeting mouse ES cells26. The Xrcc4−/− ES cells, like other NHEJ-deficient cells, are sensitive to ionizing radiation, defective in V(D)J recombination, and have elevated homology-directed DNA repair14,24,26. Because a neomycin phosphotransferase (neo) gene, which would interfere with translocation selection, was still present at the targeted Xrcc4 loci in these cells, we first expressed Cre recombinase to delete the neo gene (Supplementary Fig. 1a). Two rounds of gene targeting were then used to introduce our pCr15 translocation reporter into the neo–Xrcc4−/− cells. This reporter consists of a neo gene split into two exons, one of which is targeted to a locus on chromosome (chr.) 17 and the other to a locus on chr. 14 (Fig.1a)6. The intron segment on each chromosome is demarcated by an I-SceI site, such that cleavage of both chromosomes by I-SceI endonuclease, followed by joining of DNA ends from chrs.17 and 14 can generate a derivative chromosome 17, der(17), with a functional neo gene. Formation of a neo+ gene requires NHEJ, as the neo intron segments on chrs. 17 and 14 do not share any appreciable homology.
Figure 1.
Chromosomal translocations are suppressed by XRCC4/ligase IV. (a) Translocation reporter in Xrcc4−/− pCr15 cells. DSB formation on chromosomes 17 and 14 at the I-SceI sites, followed by interchromosomal NHEJ, results in a chromosomal translocation with a neo+ gene on der(17). FISH analysis indicates that parental pCr15 cells have normal chromosomes 17 (red) and 14 (green), whereas neo+ clones have derivative chromosomes. Vertical red bars are exons 1–5 from the targeted Pim1 locus on chr.17, and the vertical green bar is exon 20 from the Rb locus. Probes are located outside the targeting arms. HII, HincII; HIII, HindIII. (b) Translocation frequency is significantly increased in Xrcc4−/− cells, but is suppressed by Xrcc4 expression. (c) Confirmation of the genotype of the endogenous Xrcc4 alleles in wild-type (WT), Xrcc4−/− and Xrcc4-complemented cells. P, parental pCr15 cells of the indicated genotypes; t, neo+ translocation clones. (d) Western blot analysis demonstrating that transient expression of Xrcc4 restores wild-type Xrcc4 protein levels to Xrcc4−/− cells.
To determine the role of Xrcc4/ligase IV in translocation formation, we expressed I-SceI in the Xrcc4−/− pCr15 cells and selected neo+ colonies. Surprisingly, neo+ colonies were readily obtained at a frequency of 2.7 × 10−4 (Fig. 1b, Table 1). We confirmed that the neo+ colonies arose as a result of chromosomal translocation by using fluorescence in situ hybridization (FISH; Fig. 1a, Supplementary Fig. 1c, and data not shown). All 12 translocations analyzed by FISH were reciprocal, as both the expected der(17) and der(14) chromosomes were detected. The Xrcc4 genotype of these cells was verified (Fig. 1c), as was the absence of Xrcc4 protein (Fig. 1d). Transient Xrcc4 expression in the Xrcc4−/− cells restored wild-type levels of protein (Fig. 1d) and led to a 5-fold reduction in translocation frequency (Fig. 1c), indicating that the NHEJ complex Xrcc4/ligase IV is not required for NHEJ-mediated translocations but instead suppresses their occurrence.
Table 1.
Translocation frequencies are from 4 experiments. One standard deviation from the mean is indicated. A two–tailed unpaired t test was applied, with p values derived from a comparison with Xrcc4−/− cells (a) and wild-type cells (b).
| Cells | Expression vector |
Translocation frequency (× 10−4) |
p value | |
|---|---|---|---|---|
| a | b | |||
| Wild-type | – | <0.001 | ||
| I-SceI | 0.57 ± 0.11 | <0.001 | – | |
| I-SceI+Xrcc4 | 0.62 ± 0.06 | <0.001 | 0.455 | |
| I-SceI+Ku70 | 0.61 ± 0.10 | <0.001 | 0.610 | |
| Xrcc4–/– | – | <0.001 | ||
| I-SceI | 2.70 ± 0.20 | – | <0.001 | |
| I-SceI+Xrcc4 | 0.53 ± 0.12 | <0.001 | 0.641 | |
| I-SceI+Ku70 | 2.73 ± 0.23 | 0.850 | <0.001 | |
| Ku70–/– | – | <0.001 | ||
| I-SceI | 2.33 ± 0.21 | 0.043 | <0.001 | |
| I-SceI+Xrcc4 | 2.14 ± 0.19 | 0.007 | <0.001 | |
| I-SceI+Ku70 | 0.71 ± 0.02 | <0.001 | 0.046 | |
Alternative NHEJ is implicated in translocation formation
Using a similar approach, we previously demonstrated that like Xrcc4, Ku70 is also not required for translocation formation24. We compared Xrcc4/ligase IV and Ku for their role in translocation formation by directly comparing wild-type, Xrcc4−/−, and Ku70−/− pCr15 ES cells (Table 1). A consistent 5-fold higher translocation frequency was observed in the absence of Xrcc4 (p<0.0001) and a 3-fold higher translocation frequency in the absence of Ku70 (p<0.0001). The somewhat higher suppression by Xrcc4 than by Ku70 carries only a moderate level of statistical significance (p=0.043). These experiments establish that, unlike in yeast31, the canonical NHEJ factors Xrcc4/ligase IV and Ku are not essential for chromosomal translocations in mammalian cells; rather, both canonical NHEJ proteins restrain translocation formation. Our results, therefore, implicate alt-NHEJ in translocation formation.
Translocation junctions are similar in wild type and NHEJ mutants
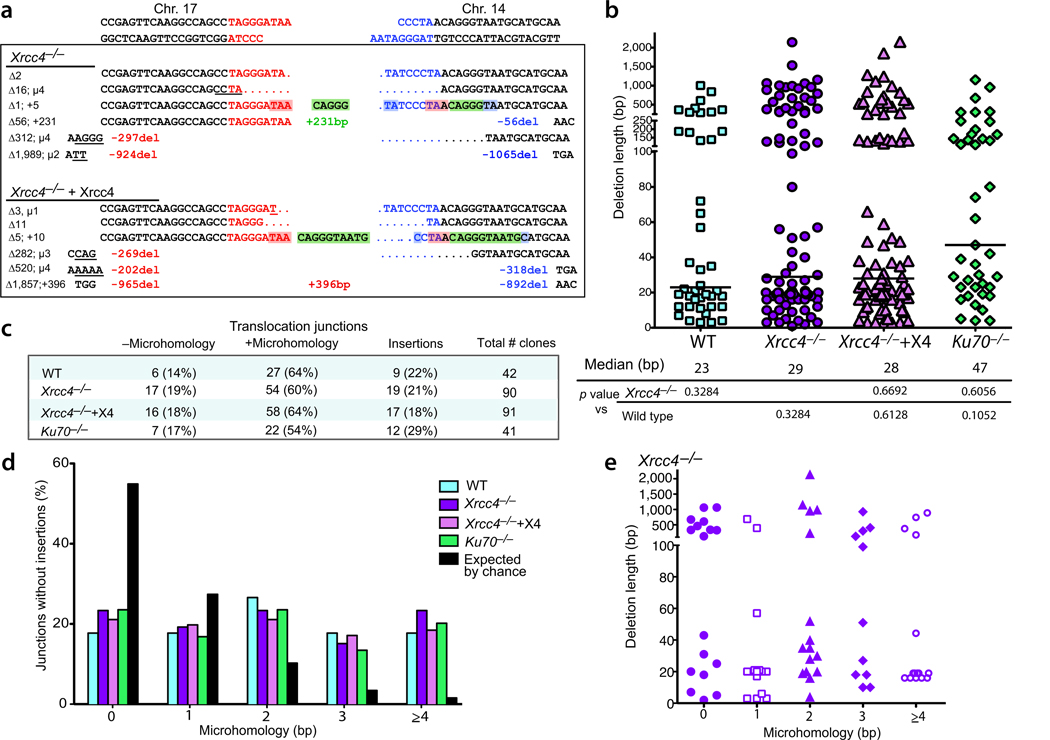
Analysis of NHEJ junctions from Xrcc4-deficient cells has previously indicated that these cells join DNA ends very differently from wild-type cells20,32,33. To gain insight into the repair mechanisms leading to translocations, der(17) breakpoint junctions were analyzed from 92 Xrcc4−/−, 92 Xrcc4-complemented Xrcc4−/−, and 43 wild-type neo+ clones. For comparison, der(17) breakpoint junctions were analyzed from 42 Ku70−/− neo+ clones. Breakpoint junctions were amplified using a series of primer pairs that span the neo intron formed upon translocation (Fig. 1d). With this approach, a total of 264 of 269 breakpoint junctions could be amplified for sequencing (Supplementary Fig. 2).
The two chromosome ends that join during translocation formation have 4-base 3′ overhangs with limited complementarity, requiring processing of the ends prior to joining. A variety of DNA end modifications were observed in all genotypes (Fig. 2a and Supplementary Fig. 2). As with patient-derived translocations6, deletions were the most commonly observed modification. Deletions were from one or both ends and were often short, encompassing only bases from the overhangs or the I-SceI recognition site, although longer deletions were also observed. The longest deletion was 2.1 kb and was split approximately equally between the two chromosome ends (1.0 and 1.1 kb). Longer deletions may have arisen but not survived the neo selection, although these would be expected to be a minority of events (~5%)6. One of the shortest deletions was missing only the terminal base of each 3′ overhang (Xrcc4−/−, Δ2; Fig. 2a); the remaining 3 bases of the 3′ overhangs were apparently filled in by a polymerase prior to joining. The lack of microhomology in this junction suggests the involvement of polymerase (pol) µ in the fill-in reaction, since this polymerase has activity even when the terminal base of the primer lacks complementarity to the template strand34,35.
Figure 2.
Translocation breakpoint junctions have similar characteristics in wild-type, Xrcc4−/− mutant and complemented cells, and Ku70−/− cells. (a) Representative der(17) translocation junction sequences obtained from Xrcc4−/− and Xrcc4-complemented cells. DNA ends generated by I-SceI on chrs.17 and 14 are indicated in red and blue, respectively. A summary of the various end modifications is presented to the left of each junction in bp: Δ, total deletion; µ, microhomology; +, insertion. Sequences are annotated as follows: del, deletion length from the DNA end; underline, microhomology; +, length of long insertion. The middle green sequences are short insertions from chr. 14; considering a template model for their insertion, the sequences in red shading (TAA) would be microhomology between the DNA ends that could anneal to act as a primer and the blue shading would represent microhomology for annealing after DNA synthesis between the 2 DNA ends (see text). (b) Deletion lengths for der(17) breakpoint junctions. Each value represents the combined deletion from both ends of an individual junction. The median deletion length for each genotype is indicated by a bar on the graph and the value is give below the graph. Deletion lengths do not differ significantly from each other (two-tailed Mann-Whitney test). +X4, transient complementation with Xrcc4. (c) Microhomology and insertion frequencies are similar for the four genotypes. (d) Distribution of microhomology lengths for der(17) breakpoint junctions. Only junctions with simple deletions (i.e., without an insertion) are included. (e) Lack of correlation between deletion length and microhomology use. Only junctions for Xrcc4−/− cells are plotted.
Overall, the median deletion length for wild-type, Xrcc4-complemented Xrcc4−/−, and Xrcc4−/− junctions ranged from 23 to 29 bp and was not significantly different for the three genotypes (Fig. 2b). For Ku70−/−, the median deletion length was longer (47 bp), although it also was not statistically different. The majority of deletions were <100 bp, but larger deletions of >250 bp were found in a portion of junctions for all 4 genotypes (Fig. 2b and Supplementary Fig. 3a). It is unclear what accounts for the range of deletions from the DNA ends prior to joining; nonetheless, the similarities found in the 4 genotypes indicate that loss of the canonical NHEJ components Xrcc4/ligase IV and Ku does not substantially alter the amount of nucleolytic processing prior to joining chromosome ends for translocation formation.
It has previously been reported that junctions formed during CSR in Xrcc4−/− cells are markedly biased toward those with microhomology compared with wild-type cells20. We determined the microhomology present at translocation junctions and found that the percent of total junctions containing microhomology was similar in Xrcc4−/− cells (60%) and wild-type and Xrcc4-complemented Xrcc4−/− cells (64%), although it was slightly lower in the Ku70−/− cells (54%) (Fig. 2c). Considering only the junctions without insertions, the percent of junctions containing microhomology was similar for all genotypes (wild-type, 82%; Xrcc4−/−, 76%; Xrcc4-complemented Xrcc4−/−, 78%; Ku70−/−, 76%). In all cases, the presence of microhomology was significantly greater than expected by chance, as only 44% of junctions would be expected to have microhomology if joining was random with respect to bp overlap (Fig. 2d). These results are consistent with a role for microhomology in NHEJ, but the Xrcc4−/− cells do not demonstrate a greater dependence on microhomology.
Consistent with the percent microhomology use, the distribution of microhomology lengths was similar between Xrcc4−/− and the other genotypes (Fig. 2d). Microhomology lengths were generally short, from 1 to 4 bp. The fraction of junctions with 1 bp of microhomology (16–21%) was lower than that expected by chance (28%); however, the fraction of junctions with 2 or more bp of microhomology was much greater than that expected by chance (Fig. 2d). For example, 22 to 27% of junctions were observed to have 2 bp of microhomology, whereas 10.5% of junctions would be expected by chance, a 2 to 2.6-fold higher level. The fold difference is even greater with 3 and ≥4 bp microhomology (~5 and 10-fold, respectively). Microhomologies of 5 to 7 bp were observed in a few junctions (6/207 junctions; 3%), although not in Xrcc4−/− junctions (Supplementary Fig. 2). No correlation was observed between deletion length and microhomology length for any of the genotypes (Fig. 2e and Supplementary Fig. 3b), indicating that joining of distant ends is not driven by microhomology to any greater extent than ends closer to the DSB.
Although we observed no difference in either the percent microhomology use or the microhomology length at the translocation breakpoint junctions in the various cell lines, previous analyses of NHEJ in several systems have demonstrated increased microhomology use in the absence of Xrcc4/ligase IV32,33. The bias previously observed may be sequence specific, for example, for repetitive switch regions20 and plasmids substrates with short repeats at or near the DNA ends32,33. Alternatively, the context of the DSB (intrachromosomal vs. interchromosomal; single DSB vs. two DSBs) may affect NHEJ pathway use. To determine if a single DSB is repaired with different microhomology characteristics, we analyzed intrachromosomal junctions after I-SceI expression in complemented and uncomplemented Xrcc4−/− cells. To maintain a similar sequence context for the joining events to those for translocations, we utilized a translocation clone derived from Xrcc4−/− cells that had by chance reconstituted an I-SceI site on der(17) (Supplementary Fig. 4a). I-SceI was expressed in these cells with or without Xrcc4 coexpression, and 72 h later pooled genomic DNA was analyzed by PCR and sequencing for imprecise NHEJ (I-SceI site loss; Supplementary Fig. 4a). We found that microhomology use differed in junctions from complemented and uncomplemented Xrcc4−/− cells (Supplementary Fig. 4b). Microhomology use in complemented Xrcc4−/− cells was not significantly different from that expected by chance (p=0.2667, one-tailed Mann-Whitney test), whereas more long microhomologies were observed in Xrcc4−/− cells, such that the distribution of microhomology use differed significantly from that expected by chance (p<0.0001). Thus, NHEJ of a single DSB is affected by Xrcc4 loss, unlike NHEJ of two DSBs during translocation. Still, 32% of junctions from Xrcc4−/− cells did not contain microhomology.
Breakpoint junction insertions can be complex
Although less frequent than deletions, insertions were also observed in a portion of the breakpoint junctions from both wild-type and mutant cells (18–29%; Fig. 2c). Approximately half of the insertions in all 4 genotypes were short (≤10 bp), while the remainder were longer, extending a few hundred bp in some cases (Fig. 2a and Supplementary Figs. 2, 3c, and 5). Some of the insertions were only 1 or a few bp, and may have been added by polymerases in a template-independent manner or by template slippage34,35. Other somewhat longer insertions appear to have been templated, for example, the +5 (CAGGG) and +10 (CAGGGTAATG) insertions derived from Xrcc4−/− mutant and complemented cells, respectively (Fig. 2a). In both cases, the terminal 3 nucleotides of the chr.17 end could have primed DNA synthesis starting 11 nucleotides into the chr. 14 end (red shading, Fig. 2a); copying of at least 7 and 11 bp from chr. 14, respectively, would have provided small microhomologies for subsequent annealing to the chr. 14 end. In such a scenario, the primer for extension and the template sequences are derived from the two der(17) chromosome ends, indicating that DNA ends in these cases would have been synapsed prior to DNA synthesis.
The longest insertions for the Xrcc4−/− mutant and Xrcc4-complemented der(17) junctions, which were 231 and 396 bp, respectively, point to the complexity of breakpoint junctions with longer insertions in terms of both the derivation of the insert and the processing of each chromosome end (Fig. 2a). The 396-bp insertion was derived from sequences starting 7.7 kb upstream of the DSB on chr. 17 and was inserted after a 965 bp deletion from the chr. 17 end and a 892 bp deletion from the chr. 14 end (Supplementary Fig. 5c). The 231 bp insertion was derived from sequences starting 21 bp upstream from the DSB on chr. 14, and, hence, from a chromosome end forming the other derivative chromosome, der(14); it was inserted with no deletion from the chr. 17 end but with a 56 bp deletion from the chr. 14 end (Supplementary Fig. 5b). Because this insertion is derived from sequences forming der(14), it must have been inserted prior to synapsis of the der(17) chromosome ends. The inserted segment from chr. 14 shows an additional complexity in that it is not contiguous; it is interrupted by 2 bp (“GA”; position 161).
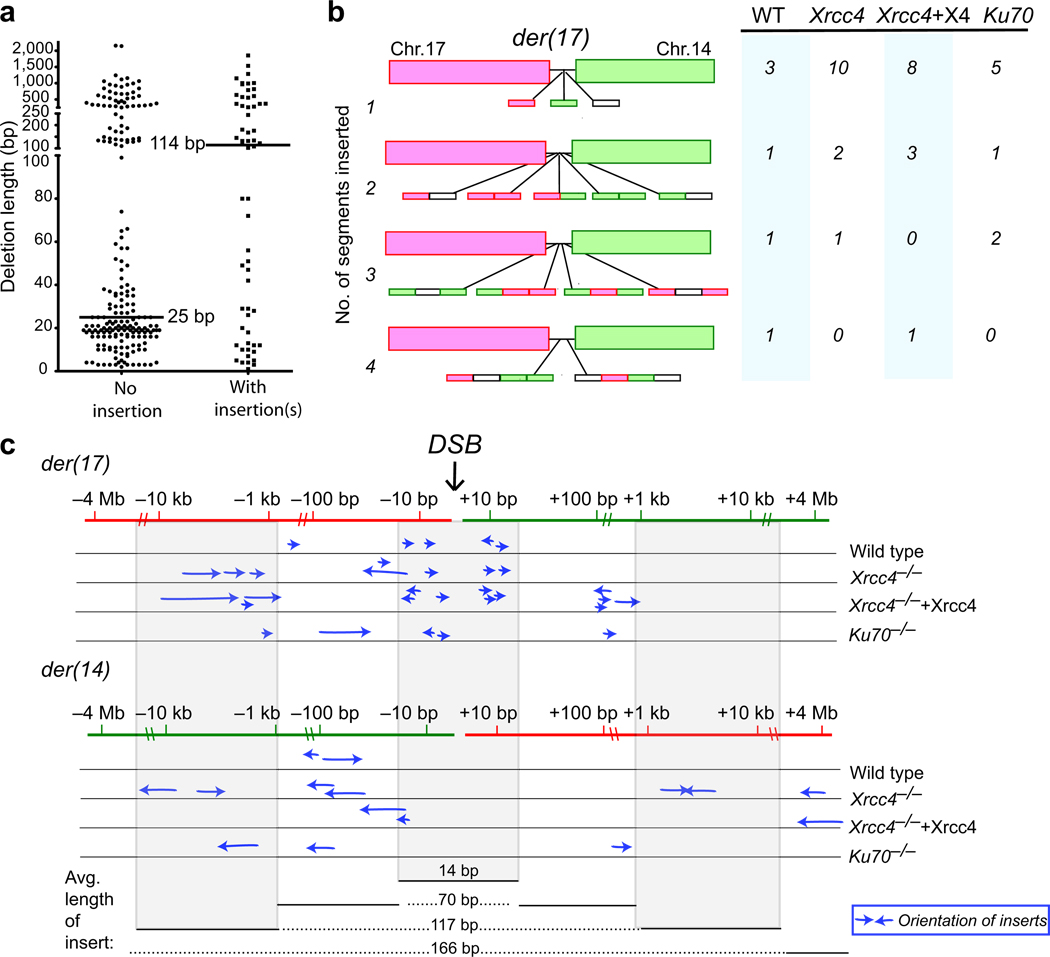
The complexity of the insertions led us to examine several parameters. We found that breakpoint junctions with insertions had a greater median deletion length than junctions without insertions (114 vs 25 bp, p=0.0127, two-tailed Mann-Whitney test; Fig. 3a), indicating that insertions were associated with more extensive processing of DNA ends. All 4 genotypes showed a similar trend (Supplementary Fig. 3d). Although most insertions had a unique origin, others were chimeric, being comprised of 2, 3, or even 4 inserts derived from different sources (Fig. 3b). This suggests some type of iterative processing of ends until joining is successful.
Figure 3.
Insertions at breakpoint junctions display complex characteristics. (a) Distribution of deletion lengths for der(17) breakpoint junctions without and with insertions. Data are combined from all four genotypes; the deletion distributions for individual genotypes are shown in Supplementary Fig. 3d. (b) Der(17) insertions are comprised of one or more distinct segments of DNA. Most insertions at the der(17) breakpoint junctions are derived from one DNA segment, although others are comprised of up to 4 segments. Most inserted segments are derived from chr. 17 (red boxes) or chr. 14 (green boxes), although some of the inserted segments are derived from other sources or are too short to be mapped precisely (white boxes). Included in this analysis are all inserted sequences >4 bp. (c) Derivation of der(17) inserts. Each arrow represents the derivation of a segment inserted at a der(17) translocation breakpoint junction and the relative orientation of the insert relative to the centromere. Inserted segments are derived from sequences adjacent to a DSB or as far away as 4 Mb; those from nearby a DSB are generally short (≤20 bp), while more distantly-derived segments tend to be larger (up to 396 bp). Note that the inserted segments can be derived from sequences used to form either der(17) or der(14). The der(17) sequences from −1.5 kb to +1.2 kb comprise the neo intron (Fig. 1a).
Most inserts were derived from the 2 chromosomes participating in the translocation (chr. 17, 24 inserts; chr. 14, 22 inserts), while the remaining 14 inserts were derived from other sources, including a segment from chr. 13, a repetitive element (LINE1), and the transfected I-SceI expression vector (Supplementary Fig. 5). Microhomology was noted at one or both ends of several of the insertions, although some insertions did not have overt microhomology. Seven inserts had single bp mutations, including point mutations, deletions, and additions, indicating the involvement of an error prone polymerase. Some very short inserts of 1 to 3 bp were also observed with the longer inserts, like the “GA” described above. These may have been derived from an error prone polymerase; the alternative is template independent polymerization. The origin of the remaining 10 inserts, which ranged from 5 to 24 bp, could not be unequivocally determined.
We mapped the inserts in the der(17) breakpoint junctions on chrs. 17 and 14 (Fig. 3c). Notably, inserts were derived from all 4 chromosome ends, indicating the proximity of the ends during insertion formation. Inserts from the ~20 bp adjacent to the der(17) ends were overrepresented (15/46, 33%), making it likely that these were inserted during the der(17) joining process itself. More distant inserts did not show this bias, as der(17) and der(14) sequences were similarly represented (16 vs. 14, respectively); thus, many of these inserts were derived at an earlier step in the joining process. A distance effect was also noted in that inserts derived further from the DSB were progressively longer on average (Fig. 3c). If the inserts are derived from a copying mechanism, polymerase processivity may be affected by proximity to a DNA end. Interestingly, the most distant inserts, from 4.7 Mb away on distal chr. 17, were derived from within ~1 kb from each other from the same gene, Psmb9. Psmb9 is a known hotspot for meiotic recombination in some strains of mice36,37, although to date this gene has not been known to participate in somatic recombination or rearrangement events.
Discussion
In this report, we examined chromosomal translocations in the absence of the NHEJ ligase. In contrast to NHEJ in other systems16,17,20,27, translocations via NHEJ occur at a significantly higher frequency in the absence of Xrcc4/ligase IV. Moreover, microhomology at translocation breakpoint junctions is unaltered, also unlike other systems20,32,33 or our results with intrachromosomal joining (Supplementary Fig. 4). These results imply a suppressive role for the canonical NHEJ pathway in preventing translocations and support a central role for alt-NHEJ in translocation formation.
The question arises as to whether alt-NHEJ is a distinct pathway or a variation of the canonical pathway. The similar results we obtained in Xrcc4/ligase IV and Ku-deficient cells, both in terms of the translocation frequency and breakpoint junction characteristics, support a role for an alternative pathway of NHEJ as the major pathway in translocation formation, rather than a variation or back-up of the canonical pathway, as it seems to be in other systems. That wild-type cells have similar junction characteristics as the two mutants further supports this view. We cannot rule out, however, that other factors, for example ligase I or ligase III38–40 could slip into the canonical pathway without perturbing its outcome.
Translocation breakpoint junctions in wild-type and NHEJ-deficient cells, as well as junctions at a single DSB in the absence of Xrcc4, show biased microhomology use: an overrepresentation of 2 or more bp of microhomology and an underrepresentation of 0 or 1 bp. Microhomology would promote annealing of two DNA ends for subsequent processing, including polymerization, and ligation. For junctions with 0 or 1 bp microhomology, another mechanism may be used to stabilize ends prior to joining. Alternatively, a polymerase like pol µ may generate chance microhomology by adding nucleotides to DNA in a template-independent manner35; this polymerase-generated microhomology would then act to promote annealing between chromosome ends, although it would not be observed in breakpoint junctions as microhomology. Interestingly, no correlation was observed between deletion length and microhomology use for any of the cell lines examined, implying that extensive nucleolytic digestion of DNA ends is not coupled to a search for microhomology.
Xrcc4/ligase IV and Ku are known to suppress two other DSB repair pathways, HR and single-strand annealing14,15,41. Both of these pathways are initiated by DSB resection to produce 3′ single-stranded DNA tails. Since both Xrcc4/ligase IV and Ku are known to protect DNA ends from nucleolytic processing42,43, it is tempting to speculate that canonical NHEJ components perform a similar role in suppressing translocations, by blocking access of DNA ends to alt-NHEJ components involved in end processing. A possibly related or additional role for the canonical NHEJ components is maintaining DNA ends from a DSB in close physical proximity through their end-binding activity to promote correct joining. Such a role has been suggested for Ku, since in its absence, DNA ends undergo long-range movements which would promote translocation formation44. Underlying these considerations are the kinetics of NHEJ, since DSB repair occurs very quickly in wild-type cells, whereas DSBs persist in Xrcc4/ligase IV deficient cells for long periods45.
It is notable in this regard that we do not see a statistically significant increase in the median deletion length at the translocation breakpoint junctions in the mutant cells compared with wild type. Thus, the increased translocation frequency in Xrcc4/ligase IV and Ku mutant cells suggests that nucleases may have access to more DNA ends for processing rather than affecting the total amount of nucleolytic processing at a particular end; alternatively, nucleases may increase resection at a DNA end, but only on one strand so as to maintain a similar deletion distribution.
Breakpoint junctions with multiple inserts provide the clearest and most complex example of iterative processing for NHEJ. For these junctions, nucleolytic processing of the DNA ends occurred, and subsequently, an insert was added to one or both ends. For junctions with three or four inserts, at least one of the ends had undergone a second round of insert addition. Since longer deletions are found in breakpoint junctions with insertions compared to those without insertions, multiple rounds of nucleolytic processing may have occurred as well. This iterative process for alt-NHEJ may facilitate joining or it may be the result of nonproductive processing events in a sort of “testing” process.
Many insertions duplicate an existing chromosomal sequence, suggesting a model whereby the insertions are added by DNA synthesis. In such a mechanism, a DNA end would prime synthesis from another chromosomal sequence acting as a template. Templated insertions have been termed “T nucleotides” to distinguish them from non-templated insertions46. In vitro, pol µ and pol λ are capable of polymerizing from DNA ends that have little complementarity (pol µ and pol λ) or no complementarity (pol µ) to the template34, and so either (or both) may be involved in these insertions. Both pol µ and pol λ interact with Ku/Xrcc4/ligase IV/DNA and, where tested, this interaction stimulates polymerization10,13. However, as we found insertions in Ku and Xrcc4-deficient cells, this interaction may not be critical for activity.
The derivation of the breakpoint junction insertions seems remarkable. Although many are derived from sequences within 10 to 100 bp from the DSB, others are derived from sequences kilobases away from the DSB. As the insertions from nearby the DSB were typically derived from the ends that form der(17), many were likely inserted during the der(17) joining process itself. Insertions from further away were nearly as likely to be derived from sequences that form der(14) as those that form der(17), suggesting that they were inserted at an earlier stage, possibly during an attempt at intrachromosomal repair. The two most distant insertions are derived from megabases away from one of the der(14) ends and from the same gene, Psmb9, a meiotic hotspot of recombination in some mouse strains36,37. The Psmb9 insertions are not from the hotspot itself located at the 3′ end of the gene, but from 5′ UTR and intron sequences. Interestingly, one report has linked sequences frequently involved in translocations in somatic cells with meiotic hotspot activity, suggesting that some regions are susceptible to cleavage (breakage) in both somatic and meiotic cells47.
In summary, alt-NHEJ appears to be the primary mediator of translocation formation in mammalian cells. Characteristics of alt-NHEJ are emerging, including a bias, but not a strict requirement, for microhomology and a role for iterative processing. Elucidating this pathway will be important for understanding mechanisms leading to oncogenic translocations.
Materials and Methods
Cell culture and gene targeting
Xrcc4−/− mouse ES cells were provided by F. Alt (Harvard)26. For construction of neo− cells (Supplementary Fig. 1a), Xrcc4−/− cells were transfected with 15 µg pCAGGS-Cre expression vector, and G418 sensitive clones were isolated. Clones were confirmed to have undergone Cre-mediated loxP recombination to remove the neo cassette at both Xrcc4 loci by PCR using primers shown in Supplementary Fig. 1b. Two sets of primers (KMO74, KMO75 and KMO77) and (KMO78 and KMO79) were used. The conditions for both PCRs, are as follows: denaturation at 95°C for 3 min, followed by 30 cycles at 95°C for 1 min, 64°C for 1 min and 72°C for 2 min.
Targeting vectors for chr.17, pC-I (Pim1), and chr. 14, rdPuroRev (Rb), were previously described6. Targeting was confirmed at the Pim1 locus using the p22 probe (a HincII-BstXI fragment) in conjunction with HincII digestion of genomic DNA48. Targeting at the Rb locus was confirmed using probe A (a PstI-PvuII fragment) in conjunction with HindIII digestion of genomic DNA49. The expression vector for I-SceI endonuclease, pCBASce, and pCAGGS were previously described50. Human XRCC4, a gift from J. Chaudhuri (MSKCC), was subcloned into the XhoI and XbaI sites of pCAGGS.
Translocation analysis
For translocation experiments, 1 × 106 cells per well were plated in 6-well plates and transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. DNA for transfection was as follows: I-SceI only, 4 µg I-SceI and 4 µg GFP expression vectors; for Xrcc4 or Ku70 complementation: 4 µg SceI, 3.5 µg GFP, and 0.5 µg Xrcc4 or 4 µg SceI, and 4 µg Ku70 expression vectors, respectively. (Note: The Xrcc4 expression vector was titrated to reach wild-type levels for complementation, as analyzed by Western blotting and resistance to ionizing radiation; data not shown). 4 h after Lipofectamine addition, each well was split into two 10 cm plates; 24 h after splitting, cells were selected in 200 µg ml−1 G418. Translocation frequency was obtained by normalizing the number of neo+ clones obtained after 7 to 10 d to the number of cells counted at 4 h post-transfection.
For translocation junction analysis, DNA was isolated from neo+ clones using the Genelute Mammalian Genomic DNA Miniprep Kit (Sigma). PCR amplification was performed with the high fidelity Herculase II Fusion DNA Polymerase (Stratagene) under the following PCR conditions: denaturation at 95°C for 3 min, followed by 30 cycles of 95°C for 1 min, 62°C for 1min, and 72°C for 3 min. PCR products were gel purified with Gel purification kit (Invitrogen). Purified PCR products were sequenced with the same primers that were used for PCR amplification at the DNA Sequencing Core Facility at MSKCC.
Intrachromosomal Junction Analysis
To analyze junctions from imprecise NHEJ at a single I-SceI site, genomic DNA was isolated from E13U18 cells 72 h after transfection of the I-SceI expression vector (with or without the Xrcc4 expression vector) and then digested in vitro with I-SceI (NEB). (Transfection conditions were the same as that for translocation experiments.) An approximately 1.5 kb fragment was then amplified using primers surrounding the I-SceI site on der(17) by Extaq enzyme (Takara inc.) with the following primers:17–754 TGACTCTGGCTTGTGGTTTG and 14–726 GCTGGATATGTGTCCCGTTT. PCR was done at an annealing temperature of 60°C and an extension time of 1.5 min, and products were cloned with TOPO TA Cloning kit (Invitrogen) and sequenced. For statistical analysis, a two-tailed Mann-Whitney test was used.
Western Blotting
Whole cell extracts were prepared with Nonidet-P40 buffer and were run on a 7.5% (w/v) Tris-HCl SDS Page and blotted using the Xrcc4 antibody clone C-20 (Santa Cruz Biotechnology), which recognizes the C terminus of the human and mouse proteins. Mouse monoclonal antibody to 〈-tubulin (Sigma) was used as a loading control.
Fluorescence in situ hybridization
FISH was performed by the Molecular Cytogenetics Core Facility at MSKCC. For visualization of translocations, the FITC (green) probe (Cambio #1189-14MF-01) was hybridized to mouse chr. 14, and the Cy3 (red) probe (Cambio #1200-17MCy3-01) was hybridized to mouse chr. 17. Images were captured using a digital FISH workstation (Metasystems).
Statistical Analysis
In order to determine which statistical test to use (i.e., Kolmogorov-Smirnoc, D’Agostino&Pearson omnibus and Shapiro-Wilk), normality tests were executed using Prism Software (GraphPad, Inc.). Because translocation frequencies showed a normal distribution, a two–tailed unpaired t test was applied for this analysis. Because the data do not assume normal distribution, a one-tailed Mann-Whitney test was used for microhomology distribution and a two-tailed Mann-Whitney test was used for other analyses. Assuming an unbiased base composition, the probability that a junction will have X nucleotides of microhomology expected by chance is calculated by the equation P(X)=(X+1)(1/4)X(3/4)2 as previously described51.
Supplementary Material
Acknowledgments
We thank Margaret Leversha and Jeremy McGuire at the MSKCC Molecular Cytogenetics Core Facility for performing the FISH analysis, Erika Brunet, David Weinstock, Yufuko Akamatsu and other members of Jasin laboratory for helpful discussions. This work was supported by R01 NIHGM54668 and a Dorothy Rodbell Cohen Cancer Research Program Grant (M.J.).
Footnotes
Author contributions statement:
D.S. performed the experiments. D.S. and M.J. designed the research and wrote the paper.
References
- 1.van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 2.Jeggo PA. Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat Res. 1998;150:S80–S91. [PubMed] [Google Scholar]
- 3.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 5.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 6.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 9.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 10.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch CA, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budman J, Kim SA, Chu G. Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV. J Biol Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 13.Ma Y, et al. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci U S A. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 21.Difilippantonio MJ, et al. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 23.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 27.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawada M, et al. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat Cell Biol. 2003;5:320–329. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- 29.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 30.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 31.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 32.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verkaik NS, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Gu J, et al. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiroishi T, Sagai T, Hanzawa N, Gotoh H, Moriwaki K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 1991;10:681–686. doi: 10.1002/j.1460-2075.1991.tb07997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillon H, de Massy B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet. 2002;32:296–299. doi: 10.1038/ng990. [DOI] [PubMed] [Google Scholar]
- 38.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 40.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 43.Smith J, et al. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riballo E, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 46.Jager U, et al. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 47.Ng SH, Maas SA, Petkov PM, Mills KD, Paigen K. Colocalization of somatic and meiotic double strand breaks near the Myc oncogene on mouse chromosome 15. Genes Chromosomes Cancer. 2009;48:925–930. doi: 10.1002/gcc.20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.te Riele H, Maandag ER, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 49.te Riele H, Maandag ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci U S A. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.