Abstract
Osterix (Osx or Sp7) is a zinc-finger-family transcriptional factor essential for osteoblast differentiation in mammals. The Osx-Cre mouse line (also known as Osx1-GFP::Cre) expresses GFP::Cre fusion protein from a BAC transgene containing the Osx regulatory sequence. The mouse strain was initially characterized during embryogenesis, and found to target mainly osteoblast-lineage cells. Because the strain has been increasingly used in postnatal studies, it is important to evaluate its targeting specificity in mice after birth. By crossing the Osx-Cre mouse with the R26-mT/mG reporter line and analyzing the progenies at two months of age, we find that Osx-Cre targets not only osteoblasts, osteocytes and hypertrophic chondrocytes as expected, but also stromal cells, adipocytes and perivascular cells in the bone marrow. The targeting of adipocytes and perivascular cells appears to be specific to those residing within the bone marrow, as the same cell types elsewhere are not targeted. Beyond the skeleton, Osx-Cre also targets the olfactory glomerular cells, and a subset of the gastric and intestinal epithelium. Thus, potential contributions from the non-osteoblast-lineage cells should be considered when Osx-Cre is used to study gene functions in postnatal mice.
Introduction
Osterix (Osx or Sp7) is a zinc finger family transcriptional factor critical for osteoblast differentiation[1]. During embryonic skeletal development, Osx is initially expressed in the perichondrium flanking the hypertrophic cartilage, where osteoblasts first arise to produce the bone collar (cortical bone). Later during development, the perichondrial Osx-expressing osteoprogenitors co-migrate with the blood vessels that invade the hypertrophic cartilage, and to generate osteoblasts responsible for depositing the trabecular bone [2], [3]. In addition, Osx is also detected in early hypertrophic chondrocytes at a relatively weak level [4]. Genetic studies have revealed the essential role of Osx in osteoblast differentiation[5]. In Osx-null embryos, cartilage elements are largely normal but osteoblast differentiation fails to complete, resulting in a complete lack of bone tissue [1]. In these embryos, Runx2 expression is relatively normal, but other osteoblast markers including Col1a1, Bsp, and osteocalcin are either absent or severely suppressed [1]. On the other hand, the expression of Osx is abolished in Runx2-null embryos[1]. Thus, Osx functions genetically downstream of Runx2 to control osteoblast differentiation[6]. In addition to its role in embryonic osteoblast differentiation, Osx also plays a critical role in the formation and function of postnatal osteoblast and osteocyte [7].
The Cre/loxP technology enables gene deletion in specific cell types and has significantly advanced our understanding of gene functions in both physiological and pathological conditions. In this system, specificity is achieved by expression of the Cre recombinase under the control of cell type-specific regulatory sequences. Osx-Cre (Osx1-GFP::Cre), a BAC transgenic mouse line expressing a GFP::Cre fusion protein from the regulatory sequence of Osx, was generated to direct gene deletion in the osteoblast lineage [8]. The initial characterization of this mouse line revealed that Cre activity is largely restricted to the osteogenic perichondrium, periosteum and osteoblast-lineage cells within the marrow cavity, but that analysis was limited to the embryo [8]. Although in recent years the Osx-Cre mouse line has been increasingly used to study the osteoblast lineage in postnatal mice [9], [10], [11], [12], [13], the targeting specificity of Osx-Cre in postnatal bones is yet to be formally evaluated.
Here we assess the cell types targeted by Osx-Cre in two-month-old mice by monitoring GFP expression from the R26-mT/mG reporter allele. The R26-mT/mG allele ubiquitously expresses a membrane-targeted red fluorescent protein (mTomato) but switches to expressing a membrane-targeted green fluorescent protein (mGFP) upon Cre recombination. We find that within the skeleton, Osx-Cre targets not only osteoblast lineage cells and a subset of chondrocytes, but also stromal cells, adipocytes and perivascular cells specifically within the bone marrow. Moreover, Osx-Cre also targets cells within the olfactory bulb, the intestine and the stomach.
Materials and Methods
Mouse strains
Osx-Cre (Osx1-GFP::Cre) and R26-mT/mG mouse lines are as previously described [8], [14]. All mouse procedures used in this study were approved by the Animal Studies Committee at Washington University.
Cryostat sections
Two-month-old mice were perfused with 4% PFA as described previously[15]. After perfusion, tibias were dissected and fixed in 4% PFA at 4°C overnight. The fixed tibias were decalcified in 14% EDTA for 3 days and then snap-frozen in OCT embedding medium. Frozen sections were cut at 8 µm thickness with a cryostat equipped with Cryojane (Leica, IL). The sections were kept at −20°C until analyses.
Immunofluorescence staining
For detection of GFP, perilipin or αSMA, immunostaining was performed on cryostat sections using a chicken polyclonal GFP antibody (1∶2500; Abcam, Cambridge, MA), or rabbit monoclonal perilipin antibody (1∶100; Cell Signaling Technology, Danvers, MA), or mouse monoclonal αSMA antibody (1∶500, Sigma, St. Louis, MO). The secondary antibodies are as follows: Alexa Fluor® 488 Goat Anti-Chicken IgG (for GFP); Alexa Fluor® 647 F(ab')2 Fragment of Goat Anti-Rabbit IgG (for perilipin), and Alexa Fluor® 647 Goat Anti-Mouse IgG2a (for αSMA) (all at 1∶250, Life Technologies, Grand Island, NY). Sections were mounted with VECTASHIELD Mounting Medium containing DAPI (VECTOR LABORATORIES, Burlingame, CA).
Quantitative analyses
All quantitative data were obtained from three independent animals. Statistical analyses were performed with student's t-test.
Results
Osx-Cre targets osteoblasts, osteocytes, bone marrow stromal cells and hypertrophic chondrocytes
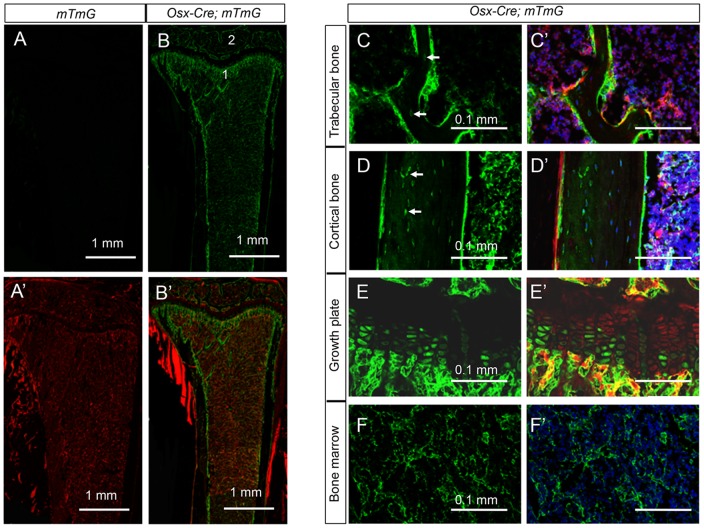
To characterize the targeting specificity of Osx-Cre in postnatal mouse bones, we generated Osx-Cre; R26-mT/mG mice (one copy each of Osx-Cre and R26-mT/mG) and analyzed GFP expression on sections of long bones at two months of age. As expected, bone sections from the control R26-mT/mG mice did not exhibit any GFP (Fig. 1A, A′), but those from Osx-Cre; R26-mT/mG mice contained many GFP-positive cells (Fig. 1B, B′). Consistent with the targeting of osteoblast-lineage cells, many GFP-positive cells were present at both primary and secondary ossification centers, as well as the cortical bone surfaces (Fig. 1B, B′). Examination at a higher magnification revealed that essentially all cells associated with the trabecular, endosteal and periosteal surfaces, as well as most osteocytes were GFP-positive (Fig. 1C, C′, D, D′). Moreover, a large population of bone marrow stromal cells expressed GFP (Fig. 1F, F′). These cells exhibited a reticular morphology and were readily distinguishable from the hematopoietic population. Finally, GFP was detected in some prehypertrophic and hypertrophic chondrocytes within the growth plate (Fig. 1E, E′), consistent with previous reports of endogenous Osx expression in these cells [16]. Thus, in addition to osteoblasts and osteocytes, Osx-Cre also targets the bone marrow stromal cells and the growth plate hypertrophic chondrocytes in postnatal mice.
Figure 1. Osx-Cre targets osteoblast lineage cells, hypertrophic chondrocytes and bone marrow stromal cells.
(A–B) Confocal images for direct fluorescence from EGFP (A, B) or EGFP/tdTomato (A′, B′) on longitudinal tibial sections from two-month-old R26-mT/mG (A, A′) or Osx-Cre; R26-mT/mG mice (B, B′); 1: chondro-osseous junction of primary ossification center; 2: secondary ossification center. (C–F, C′–F′) Higher magnification images for EGFP (C–F) or EGFP/tdTomato: trabecular bone (C, C′), cortical bone (D, D′), growth plate (E, E′) and bone marrow (F, F′). Red: membrane-targeted tdTomato; Green: membrane-targeted EGFP; Blue: DAPI. Arrow: osteocyte.
Osx-Cre targets adipocytes specifically in the bone marrow
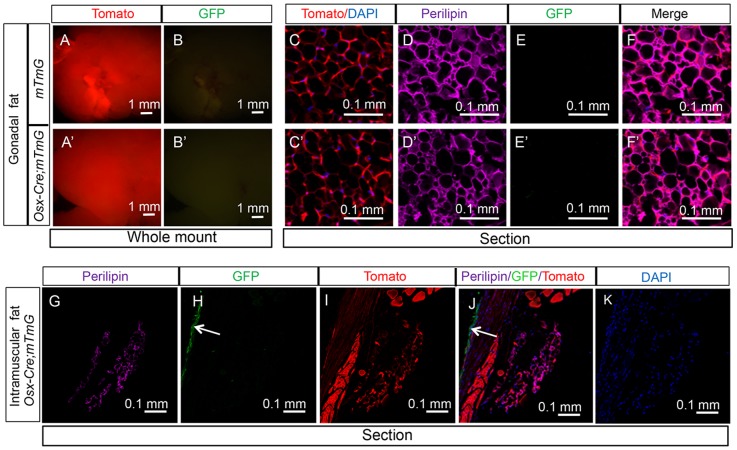
Within the bone marrow, in addition to the reticular stromal cells, we also detected other GFP-positive cells that appeared to be adipocytes. To confirm this observation, we performed double immunofluorescence experiments with antibodies against GFP and the adipocyte-specific marker perilipin. Indeed, on bone sections from the Osx-Cre; R26-mT/mG mice, more than 90% of the perilipin-positive adipocytes present within the bone marrow also stained for GFP (Fig. 2). To determine whether Osx-Cre also targets adipocytes outside the bone marrow, we examined the whole-mount gonadal fat depot from Osx-Cre; R26-mT/mG mice under a fluorescence microscope but didn't detect any GFP (Fig. 3A–B). Double immunostaining of sections from the gonadal fat depot confirmed that the perilipin-positive adipocytes were GFP-negative (Fig. 3C–F). Similarly, adipose tissues adjacent to the periosteum and associated with the skeletal muscle did not express GFP (Fig.3G–J). Thus, Osx-Cre effectively targets adipocytes specifically within the bone marrow environment.
Figure 2. Osx-Cre marks adipocytes in bone marrow.
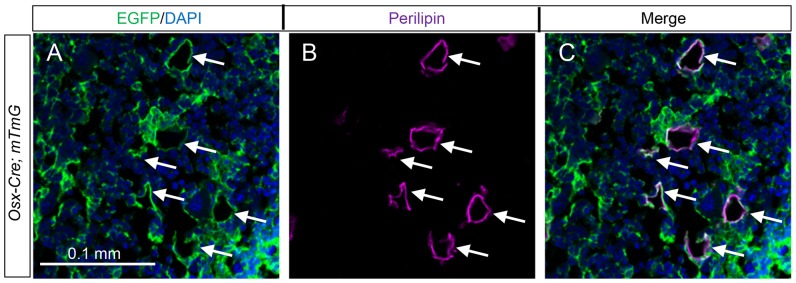
(A–B) Double immunostaining for EGFP (A) and perilipin (B) on longitudinal sections of tibias from two-month-old Osx-Cre; R26-mT/mG mice. (C) Co-localization of EGFP and perilipin. Arrows denote co-expression of GFP and perilipin. Green: EGFP; magenta: perilipin; blue: DAPI.
Figure 3. Osx-Cre does not mark non-bone marrow adipocytes.
(A–B) Images for direct fluorescence from tdTomato (A) or EGFP (B) in whole-mount gonadal fat depots from two-month-old Osx-Cre; R26-mT/mG mice. (C–F) Direct fluorescence for tdTomato (C) and immunofluorescence for perilipin (D) and EGFP (E) on sections of gonadal fat depots from two-month-old Osx-Cre;R26-mT/mG mice. (G–K) Imaging of longitudinal sections of an intramuscular fat depot associated with a tibia from two-month-old Osx-Cre;R26-mT/mG mice. G: perilipin immunofluorescence; H: EGFP immunofluorescence; I: direct fluorescence for tdTomato; J: merged view of G–I; K: DAPI staining. Arrow: GFP-positive periosteum.
Osx-Cre targets perivascular cells specifically in the bone marrow
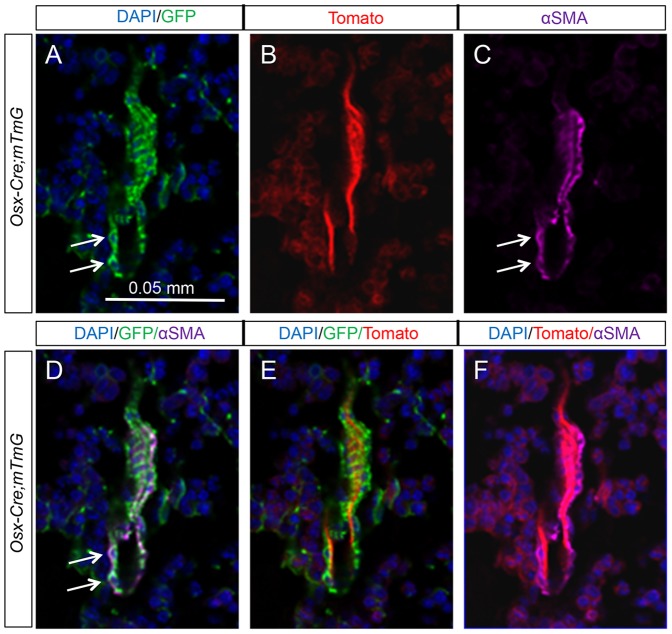
Our initial examination of the long bone sections revealed that certain GFP-positive cells appeared to associate directly with blood vessels. To confirm the identity of these cells as perivascular smooth muscle cells, we performed double immunofluorescence experiments with antibodies against GFP and αSMA. Direct visualization of tdTomato under a fluorescence microscope revealed that the endothelial cells expressed a strong signal, whereas the αSMA-positive cells were immediately adjacent to the endothelial cells as expected (Fig. 4F). Importantly, in three Osx-Cre; R26-mT/mG mice analyzed, nearly all αSMA-positive perivascular smooth muscle cells co-expressed GFP (Fig. 4D) (93.9±2.8%, n = 3), and 100% of the ten bone marrow blood vessels observed were found to associate with GFP-positive cells. In contrast, the blood vessels abundantly present in the gonadal adipose tissue did not contain GFP-positive cells (Fig. 3C′, E′). Thus, Osx-Cre targets a high percentage of the perivascular smooth muscle cells specifically within the bone marrow.
Figure 4. Osx-Cre marks perivascular smooth muscle cells in bone marrow.
(A–C) Confocal images of EGFP (A), tdTomato (B), and αSMA (C) on longitudinal sections of tibias from two-month-old Osx-Cre; R26-mT/mG mice. (D–F) Merged images. Arrow: co-expression of EGFP and αSMA. EGFP and αSMA: immunofluorescence; tdTomato: direct fluorescence. DAPI stains DNA blue.
Osx-Cre targets cells beyond the skeleton
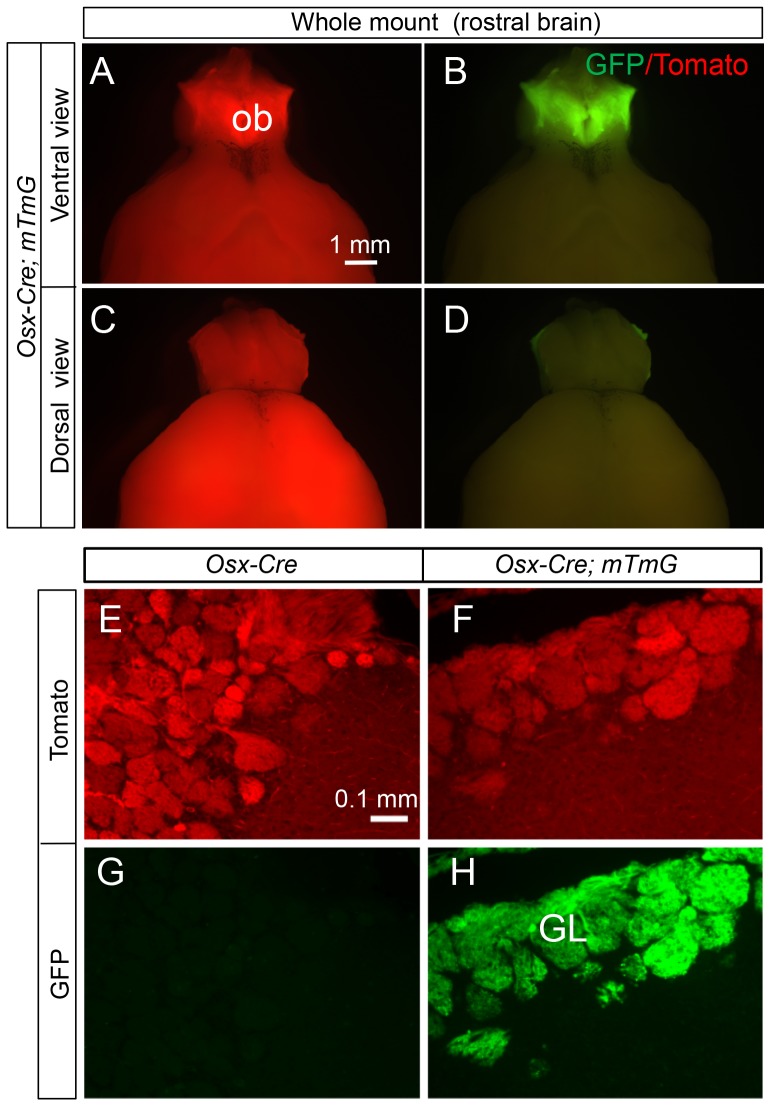
We next examined all tissues of the Osx-Cre; R26-mT/mG mouse for potential targeting by Osx-Cre. We first ensured that no GFP was detected in any of the organs of the control R26-mT/mG mice. In Osx-Cre; R26-mT/mG mice, we found no GFP in liver, kidney, pancreas, heart, lung, adrenal gland, thymus, thyroid or skeletal muscle. Much of the brain was also negative, but the olfactory bulb expressed strong GFP as evident from the ventral view of the rostral brain (Fig. 5A–D). Sagittal sections through the bulb revealed that Osx-Cre targeted essentially all cells of the glomerular layer (Fig. 5E, F). In addition, a strong GFP signal was observed in whole-mount samples of the stomach, small and large intestines from Osx-Cre; R26-mT/mG but not R26-mT/mG littermates (Fig. 6 A–D, data not shown). Cross-section of the small intestine revealed that GFP was non-uniformly expressed by enterocytes along the villi, whereas cells in the crypts were largely negative (Fig. 6E–G). Similarly, cross sections through the stomach indicated mosaic GFP expression in the epithelium including parietal cells (Fig. 6 H–J). The precise identity of the targeted intestinal or gastric cells was not further pursued in the present study.
Figure 5. Osx-Cre targets olfactory glomerular cells.
(A–D) Direct fluorescence of whole-mount rostral brain from a two-month-old Osx-Cre; R26-mT/mG mouse. ob: olfactory bulb. (E–H) Direct fluorescence of tdTomato (E, F) and EGFP (G, H) on sagittal sections through the olfactory bulb of Osx-Cre (E, G) or Osx-Cre; R26-mT/mG (F, H) mice at two months of age. GL: glomerular layer.
Figure 6. Osx-Cre targets gastric and intestinal epithelia.
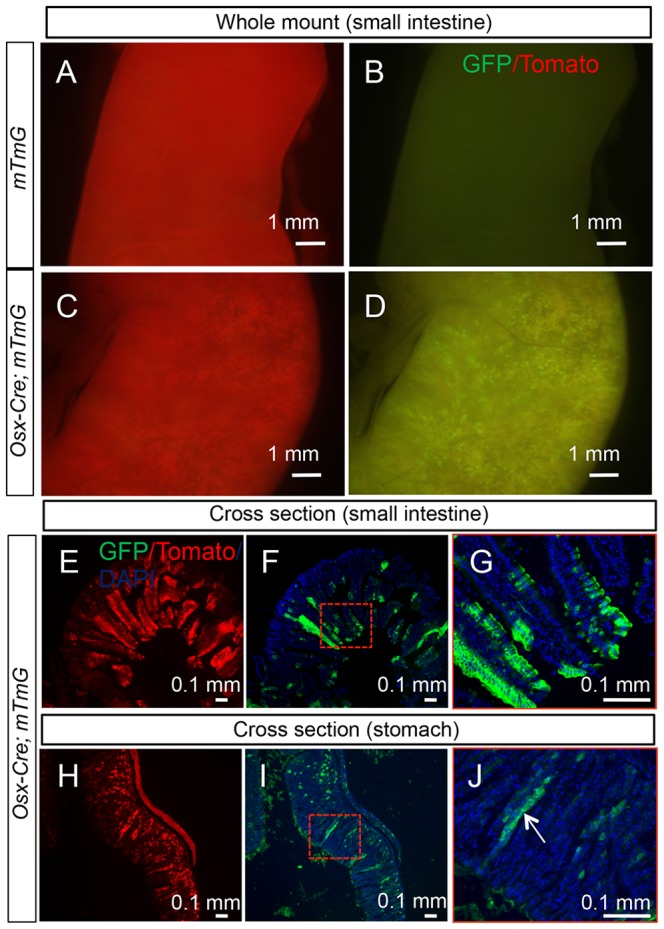
(A–D) Direct fluorescence of tdTomato (A, C) and EGFP (B, D) in whole-mount small intestine of two-month-old R26-mT/mG (A, B) or Osx-Cre; R26-mT/mG mice (C, D). (E–G) Direct fluorescence on cross sections of small intestine from a two-month-old Osx-Cre; R26-mT/mG mouse. Red-boxed area in F shown at a higher magnification in G. (H–J) Direct fluorescence on stomach sections from a two-month-old Osx-Cre; R26-mT/mG mouse. Red-boxed area in I shown at a higher magnification in J. White arrow denotes parietal cells.
Discussion
Osx-Cre and bone marrow mesenchymal stem cells
Evidence suggests that mesenchymal stem cells (MSCs) residing within the adult bone marrow produce osteoblasts, adipocytes and bone marrow stromal cells necessary for postnatal tissue homeostasis [17], [18]. Moreover, accumulating evidence supports that mesenchymal stem cells reside within a perivascular niche [18], [19], [20], [21], [22]. Our present work demonstrates that Osx-Cre targets not only osteoblasts and osteocytes, but also the bone marrow stromal cells and perivascular smooth muscle cells, even though the later cell types do not actively express the GFP::Cre protein. These results raise the possibility that Osx-Cre targets a common progenitor of the aforementioned cell types, perhaps the bone marrow MSC. Alternatively, Osx-Cre may mark a diverse group of progenitors each producing a single mature cell type (e.g., stromal or perivascular cell). In either case, the progenitors do not appear to continuously express Osx or Osx-Cre throughout postnatal life, as cells actively expressing Osx postnatally, when marked with Osx-CreER in response to tamoxifen, failed to sustain the turnover of either osteoblasts or stromal cells [15], [23]. Overall, the current data support the model that embryonic Osx-expressing cells give rise to the bone marrow mesenchymal progenitors in postnatal mice. Additional experiments are necessary to test this hypothesis formally.
Organ-specific origin of bone marrow adipocytes and perivascular cells
Bone marrow adipose tissue is believed to be metabolically different from non-marrow peripheral fat depots [24]. Our data showed that bone marrow but not other adipocytes are derived from Osx-lineage cells, indicating a distinct cell origin of the bone marrow adipocytes. Similarly, perivascular smooth muscle cells specifically associated with bone marrow but not other blood vessels are derived from Osx-expressing progenitors. While this work was in review, others using the Ai9 Cre reporter mouse reported similar findings about the targeting of bone marrow adipocytes and perivascular smooth muscle cells by Osx-Cre, thus allaying the concern over limitations of a single Cre reporter [25]. Overall, these findings support the view that mesenchymal cell types in different organs may be derived from organ-specific stem/progenitor cells that reside locally.
Limitation of Osx-Cre as a tool for studying osteoblast biology
Our data clearly indicate that Osx-Cre, when activated in the embryo, targets more than osteoblast-lineage cells in postnatal mice. These findings echo the increasing concern that many Cre strains exhibit some degree of unintended recombination activity [26]. Whereas the relationship between marrow fat and bone is increasingly appreciated [24], the potential influence of stromal cells and perivascular cells on bone is largely unknown. In addition, Osx-Cre targets olfactory glomerular cells and the GI tract epithelia. Targeting of the olfactory bulb is consistent with endogenous Osx expression as previously reported in this organ [27]. On the other hand, it is not clear at present whether Osx is normally expressed in the GI tract, or the mosaic Cre activity there simply reflects a phenomenon specific to the Osx-Cre transgene. Furthermore, it is not known whether Osx-Cre-mediated gene deletion in the olfactory bulb or the GI track affects bone physiology. Nonetheless, caution needs to be taken when one interprets postnatal bone phenotypes caused by gene deletion with Osx-Cre beginning in the embryo.
Acknowledgments
We thank Dr. Jason Mills (Washington University School of Medicine) for his assistance in identifying the targeted cell types in the stomach.
Funding Statement
The work is funded by National Institutes of Health grants DK065789 and AR055923 to FL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, et al. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29. [DOI] [PubMed] [Google Scholar]
- 2. Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell 19: 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Long F (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nature reviews Molecular cell biology 13: 27–38. [DOI] [PubMed] [Google Scholar]
- 4. Hilton MJ, Tu X, Cook J, Hu H, Long F (2005) Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132: 4339–4351. [DOI] [PubMed] [Google Scholar]
- 5. Long F (2011) Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13: 27–38. [DOI] [PubMed] [Google Scholar]
- 6. Long F, Ornitz DM (2013) Development of the endochondral skeleton. Cold Spring Harb Perspect Biol 5: a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, et al. (2010) Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A 107: 12919–12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodda SJ, McMahon AP (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133: 3231–3244. [DOI] [PubMed] [Google Scholar]
- 9. Davey RA, Clarke MV, Sastra S, Skinner JP, Chiang C, et al. (2012) Decreased body weight in young Osterix-Cre transgenic mice results in delayed cortical bone expansion and accrual. Transgenic Res 21: 885–893. [DOI] [PubMed] [Google Scholar]
- 10. Xian L, Wu X, Pang L, Lou M, Rosen CJ, et al. (2012) Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med 18: 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joeng KS, Long F (2013) Constitutive activation of Gli2 impairs bone formation in postnatal growing mice. PLoS ONE 8: e55134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Canalis E, Parker K, Feng JQ, Zanotti S (2013) Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology 154: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Razidlo DF, Whitney TJ, Casper ME, McGee-Lawrence ME, Stensgard BA, et al. (2010) Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS ONE 5: e11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605. [DOI] [PubMed] [Google Scholar]
- 15. Chen J, Long F (2013) beta-catenin promotes bone formation and suppresses bone resorption in postnatal growing mice. J Bone Miner Res 28: 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, et al. (2005) Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 132: 49–60. [DOI] [PubMed] [Google Scholar]
- 17. Caplan AI, Correa D (2011) The MSC: an injury drugstore. Cell Stem Cell 9: 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bianco P, Robey PG, Simmons PJ (2008) Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morikawa S, Mabuchi Y, Kubota Y, Nagai Y, Niibe K, et al. (2009) Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med 206: 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, et al. (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3: 301–313. [DOI] [PubMed] [Google Scholar]
- 21. Hirschi KK, D'Amore PA (1996) Pericytes in the microvasculature. Cardiovasc Res 32: 687–698. [PubMed] [Google Scholar]
- 22. Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, et al. (2007) Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 131: 324–336. [DOI] [PubMed] [Google Scholar]
- 23. Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, et al. (2012) Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fazeli PK, Horowitz MC, MacDougald OA, Scheller EL, Rodeheffer MS, et al. (2013) Marrow fat and bone—new perspectives. J Clin Endocrinol Metab 98: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Strecker S, Wang L, Kronenberg MS, Wang W, et al. (2013) Osterix-cre labeled progenitor cells contribute to the formation and maintenance of the bone marrow stroma. PloS one 8: e71318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, et al. (2012) Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nature communications 3: 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park JS, Baek WY, Kim YH, Kim JE (2011) In vivo expression of Osterix in mature granule cells of adult mouse olfactory bulb. Biochemical and biophysical research communications 407: 842–847. [DOI] [PubMed] [Google Scholar]