Abstract
PprA is known to contribute to Deinococcus radiodurans' remarkable capacity to survive a variety of genotoxic assaults. The molecular bases for PprA's role(s) in the maintenance of the damaged D. radiodurans genome are incompletely understood, but PprA is thought to promote D. radiodurans's capacity for DSB repair. PprA is found in a multiprotein DNA processing complex along with an ATP type DNA ligase, and the D. radiodurans toposiomerase IB (DraTopoIB) as well as other proteins. Here, we show that PprA is a key contributor to D. radiodurans resistance to nalidixic acid (Nal), an inhibitor of topoisomerase II. Growth of wild type D. radiodurans and a pprA mutant were similar in the absence of exogenous genotoxic insults; however, the pprA mutant exhibited marked growth delay and a higher frequency of anucleate cells following treatment with DNA-damaging agents. We show that PprA interacts with both DraTopoIB and the Gyrase A subunit (DraGyrA) in vivo and that purified PprA enhances DraTopoIB catalysed relaxation of supercoiled DNA. Thus, besides promoting DNA repair, our findings suggest that PprA also contributes to preserving the integrity of the D. radiodurans genome following DNA damage by interacting with DNA topoisomerases and by facilitating the actions of DraTopoIB.
Introduction
Deinococcus radiodurans is extremely resistant to many abiotic stresses including ionizing radiation, UV and other DNA damaging agents [1]. After exposure to ordinarily lethal doses of γ radiation, the D. radiodurans genome is shattered into numerous double strand breaks (DSBs) and single strand breaks [2]. The genome is subsequently reassembled back to its full length with high fidelity by an extended synthesis dependent strand annealing (ESDSA) DSB repair mechanism [3], [4]. Besides the highly efficient DSB repair mechanism, strong antioxidant mechanisms [5], [6], [7] are also thought to contribute to the remarkable resistance of this bacterium to genotoxic assaults. Another distinctive feature of this bacterium is its highly condensed toroidal genome; however, it remains unclear whether the compactness and shape of the D. radiodurans genome contribute to its extraordinary radiation resistance [8], [9]. Also, although D. radiodurans encodes both subunits of DNA TopoII and a type IB DNA topoisomerase (DraTopoIB) [10], the roles of these enzymes in D. radiodurans' resistance to DNA damage are not known.
A mutation making D. radiodurans hypersensitive to radiation and other DNA damaging agents was mapped to a locus named pprA (a pleiotropic protein involved in radiation resistance, ORF DR_A0346 in D. radiodurans R1) [10]. The PprA protein binds to broken double stranded DNA (dsDNA) and protects it from exonuclease degradation similar to the eukaryotic Ku protein [11]. PprA stimulates both ATP and NAD dependent DNA ligase activities in vitro [11]. It was found to be the part of a multiprotein DNA processing complex comprised of 24 proteins including the ATP type DNA ligase (LigB), DraTopoIB (DR_0690) and 11 hypothetical polypeptides of D. radiodurans [12]. Subsequently, it was shown that PprA could restore the DNA end joining activity of LigB, which was otherwise inactive in purified form [13], [14]. The involvement of PprA in Deinococcus resistance to different DNA damaging agents including radiations and mitomycin C (MMC) has been demonstrated, but not to the Topo II inhibitor nalidixic acid (Nal), which also damages DNA.
DNA topoisomerases are ubiquitous enzymes that help cells to maintain the correct topology of their DNA. Routine cellular processes, including DNA replication, transcription and recombination alter DNA topology, and topoisomerases are essential for restoring proper topology and maintaining genome integrity. Topoisomerases are typically classified into type I (TopoI) and type II (TopoII) based on their substrate preferences (the number of nicks in DNA), subunit structures, and cofactor requirements. Phylogenetically, these enzymes are grouped into TopoIA, IB and IC, and Topo IIA and IIB, respectively [15], [16]. TopoI subtypes are highly divergent in terms of both structure and function [17], [18]. D. radiodurans encodes a Topo IB that is structurally similar to the TopoIB of poxviruses [19], and this class of enzymes forms a transient covalent link 3′ of the DNA break. Topo IB can relax both positive and negative superturns in vitro. In Saccharomyces cerevisiae, genetic analyses suggest that Topo IB plays a major role in DNA replication, transcription and genome structure maintenance, processes which are ordinarily thought to be accomplished by TopoII [20]. Although, DraTopoIB relaxes both negatively and positively supercoiled DNA in vitro, its activity is resistant to camptothecin, a compound that inhibits the activity of nuclear encoded TopoIB [19].
Here we report that a D. radiodurans pprA mutant is even more sensitive to inhibition of TopoII activity by Nal than to γ radiation. In normal growth conditions, in the absence of exogenous genotoxic insults, the growth of the pprA mutant was indistinguisable from that of the wild type. However, after exposure to either Nal or γ radiation, the mutant exhibited marked growth arrest and an elevated fraction of anucleate cells compared to the wild type. In addition, we found that PprA interacts with DraTopoIB and DraGyrA and could enhance the relaxation of supercoiled DNA by recombinant DraTopoIB in vitro. Nal treatment reduced the expression of both dratopoIB and dragyrA genes, particularly in cells lacking PprA. Collectively, our observations suggest that in addition to its known role in enhancing DSB repair, PprA promotes the maintenance and recovery of the damaged D. radiodurans genome by interacting with topoisomerases in this bacterium.
Materials and Methods
Bacterial strains, plasmids and media
Deinococcus radiodurans R1 (ATCC13939) was a gift from Professor J. Ortner, Germany [21] and the pprA::cat mutant was a gift from Prof. I. Narumi, JAERI Japan. Strains were maintained in TGY (0.5% Bacto Tryptone, 0.3% Bacto Yeast Extract, 0.1% Glucose) medium at 32°C in the presence of appropriate antibiotics as required. The E. coli strain HB101 was used for maintaining cloned genes on plasmids while strain E. coli BL21 (DE3) pLysS was used for protein expression. E. coli and D. radiodurans were grown as batch cultures in LB broth or TGY broth, as required with shaking at 180 rpm. The shuttle expression vector p11559 [22] and its derivative pVHS559 [23] were maintained in E. coli strain HB101 in presence of spectinomycin (40 µg/ml), while in D. radiodurans these vectors were maintained in presence of spectinomycin (75 µg/ml). All recombinant techniques were as described earlier [24].
Construction of DraTopoIB expression plasmid
Genomic DNA of D. radiodurans R1 was prepared as described in [25]. The 1041 bp coding sequence of DraTopoIB (DR_0690) was PCR amplified from genomic DNA using topoIF (5′ CGCGGATCCATGCCGAGCCGCACCGAA′) and topoIR (5′ CCCAAGCTTTCATTTAGCGCCCGGCC3′) primers containing appropriate restriction enzyme sites at their respective 5′ ends. The PCR product was inserted into the BamHI and HindIII sites of the pET28a+ expression vector. The resulting recombinant plasmid, pETtopoIB, was used for the expression of recombinant DraTopoIB in E. coli. The PprA expression plasmid pETpprA was constructed as described earlier [26]. For construction of pVHSpprA, the pprA coding sequence was PCR amplified using pprAF (5′ CGCGGTACATATGGTGCTACCCCTGGCCTT 3′) pprAR (5′ CCGCTCGAGTCAGCTCTCGCGCAGGCCGT3′) primers and cloned into the NdeI-XhoI sites of pVHS559 [23] to yield pVHSpprA.
Nalidixic acid and γ radiation treatment
Wild type D. radiodurans, the pprA mutant, and the pprA mutant expressing wild type PprA on pVHSpprA under the control of an IPTG inducible promoter were grown in TGY medium and serial dilutions of these cells were spotted on TGY agar plates supplemented with Nal (20 µg/ml). These cells were also exposed to the indicated doses of γ radiation as described earlier [27] at a dose rate of 4.16 kGy/h in Gamma chamber (GC 5000, 60Co, Board of Radiation and Isotopes Technology, India). Different dilutions of treated cultures were spotted on TGY agar plates. The plates were incubated at 32°C for 48 h and growth was monitored.
Microscopy techniques
Fluorescence microscopic studies were carried out as described earlier [23], [28]. Cells were stained with Nile red (4 µg/ml) and 4,6 diamdino2-phenylodine dihydroide chloride (DAPI) (0.2 µg/ml), mounted on 1.0% agarose-coated slides and observed under Axio Imager CM5 microscope. Nile red and DAPI fluorescence was recorded at 516 nm and 350 nm, respectively. Images were analyzed using Axiovision 4.8 software, modified by Image J and Adobe photoshop CS3 software. Cells lacking DAPI stained were counted and statistical analysis was carried out using Prism Software and graphs were plotted.
Protein purification and topoisomerase assay
Recombinant PprA and DraTopoIB expressed in E. coli BL21 (DE3) pLysS from pETpprA and pETtopoIB respectively, were purified from soluble fractions by metal affinity chromatography using NiNTA matrix (Qiagen Inc, Germany) followed by Q-sepharose anion exchange chromatography (GE Healthcare) using protocols as described earlier [29]. The DNA relaxation activity of DraTopoIB was monitored as described in [19]. In brief, 0.5 µM DraTopoIB was incubated in a reaction mixture containing 50 mM TrisHCl (pH 7.6), 100 mM NaCl, 5 mM MgCl2 and 500 ng supercoiled pUC18 DNA, in the presence of increasing concentrations of purified PprA as specified in the figure 4 legend. The reaction was carried out at 37°C for 30 min and quenched by heating at 65°C for 15 min in presence of 0.1%SDS. The products were analysed on 1% agarose gels, stained with ethidium bromide and DNA band intensity was measured densitometrically.
In vivo protein –protein interaction
We used the Escherichia coli bacterial two-hybrid system as described in [30] to explore in vivo protein-protein interactions between PprA and topoisomerases from Deinococcus radiodurans. The pprA gene was cloned in both pKNT25 and pUT18 at BamHI and KpnI sites to generate pUTpprA and pKNTpprA. The dratopoIB gene was also introduced into the BamHI and KpnI sites in these vectors. Similarly, deinococcal DNA gyrase A (dragyrA) gene was cloned at BamHI and KpnI in pUT18 to generate pUTgyrA, while DNA gyrase B (dragyrB) gene was cloned at BamHI and KpnI sites to generate pKNTgyrB. For evaluating the interaction of PprA with DraTopoIB and DraGyrA and/or DraGyrB, these proteins were co-expressed in E. coli strain BTH101 as discussed in [30] and isolated colonies were spotted on LB plate containing 100 µg/ml ampicillin, 50 µg/ml kanamycin, 200 mg/L X-gal and 0.5 mM IPTG and incubated at 30°C overnight and documented as described in [30]. For measuring the levels of β-galactosidase activity in different samples, these cells were grown and beta galactosidase activity was measured using ONPG colour substrate and the enzymatic activity was calculated as given in [31].
Gene expression studies
For monitoring the levels of expression of draTopoIB and dragyrA genes, quantitative real-time PCR (RT-PCR) was performed as described previously [32] with the RNA isolated from wild type and mutants grown for 4 h in the presence and absence of Nal (20 µg/ml) and treated with RNase free DNase I (Roche) and subsequently purified by phenol chloroform extraction. First-strand cDNA synthesis was carried out in 20 µl reaction containing 1.5 µg of RNA sample using SuperScript III Reverse Transcriptase kit (Invitrogen, Inc.) mixed with random hexamers as per manufacturer protocol and as described in [32]. RT-PCR was carried out using a Corbett rotor gene 3000 PCR machine with gene specific primers and cDNA as template. The primers used included, TIRTF (5′ CAGAAGTTCCGCTATGTCCA 3′) and TIRTR (5′ GGTGATAGCGGTACTGCA 3′) primers for Topoisomerase IB gene, T2ARTF (5′ GCGATGAACGTCATCGT 3′) and T2ARTR (5′ GTGTAGCGCATGTTCCA 3′) primers for gyrase A subunit of DNA TopoII, and gapF (5′-GAAGGGGCCTCCAAGCACAT-3′) and gapR (5′-TTGTACTTGCCGTTCGCGGCT-3′) for the dr1343 reference gene. Real-time signal detection of RT-PCR product was done using SyBr green 2X master mix kit (Sigma), as per manufacturer's instructions. Levels of transcripts were estimated and normalized by dividing with the levels of reference gene dr1343 (Glyceraldehyde 3-phosphate dehydrogenase (GAP) transcript.
Co-immunoprecipitation
Co-immunoprecipitation of DraTopoIB with PprA protein was carried out by mixing ∼2 mg of protein equivalent cell free extract of cells expressing recombinant (His)-PprA and (His)-DraTopoIB separately and incubating at 4°C for 3 h with gentle mixing. The mixture was further incubated with PprA specific antibody [11] in binding buffer (140 mM NaCl, 8 mM sodium phosphate, 2 mM potassium phosphate and 10 mM KCL, pH 7.4) at 4°C overnight as described in [33] with slow shaking. Subsequently, Protein G agarose beads were added for 3 h with gentle shaking. The antigen-antibody complexes was precipitated by centrifugation at 14,000 g for 5 min, and the pellet was washed with pre-chilled binding buffer 3 times (800 µl each) and eluted with 500 mM NaCl in binding buffer. Eluted proteins were precipitated with 2.5 volume ice-chilled acetone and dissolved in 2× Laemmili buffer. Proteins were separated on 10% SDS-PAGE, transferred to PVDF membrane and western blotting was done with anti-His antibodies to detect both proteins.
Data presented without standard deviations are illustrative of typical experiments, where variation among replicates was less than ∼15%. All experiments were carried out at least three times.
Results
PprA promotes D. radiodurans resistance to nalidixic acid
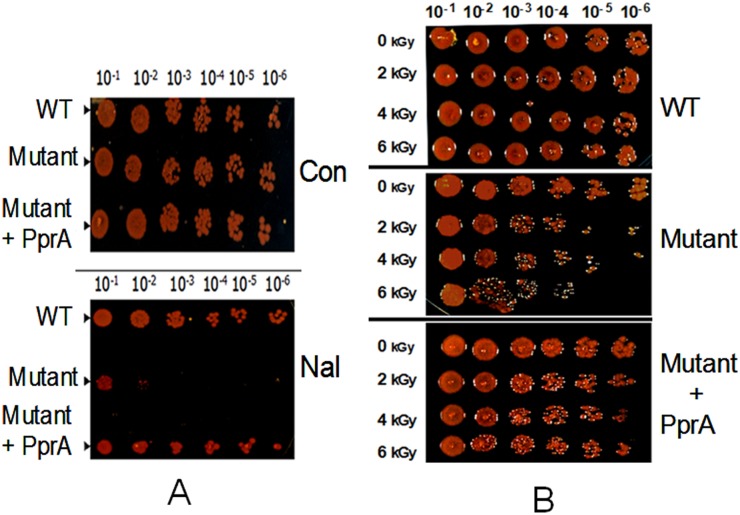
Nalidixic acid (Nal) is a quinolone antibiotic that inhibits DNA topoisomerase II and IV, enzymes that function in a variety of DNA transactions that are important for maintaining the topology of the genome and decatenating intertwined circular chromosomes [16], [34], [35]. Since PprA is known to augment D. radiodurans' resistance to DNA-damaging γ radiation, we wondered whether this pleiotropic protein also contributes to the organism's response to treatment with Nal, which also damages DNA [36] and affects genome topology through the inhibition of TopoII activity. A D. radiodurans R1 pprA mutant (pprA::cat) proved to be markedly more susceptible to Nal (20 µg/ml) than the isogenic wild type strain (Figure 1A). Nal resistance was restored to the pprA mutant by provision of pprA in trans, establishing that PprA promotes D. radiodurans' resistance to this DNA damaging agent. Notably, the reduction in the survival of the pprA mutant following Nal treatment was even more pronounced than that following 6 kGy γ radiation (Figure 1B), a dose that produces nearly 200 DSBs and 3000 single strand breaks in D. radiodurans [2]. The larger magnitude of the killing of the pprA mutant following Nal treatment raises the possibility that PprA contributes to D. radiodurans' survival after DNA damage through mechanisms besides its known role in augmenting DSB repair.
Figure 1. PprA promotes D radiodurans resistance to nalidixic acid.
Different dilutions of exponentially growing cells of D. radiodurans (WT), a pprA:cat mutant (Mutant) and the mutant expressing PprA in trans (Mutant +PprA) were spotted on TGY agar plates in absence (Con) and presence of nalidixic acid (Nal) (20 µg/ml) (A). Similarly, these cells were treated with different doses of γ radiation and different dilutions were spotted on TGY agar plate and growth was monitored (B).
PprA promotes D. radiodurans' growth after exposure to γ radiation or nalidixic acid
Genotoxic treatments lead to varied periods of D. radiodurans growth arrest during which damaged DNA is repaired and the integrity of its multipartite genome is restored. In wild type cells following exposure to either 6 kGy γ radiation or Nal (20 µg/ml), there is a ∼6 hr and ∼9 hr lag respectively before growth resumes (Figure 2). Inactivation of pprA led to prolonged periods of growth arrest after γ radiation treatment (up to ∼12 hr) and especially after Nal treatment (up to ∼18 hr). Notably, PprA does not appear to augment D. radiodurans growth in the absence of DNA damage, as wild type D. radiodurans and the pprA mutant exhibit very similar growth curves (Figure 2). Collectively, these observations suggest that pprA makes a critical contribution to D. radiodurans recovery and/or growth following damage to its DNA, but it is not required for growth in the absence of exogenous genotoxic insult.
Figure 2. PprA promotes D. radiodurans growth following DNA damage.
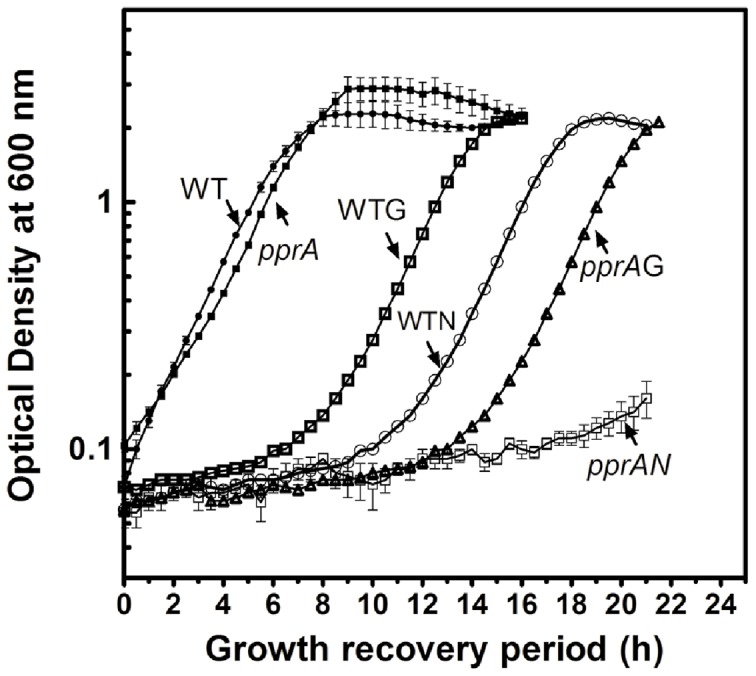
D. radiodurans (WT) and a pprA::cat mutant (pprA) were treated with nalidixic acid (20 µg/ml) for 2 h (WTN, pprAN) and γ radiation (6 kGy) (WTG, pprAG). These cells were washed and resuspended in fresh TGY medium, and growth at 30°C was monitored as optical density 600 nm.
PprA promotes maintenance of the D. radiodurans genome after exposure to DNA damage
In order to begin to explore the reasons for the extended delays in the growth of the pprA mutant following γ radiation and Nal treatments, we used DAPI staining to assess the state of the D. radiodurans nucleoid after exposure to these agents. Under normal growth conditions in TGY medium, it was difficult to detect anucleate cells with DAPI staining in both wild type and pprA mutant cells (Figure 3). However, there was a marked increase in the frequency of anucleate pprA mutant cells following their exposure to Nal (20 µg/ml) for 2 h, or γ radiation (6.0 kGy): 6.7±0.8% and 23.9±1.6% respectively. In contrast, the frequency of anucleate wild type cells increased to only 1.0±0.2% and 2.3±0.2% when wild type cells were subjected to these 2 treatments. Thus, pprA appears to specifically promote the integrity of the D. radiodurans genome after the organism is exposed to exogenous genotoxic conditions and not during routine growth.
Figure 3. PprA promotes D. radiodurans genome maintenance following DNA damage.
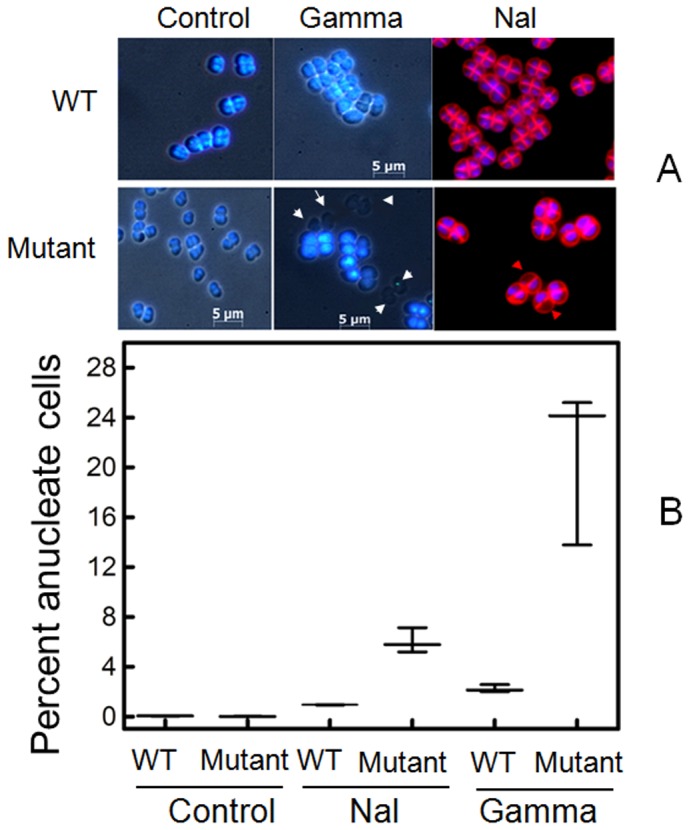
Both wild type (WT) and pprA mutant (Mutant) D. radiodurans cells were treated with nalidixic acid (Nal) for 2 h and 6 kGy γ radiation (gamma) and stained with DAPI to detect anucleate cells. Representative micrographs are shown in (A), where anucleate cells, lacking DAPI fluorescence, are indicated with arrows. The percent anucleate cells in each condition (∼500 cells/sample) is plotted in (B).
Recombinant PprA interacts with recombinant DraTopoIB and stimulates its activity
PprA was found to be present along with several other proteins, including LigB (DR_B0100), DncA (DR_2417), a nuclease [29], and DraTopoIB (DR_0690) in a multiprotein ‘DNA-processing complex’ that is thought to play a critical role in this organism's remarkable resistance to γ radiation. Earlier, PprA interaction with LigB (DR_B0100) and T4 DNA ligase and stimulation of ligase activity of these enzymes has been demonstrated [11], [14]. Since DraTopoIB is known to play an essential role in the maintenance of proper DNA topology by relaxing the positive superturns accumulated during DNA replication, transcription, and recombination [19], [35], [37], we speculated that the PprA – topoisomerase interaction might also be important for maintenance/restoration of the integrity of the D. radiodurans genome following exposure to genotoxic agents. To begin to explore this idea, we tested whether PprA modulated the activity of purified recombinant DraTopoIB using a supercoiled DNA relaxation assay, where conversion of a supercoiled plasmid DNA (CC) to a relaxed molecule (OC) was monitored. On its own, PprA had no effect on the CC to OC conversion (Figure 4A, lane 3), whereas DraTopoIB alone could mediate the conversion of the CC form to the OC form (Figure 4A, lane 2). However, addition of PprA to DraTopoIB markedly increased the ratio of OC to CC (Figure 4B, lanes 4–8), indicating that PprA enhanced the activity of DraTopoIB. The identity of the DNA band located between the OC and CC forms of the DNA, which increased in abundance as more PprA was added to the reaction is unknown.
Figure 4. Effect of purified PprA on D. radiodurans Type IB topoisomerase activity.
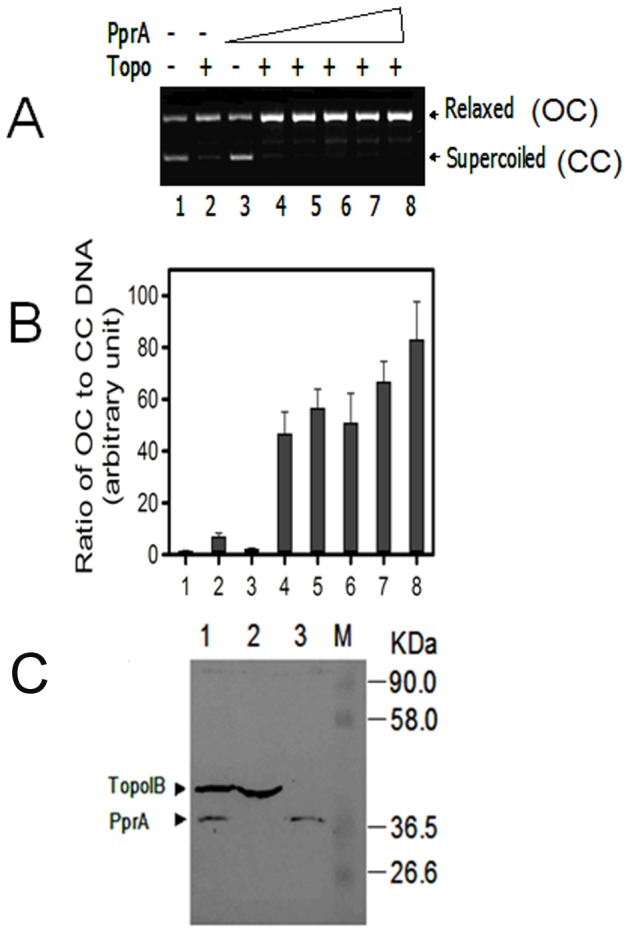
Plasmid DNA having both supercoiled (CC) and relaxed (OC) species was incubated with purified recombinant DraTopoIB (Topo) alone and with increasing molar ratio (1, 2, 3, 4 and 5) of purified PprA. Products were analysed on 1% agarose gels (A). Supercoiled (CC) and relaxed (OC) DNA band intensities were quantified and the ratios of OC to CC were plotted (B). In (C), cell free extracts of E. coli expressing PprA and DraTopoIB from pETpprA and pETtopoIB plasmids respectively were immunoprecipitated with anti PprA antibodies (1). This immunoprecipitate and extracts of cells expressing pETtopoIB (2) and pETpprA (3) independently were immunoblotted with antibodies against (His)6 tag and the sizes of these immunostained bands were compared to the molecular weight marker (M).
Interaction between plasmid-expressed PprA and DraTopoIB in E. coli was confirmed with co-immunoprecipitation experiments. When cell free extracts from E. coli co-expressing His-PprA and DraTopoIB-His were immunoprecipitated with polyclonal antibodies against PprA, two protein bands with sizes corresponding to DraTopoIB and PprA were detected on immunoblots with monoclonal antibodies against the His tag (Figure 4C lane 1). Mixture of cell free extracts having both DraTopoIB and PprA proteins when processed without PprA antibodies did not yield immunosignals with (His)6 antibodies (data not shown)). Together, these observations suggest that PprA and DraTopoIB do not need additional D. radiodurans-specific factors to enable their interaction. Surprisingly, the amount of DraTopoIB was higher than PprA in the immunoprecipitates, raising the possibility that DraTopoIB is present in higher stiochiometric ratio than PprA in the complex.
PprA influences expression of dragyrA and dratopoIB and interacts with DraGyrA in addition to DraTopoIB
To begin to address how the absence of pprA renders D. radiodurans extremely sensitive to Nal, we analyzed the effect of Nal on expression of the dratopoIB and dragyrA genes in the wild type and pprA mutant backgrounds. Nal affected expression of both dratopoIB and dragyrA genes and this effect was more prominent in the absence of PprA (Figure 5A). For example, wild type cells treated with Nal showed ∼6 and ∼1.5 fold reduction in the abundance of dratopoIB and dragyrA transcripts, respectively (Figure 5A, compare lane 1 and 2). However, in Nal treated pprA mutant cells, the abundance of these transcripts decreased by ∼50 fold for dratopoIB and ∼3.3 fold for dragyrA as compared to the respective untreated controls (Figure 5A, compare lane 1 and 2 with lane 3 and 4), indicating that Nal-induced effects on expression of these genes were amplified in absence of PprA.
Figure 5. PprA modulates nalidixic acid's effect on gene expression and PprA interactions with DNA topoisomerases.
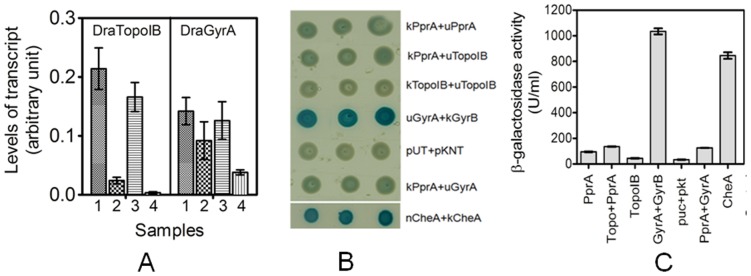
In (A) the effect of nalidixic acid on expression of dratopoIB and dragyrA genes was monitored by RT-PCR. Total RNA was extracted from both wild type (1, 2) and pprA mutant (3, 4) cells grown under normal (1, 3) and in the presence (2, 4) of nalidixic acid (20 µg/ml), and converted to cDNA as described materials and methods. RT-PCR was carried out and levels of expression of both genes were normalized to those of a reference gene, dr1343, encoding glyceraldehydes 3-P dehydrogenase. In (B) E. coli strain BTH101 cells expressing different proteins on plasmids pUTpprA (uPprA) and pKNTpprA (kPprA), pUTtopoIB (uTopoIB), pKNTtopoIB (kTopoIB), pUTgyrA (uGyrA), pKNTgyrB (kGyrA), pUTcheA (uCheA) and pKNTcheA (kCheA) in different combinations were spotted on LB agar plates supplemented with IPTG and X gal. Plates were incubated at 30°C overnight. Results were compared with E. coli strain BTH101 harboring pUT18 and pKNT25 vectors (pUT+pKNT). Similarly in (C) these six strains were induced with 200 µM IPTG for five hours and levels of β -galactosidase activity was monitored on ONPG substrate and blue color product was measured spectrophotometrically. Enzyme activity (U/ml) was calculated as described in materials and methods.
We used the bacterial two hybrid (BTH) system to further evaluate PprA interactions with deinococcal gyrase (TopoII) and DraTopoIB. In these experiments, PprA and the subunits of these topoisomerases were introduced into the pUT18 and pKNT25 expression vectors [30] and β-galactosidase expression was monitored as a way to assess protein interactions as described earlier. We used the previously demonstrated CheA-CheA interaction [38] as a positive control for these experiments. Notably co-expression of DraGyrA and DraGyrB yielded as strong a signal as CheA/CheA in this reporter system (Figure 5BC), consistent with the known interactions of these 2 gyrase subunits in other organisms and also validating that the BTH can be used to detect interactions of D. radiodurans proteins. Although the signal was not as strong as from the DraGyrA and DraGyrB interaction, co-expression of PprA with either DraTopoIB or DraGyrA also yielded β-galactosidase expression that was greater than the vector controls (pUT18 and pKNT25) (Figure 5BC), strongly suggesting that PprA can interact with both of these topoisomerases. PprA was also found to interact with itself in this system, consistent with our unpublished findings that this protein oligomerizes. Together these observations suggest that PprA interacts with both TopoII and TopoIB in D. radiodurans.
Discussion
Our findings underscore the idea that PprA, a protein identified as important for D. radiodurans' remarkable resistance to radiation more than a decade ago [39], has several activities that contribute to the organism's survival in response to genotoxic insults. Previous studies have demonstrated that PprA promotes D. radiodurans DSB repair by stimulating the DNA end joining activity of DNA ligases [11], [14]. In addition to PprA's contribution to DNA repair, recent work suggests PprA plays a role in D. radiodurans cell division and genome segregation [40], though the molecular bases linking PprA to these cellular processes were not defined. Here, we found that PprA stimulates the DNA relaxation activity of DraTopoIB and contributes to D. radioduran's resistance to the TopoII inhibitor Nal and to the maintenance of the damaged genome in this bacterium. Furthermore, we observed that PprA interacts with both types of topoisomerases and it insulated cells from the Nal induced reduction in expression of dragyrA as well as dratopoIB genes in D. radiodurans. Thus, our observations strongly suggest that interactions between PprA and the topoisomerases that control the topology of the D. radiodurans' genome play a heretofore-unappreciated role in PprA's contribution to D. radiodurans' resistance to genotoxic assaults.
It is tempting to speculate that both Topo II, which is inhibited by Nal, and DraTopoIB, which is stimulated by PprA, contribute to maintaining the integrity of the D. radiodurans genome and hence its survival after its DNA has been damaged. While speculative, this model could explain why the pprA mutant is particularly sensitive to damage caused by Nal; in this setting, there is little topoisomerase activity because TopoII is inactivated by the antibiotic as well as its level is reduced, and also DraTopoIB expression as well as activity is reduced due to the absence of PprA. Despite the higher frequency of anucleate cells in the pprA mutant after γ radiation vs Nal treatment, the latter treatment was even more toxic to D. radiodurans than the former (see Figure 1 and Figure 2). The explanation for this discrepancy is not known, but it appears that the net effect of inhibition of both TopoII and DraTopoIB is more toxic to the D. radiodurans cell than the extensive DNA damage caused by γ radiation.
Mechanistically, it is hard to visualize how PprA stimulates two distinct DNA metabolic processes in vivo (end joining and relaxation). However, we know that a multiprotein complex comprised of LigB, DraTopoIB and PprA also exhibited the capacity for both relaxation of superhelical molecules and DNA end joining activity [12]. These two DNA transactions are differentially regulated by ATP in solution. Therefore, we may speculate that PprA has the unique ability to interact with and stimulate other proteins' functions, but protein specific functions depend upon cofactors requirements which may be favorable for one activity and unfavorable for another.
In summary, our findings suggest that PprA contributes to the maintenance of D. radiodurans DNA topology, likely through topoisomerases, in addition to its well-known role in DSB repair. Although, PprA interacts with the GyrA subunit of TopoII, it remains to be seen whether PprA modulates TopoII activity. PprA's facilitation of DNA repair and maintenance of DNA topology both promote the integrity of the D. radiodurans genome. Future studies can test whether these 2 distinct activities depend on PprA's capacity to preferentially bind DNA containing breaks. Regardless of the mechanism(s), it is important to emphasize that the pprA mutant only exhibited detectable phenotypes (growth arrest and increased anucleate cells) following exposure to DNA damaging conditions. Thus, PprA's pleiotropic functions maintaining the integrity of the D. radiodurans genome do not appear to be active under normal growth conditions.
Acknowledgments
Authors are grateful to Dr S. K. Apte, for his constructive criticism and encouragement while pursuing this work. We thank Professor I. Narumi for pprA::cat mutant and Professor Suzanne Sommer for p11559.
Funding Statement
This study was supported by the Bhabha Atomic Research Centre, Harvard Medical School and HHMI. HSM is grateful to Fulbright Commission, United States- India Education Foundation (USIEF) for Fulbright-Nehru Senior Research Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Slade D, Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans. . Microbiol Mol Biol Rev 75: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Battista JR (2000) Radiation resistance: the fragments that remain. Curr Biol 10: R204–205. [DOI] [PubMed] [Google Scholar]
- 3. Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, et al. (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans . Nature 443: 569–573. [DOI] [PubMed] [Google Scholar]
- 4. Slade D, Lindner AB, Paul G, Radman M (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans . Cell 136: 1044–1055. [DOI] [PubMed] [Google Scholar]
- 5. Tian B, Hua Y (2010) Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol 18: 512–520. [DOI] [PubMed] [Google Scholar]
- 6. Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, et al. (2010) Small-molecule antioxidant proteome-shields in Deinococcus radiodurans . PLoS One 5: e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rajpurohit YS, Gopalakrishnan R, Misra HS (2008) Involvement of a protein kinase activity inducer in DNA double strand break repair and radioresistance of Deinococcus radiodurans . J Bacteriol 190: 3948–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levin-Zaidman S, Englander J, Shimoni E, Sharma AK, Minton KW, et al. (2003) Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299: 254–256. [DOI] [PubMed] [Google Scholar]
- 9. Zimmerman JM, Battista JR (2005) A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol 31: 15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286: 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Narumi I, Satoh K, Cui S, Funayama T, Kitayama S, et al. (2004) PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol Microbiol 54: 278–285. [DOI] [PubMed] [Google Scholar]
- 12. Kota S, Misra HS (2008) Identification of a DNA processing complex from Deinococcus radiodurans. . Biochem Cell Biol 86: 448–458. [DOI] [PubMed] [Google Scholar]
- 13. Blasius M, Buob R, Shevelev IV, Hubscher U (2007) Enzymes involved in DNA ligation and end-healing in the radioresistant bacterium Deinococcus radiodurans. . BMC Mol Biol 8: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kota S, Kamble VA, Rajpurohit YS, Misra HS (2010) ATP-type DNA ligase requires other proteins for its activity in vitro and its operon components for radiation resistance in Deinococcus radiodurans in vivo . Biochem Cell Biol 88: 783–790. [DOI] [PubMed] [Google Scholar]
- 15. Forterre P, Gribaldo S, Gadelle D, Serre MC (2007) Origin and evolution of DNA topoisomerases. Biochimie 89: 427–446. [DOI] [PubMed] [Google Scholar]
- 16. Baker NM, Rajan R, Mondragon A (2009) Structural studies of type I topoisomerases. Nucleic Acids Res 37: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng C, Kussie P, Pavletich N, Shuman S (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92: 841–850. [DOI] [PubMed] [Google Scholar]
- 18. Taneja B, Patel A, Slesarev A, Mondragon A (2006) Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J 25: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krogh BO, Shuman S (2002) A poxvirus-like type IB topoisomerase family in bacteria. Proc Natl Acad Sci USA 99: 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garinther WI, Schultz MC (1997) Topoisomerase function during replication-independent chromatin assembly in yeast. Mol Cell Biol 17: 3520–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schaefer M, Schmitz C, Facius R, Horneck G, Milow B, et al. (2000) Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between biological effect and singlet-oxygen production. Photochem Photobiol 71: 514–523. [DOI] [PubMed] [Google Scholar]
- 22. Lecointe F, Coste G, Sommer S, Bailone A (2004) Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans . Gene 336: 25–35. [DOI] [PubMed] [Google Scholar]
- 23. Charaka VK, Misra HS (2012) Functional characterization of chromosome I partitioning system in Deinococcus radiodurans for its role in genome segregation. J Bacteriol 194: 5739–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual third ed., Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- 25. Battista JR, Park MJ, McLemore AE (2001) Inactivation of two homologous proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43: 133–139. [DOI] [PubMed] [Google Scholar]
- 26. Kota S, Misra HS (2006) PprA: A protein implicated in radioresistance of Deinococcus radiodurans stimulates catalase activity in Escherichia coli. Appl Microbiol Biotechnol 72: 790–796. [DOI] [PubMed] [Google Scholar]
- 27. Misra HS, Khairnar NP, Kota S, Shrivastava S, Joshi VP, et al. (2006) An Exonuclease I sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 59: 1308–1316. [DOI] [PubMed] [Google Scholar]
- 28. Ringgaard S, Schirner K, Davis BM, Waldor MK (2011) A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev 25: 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das AD, Misra HS (2012) DR2417, a hypothetical protein characterized as a novel beta-CASP family nuclease in radiation resistant bacterium, Deinococcus radiodurans, Biochim Biophys Acta. 1820: 1052–1061. [DOI] [PubMed] [Google Scholar]
- 30. Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karimova G, Dautin N, Ladant D (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187: 2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou L, Lei XH, Bochner BR, Wanner BL (2003) Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185: 4956–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajpurohit YS, Misra HS (2013) Structure-function study of deinococcal serine/threonine protein kinase implicates its kinase activity and DNA repair protein phosphorylation roles in radioresistance of Deinococcus radiodurans . Int J Biochem Cell Biol 45: 2541–2552. [DOI] [PubMed] [Google Scholar]
- 34. Drlica K, Zhao X (1997) DNA gyrases, topoisomerase IV and the quinolones. Microbiol Mol Biol Rev 61: 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Champoux JJ (2001) DNA topoisomerase: structure, function and mechanism. Ann Rev Biochem 70: 369–413. [DOI] [PubMed] [Google Scholar]
- 36. Hiraku Y, Kawanishi S (2000) Distinct mechanisms of guanine –specific DNA photodamage induced by Nalidixic acid and fluoroquinolone antibacterials. Arch Biochem Biophys 382: 211–218. [DOI] [PubMed] [Google Scholar]
- 37. Corbett KD, Berger JM (2004) Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct 33: 95–118. [DOI] [PubMed] [Google Scholar]
- 38. Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, et al. (1996) Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J Biol Chem 271: 939–945. [DOI] [PubMed] [Google Scholar]
- 39. Narumi I (2003) Unlocking radiation resistance mechanisms: still a long way to go. Trends Microbiol 11: 422–425. [DOI] [PubMed] [Google Scholar]
- 40. Devigne A, Mersaoui S, Bouthier-de-la-Tour C, Sommer S, Servant P (2013) The PprA protein is required for accurate cell division of γ-irradiated Deinococcus radiodurans bacteria. DNA Repair 12: 265–272. [DOI] [PubMed] [Google Scholar]