Abstract
Context
Fetal growth involves highly complex molecular pathways. IGF2 is a key paternally expressed growth hormone that is critical for in utero growth in mice. Its role in human fetal growth has remained ambiguous, as it has only been studied in term tissues. Conversely the maternally expressed growth suppressor, PHLDA2, has a significant negative correlation between its term placental expression and birth weight.
Objective
The aim of this study is to address the role in early gestation of expression of IGF1, IGF2, their receptors IGF1R and IGF2R, and PHLDA2 on term birth weight.
Design
Real-time quantitative PCR was used to investigate mRNA expression of IGF1, IGF2, IGF1R, IGF2R and PHLDA2 in chorionic villus samples (CVS) (n = 260) collected at 11–13 weeks' gestation. Expression was correlated with term birth weight using statistical package R including correction for several confounding factors.
Results
Transcript levels of IGF2 and IGF2R revealed a significant positive correlation with birth weight (0.009 and 0.04, respectively). No effect was observed for IGF1, IGF1R or PHLDA2 and birth weight. Critically, small for gestational age (SGA) neonates had significantly lower IGF2 levels than appropriate for gestational age neonates (p = 3·6×10−7).
Interpretation
Our findings show that IGF2 mRNA levels at 12 weeks gestation could provide a useful predictor of future fetal growth to term, potentially predicting SGA babies. SGA babies are known to be at a higher risk for type 2 diabetes. This research reveals an imprinted, parentally driven rheostat for in utero growth.
Introduction
Fetal growth involves a complex interaction between genes and the environment, with such significant complexity that many of the molecular pathways remain to be elucidated. Normal fetal growth depends on the successful nutrient exchange between the mother and the fetus via the placenta. When this critical balance is impaired it can result in a small for gestational age (SGA) baby [1]. SGA neonates have a reported incidence of ∼6% of pregnancies in developed countries and as high as 40% in some developing countries [2], [3]. SGA is also associated with an increased risk of neonatal death. Although the survivors do exhibit catch up growth, it is important to note that they remain at a higher risk for common, late onset chronic diseases such as type 2 diabetes (T2D) and cardiovascular disease [4]. Hales et al., (1991) reported a graded inverse association between birth weight and T2D risk with the highest risks of T2D occurring at the lowest levels of birth weight [5]. In a more recent meta-analysis, an increased risk for T2D (OR = 1.47) is seen for SGA versus appropriate for gestational age (AGA) neonates [6].
One group of growth genes of particular interest are the imprinted genes that are found almost exclusively in Eutherian (placental) mammals. Genomic imprinting defines allele-specific, differential expression of a gene, according to its parent of origin. If the paternal allele is expressed, the maternal allele is imprinted (silenced) and vice versa. The “parental conflict hypothesis” is still the most widely accepted explanation for the evolution of genomic imprinting [7]. It has been suggested that expression of the father's genes enhance fetal growth improving the success of the paternal genome to be passed on. In contrast, the mother's genome limits fetal growth, distributing equal resources to each of her offspring, whilst ensuring her own survival post birth allowing her to reproduce again. Studies of imprinted genes generally support this model, with one of the most striking examples being the reciprocal imprinting effects and associated growth patterns for mouse insulin-like growth factor 2 (Igf2) and its chelating receptor Igf2r [8]. In transgenic mice, loss of function of the paternally-expressed Igf2 results in a 40% reduction in birth weight which contrasts with loss of the maternally-expressed Igf2r, resulting in a 30% increase in birth weight [9], [10]. In addition, IGF2 has been implicated in two imprinted human growth disorders, the overgrowth, Beckwith-Wiedemann syndrome (BWS) [11] and the growth restricting Silver-Russell syndrome (SRS) [12].
Pleckstrin homology-like domain family A member 2 (PHLDA2) is maternally expressed, paternally imprinted, in both humans and mice [13]. Involvement of Phlda2 in growth and development of the placenta was demonstrated by knockout mouse models that were associated with a significant increase in placental size during mid to late gestation [14]. Studies that have investigated the human placental expression of PHLDA2 report increased expression in SGA pregnancies and a negative association with birth weight [15], [16], [17].
Insulin-like growth factor 1 (IGF1) and its primary binding receptor IGF1R are not imprinted. IGF1 exerts its growth properties on almost every cell in the body. A homozygous partial deletion of Igf1r in mice stunted height and weight, as well as disrupted the pubertal growth spurt. A complete inactivation of Igf1r is lethal in the neonatal period [18]. In contrast to Igf2 deficient mice, restriction of growth was seen to continue into the postnatal period in the Igf1 mutants [19].
The analyses of human placental expression of IGF1, IGF2, IG1R and IGF2R have previously been confined to samples obtained at the time of birth. IGF1 under-expression was observed in term placental samples from SGA pregnancies [20], while mutations in the IGF1R gene led to abnormalities in the function of IGF1 receptors that may also slow down intrauterine and subsequent growth in humans [21].
Studies on IGF2 have reported conflicting results. Some demonstrate no correlation between IGF2 expression and birth weight [16] whereas others have variably shown that in SGA pregnancies compared to controls, IGF2 expression is either increased [22], decreased [15], [23], [24], or similar [25], including at the protein level [26]. Moreover, studies have either shown no significant relationship between IGF2 cord serum levels and size at birth [27], or a positive effect on birth weight [28]. In others, IGF2 cord blood levels were significantly correlated with birth weight only when its interaction with IGF2R was taken into account [29].
The profound role of the IGF1 and IGF2 pathways in the regulation of fetal growth have been established largely based on experiments using the mouse as a model [9], [10], [18]. Analysis in humans has been hampered by the lack of available tissue to study during the course of pregnancy and has therefore relied on the use of term placenta, at a time when this tissue has become redundant and therefore the levels of gene expression may no longer reliably reflect the needs of the growing baby. To fully assess the role of these growth factors and their receptors, we have focused on an earlier developmental time point when these genes are much more likely to measure functionally relevant expression levels.
Methods
Study population
Chorionic villus sampling (CVS) was performed at 11–13 weeks of gestation in 260 singleton pregnancies that subsequently resulted in normal live births at term. The samples were collected from women undergoing CVS for prenatal diagnosis of chromosomal defects at King's College Hospital London. The excess tissue samples used for this study were obtained from women agreeing to participate in research, which was approved by the King's College Hospital Ethics Committee.
Demographic characteristics were recorded including maternal age, racial origin, smoking status, parity and body mass index as well as pregnancy outcomes such as gestational age at delivery, sex and birth weight. Other birth parameters such as placental weight were not available, nor were blood or tissue samples from the newborn baby. The pregnancies were subdivided according to the birth weight of neonates into small for gestational age (SGA) with birth weight <10th percentile, large (LGA) with birth weight >90th percentile and appropriate (AGA).
Preparation of DNA and RNA from chorionic villus samples
DNA and RNA were extracted using the iPrep Purification Instrument (Invitrogen), either by use of the iPrep™ ChargeSwitch® gDNA Tissue Kit, or iPrep™ PureLink™ Total RNA and Trizol® Plus RNA kit including DNAse treatment, according to the manufacturer's instructions.
Reverse transcription
Reverse transcriptase (RT) methodology was based on a standard protocol using M-MLV reverse transcriptase and random primer hexamers (Promega). Primers for the housekeeping gene β-actin (ACTB) were used to check the integrity of the cDNA and ensure no DNA contamination. The forward (F) and reverse (R) primers spanned an intron and the sequences are: β-actin.F- gtcttcccctccatcgtg and β-actin.R- ggtcatcttctcgcggttg.
Polymerase Chain Reaction (PCR)
Genomic DNA and cDNA from CVS were amplified by PCR before sequencing. Primers (5′ to 3′ sequence) used for genomic DNA are IGF2.F- aacaccccacaaaagctcag; IGF2.R- tgcatggattttggttttca; IGF2R.F- gaaacacaaaacctacgacc; IGF2R.R- agaacccaaaagagccaacc; PHLDA2.F- caaaccccgcacgccatgag and PHLDA2.R- ctgtgcccattgcaaataaatc. The same primers were used for cDNA with the exception of IGF2R.R- cctttggagtacgtgacaac. 20 ul reactions were set up and thermal cycling conditions were 94°C for 5 min, 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec for 35 cycles, and 72°C for 2 min.
Imprinting analysis
Imprinting analysis of IGF2, IGF2R and PHLDA2 in CVS was carried out by DNA sequence analysis of expressed single nucleotide polymorphisms (SNP) in CVS gDNA after amplification using specific primers (IGF2.F- aacaccccacaaaagctcag; IGF2R.R- cctttggagtacgtgacaac and PHLDA2.F- caaaccccgcacgccatgag). Corresponding cDNA samples were screened in patients heterozygous for the IGF2 A/G (rs680), IGF2R A/G (rs1805075) and PHLDA2 A/G (rs1056819) SNPs (http://www.ncbi.nlm.nih.gov/projects /SNP/). Sequencing reactions were prepared according to the manufacturer's instructions (Applied Biosystems) using the ABI Prism Big Dye terminator cycle sequencing ready reaction kit (BDT v1.1). Sequencing products were run on an ABI Prism 3730 DNA analyzer, and the read-out was analysed with Sequencher™ v4.8 (Gene Codes Corporation).
Real-time quantitative PCR
The quantitative expression analysis of the genes of interest, IGF1, IGF2, IGF1R, IGF2R and PHLDA2 as well as the endogenous control gene L19 (a housekeeping gene ubiquitously expressed in the placenta) [16] was determined by real-time quantitative PCR (RTqPCR) with SYBR Green (ABI) using the StepOnePlus Real-Time PCR System (ABI). Primers (5′ to 3′ sequence) used for quantitative analysis are IGF1.F- ggaggctggagatgtattgc; IGF1.R- acttgcttctgtcccctcct; IGF2.F- cgagagggacgtgtcgacc; IGF2.R- ggactgcttccaggtgtcata; IGF2R.F- ccggcgtgctctgga; IGF2R.R- ccagagggtcacagtggaaga; IGF1R.F- ccaagggtgtggtgaaagat; IGF1R.R- tccatgatgaccagtgttgg; PHLDA2.F- ccatccccgcagcccaaacc; PHLDA2.R- ccacgtcctagcctgggtcc; L19.F- gcggaagggtacagccaat and L19.R- caggctgtgatacatgtggcg. All primer sets were free of primer-dimer products. The RTqPCR assays were run in triplicate for each sample, with the gene of interest and housekeeping gene run on the same 96-well plates. A control pool of CVS cDNA was also included on each plate. Amplification conditions include initial incubation at 95°C for 10 min and repetitive denaturation at 95°C for 15 sec and annealing at 60°C for 1 min for 40 cycles.
After amplification, quantitative expression levels were obtained using the StepOne software (version 2.1). All triplicate cycle threshold (CT) values were within 1 CT of each other. The quantitative values for each triplicate were averaged and the relative expression of the genes of interest was determined by a ratio of their expression to that of the L19 housekeeping gene for the same sample.
Statistical analysis
Standard multiple linear regression models were used to examine the correlation of IGF1, IGF2, IGF2R, IGF1R and PHLDA2 relative expression values to birth weight after adjustment for gestational age at delivery, sex, parity, maternal age, smoking status and maternal body mass index. Any outliers more than 2 standard deviations from the mean were removed. The analysis of variance (one-way ANOVA) was used to compare the gene expression means between the three different categorical birth weight groups (SGA, AGA, LGA) taking into consideration any confounding factors. Statistical package “R” (version 2.13.0) was used for the analyses and for calculating adjusted R-squared (R2) values. Significance was defined as p<0.05.
Results
In this study, we have investigated the relationship between IGF1, IGF2, IGF2R, IGF1R and PHLDA2 mRNA levels in first-trimester placental tissue with the birth weight of the resultant term new born babies. We correlate this data taking into account the carefully recorded birth parameters that may have a confounding effect (gestational age, sex, parity, maternal BMI and smoking status; figure S1). Regression analysis was performed after standardization to the endogenous control gene L19, in relation to birth weight corrected for confounding factors.
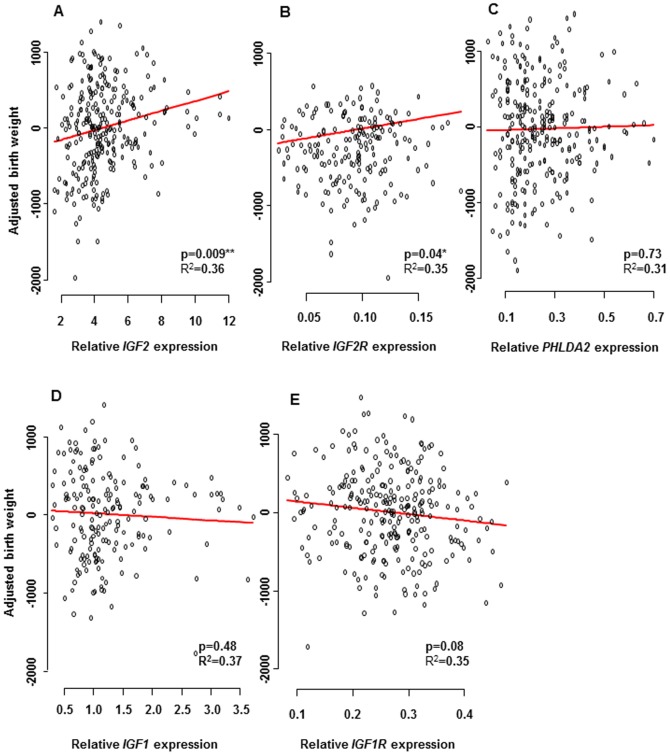
Significant associations were observed for expression of IGF2 (p = 0.009) and IGF2R (p = 0.04) between CVS tissue and birth weight (figure 1A–B). Importantly the range of IGF2 in relative expression units was from 1.5–12 with one relative expression unit being equivalent to a change in birth weight of 63 g. No association was found for PHLDA2 (p = 0.73), IGF1 (p = 0.48) or IGF1R (p = 0.08) between CVS tissue and birth weight (figure 1C–E).
Figure 1. mRNA expression levels of IGF2, IGF2R, PHLDA2, IGF1 and IGF1R in chorionic villi.
The expression levels of IGF2, IGF2R, PHLDA2, IGF1 and IGF1R after standardization to the endogenous control gene L19, in relation to birth weight corrected for parity, sex, GA at term, maternal BMI and smoking status. Significant associations were observed for CVS expression of A) IGF2 (p = 0.009) and B) IGF2R (p = 0.04). No significant association was found for C) PHLDA2 (p = 0.73), for D) IGF1 (p = 0.48) or for E) IGF1R (p = 0.08).
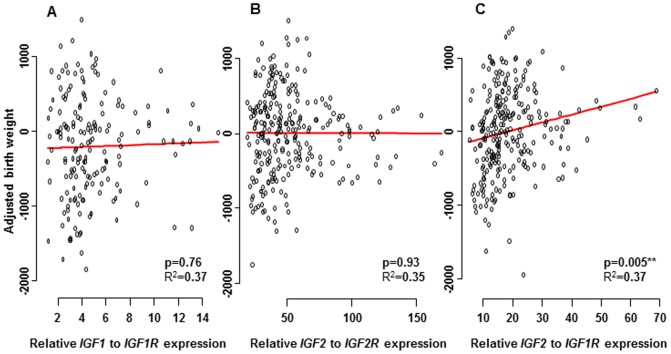
As both IGF1 and IGF2 bind to the IGF1R and only IGF2 binds to the IGF2R we decided to look at this relationship between the ligands and their receptors and any correlation they might have with birth weight. The ratio of IGF2 to IGF1R (p = 0.005) was significantly associated with birth weight but no association was found for the ratio of IGF1 to IGF1R (p = 0.76) or the ratio of IGF2 to IGF2R (p = 0.93) between CVS tissue and birth weight (figure 2).
Figure 2. mRNA expression levels of IGF1/IGF1R, IGF2/IGF2R and IGF2/IGF1R in chorionic villi.
The expression levels of IGF1/IGF1R, IGF2/IGF2R and IGF2/IGF1R after standardization to the endogenous control gene L19, in relation to birth weight corrected for parity, sex, GA at term, maternal BMI and smoking status. No significant association was found for A) the ratio of IGF1 to IGF1R (p = 0.76), or for B) the ratio of IGF2 to IGF2R (p = 0.93). Significant association was observed for CVS expression of C) the ratio of IGF2 to IGF1R (p = 0.005) and birth weight.
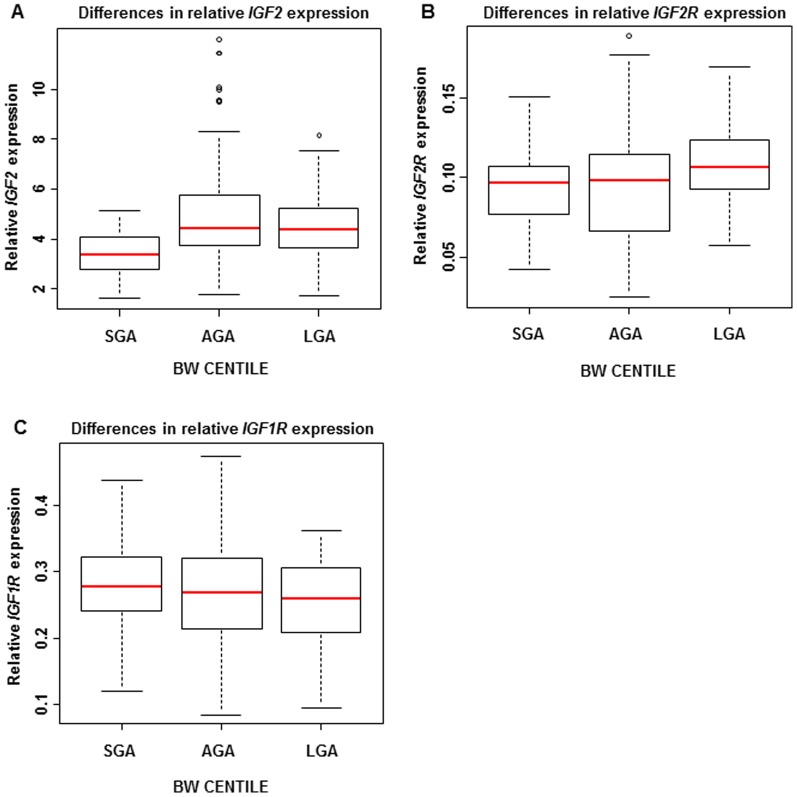
To investigate the whole growth spectrum we subdivided the pregnancies into small for gestational age (SGA; n = 50) with birth weight <10th percentile, large (LGA; n = 65) with birth weight >90th percentile and appropriate (AGA; n = 145). In the SGA group compared to the AGA group, IGF2 expression was reduced by statistically significant amounts (p = 3.6×10−7), despite no significant differences in the expression levels of individual receptors (IGF2R, p = 0.76 or IGF1R, p = 0.15). In the LGA group IGF2R expression was found to be higher than in the AGA group (p = 0.02) and IGF1R expression was lower than in the SGA group (p = 0.05) (figure 3).
Figure 3. mRNA expression levels of IGF2, IGF2R, and IGF1R in chorionic villi according to BW centile.
Relative IGF2, IGF2R, and IGF1R expression levels in the pregnancies with small (SGA), appropriate (AGA) and large (LGA) neonates. In the SGA group, compared to the AGA group, A) IGF2 expression was lower (p = 3·6×10−7), but there was no significant difference in the expression levels of B) IGF2R (p = 0·76) or C) IGF1R (p = 0·15). In the LGA group B) IGF2R expression was higher than in the AGA group (p = 0·02) and C) IGF1R expression was lower than in the SGA group (p = 0·05).
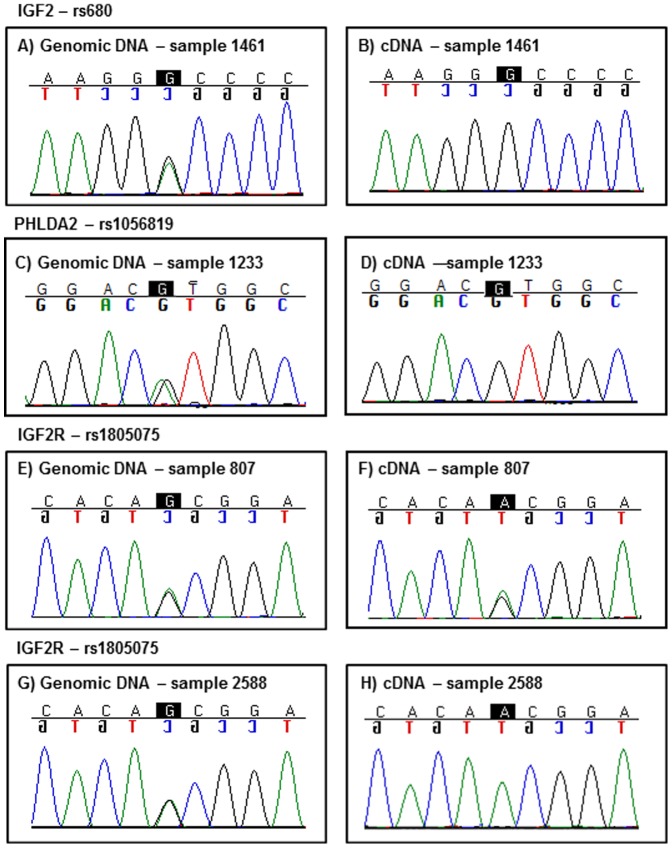
Given that IGF2 and PHLDA2 are both known to be imprinted in mouse and human [8], [13], [30], we wanted to confirm that monoallelic expression was maintained in CVS material, since reversion to biallelic expression may impact on mRNA levels. Furthermore, IGF2R in the human population is reported to be monoallelically expressed (i.e. imprinted) in only 10% of individuals, which is in contrast to the mouse, where it is fully imprinted [31]. We therefore wished to confirm its imprinting status in CVS material and to investigate for any potential skewing. The transcribed SNPs rs680 (IGF2), rs1805075 (IGF2R) and rs1056819 (PHLDA2) were used to report on allele-specific expression and thus determine imprinting status of IGF2, IGF2R and PHLDA2. Genomic DNA extracted from the CVS from 200 patients were tested for heterozygosity at these SNPs, with 40 samples found to be informative at rs680 for IGF2, 24 samples at rs1805075 for IGF2R and 21 samples at rs1056819 for PHLDA2. The 40 informative IGF2 individuals and the 21 informative PHLDA2 individuals were all found to be monoallelically expressed. For the 24 samples informative for IGF2R, 21 (88%) were biallellically expressed for IGF2R and 3 (12%) were monoallelic, which is similar to the expected population frequency (figure 4). Thus, skewed monoallelic/biallelic expression was unlikely to be a factor reflected in the relative expression levels associated with birth weight.
Figure 4. Imprinting analysis of IGF2, PHLDA2 and IGF2R.
Representative sequencing chromatograms are shown from one of the 40 informative CVS samples for the IGF2 A/G polymorphism (rs680) (A, B), one (from 21) for the PHLDA2 A/G polymorphism (rs1056819) (C, D) and two (from 24) for the IGF2R A/G polymorphism (rs1805075) (E, F, G, H). Heterozygous genomic CVS DNA sequence is shown in A), C), E) and G). While monoallelic expression from the corresponding cDNA samples are shown in B), D) and H). Monoallelic expression in cDNA was found in all informative IGF2 and PHLDA2 samples as well as 12% of IGF2R samples. Biallelic expression, as shown in F), was found in the majority (88%) of informative cDNA samples for IGF2R.
Discussion
Previous studies have linked IGF2 expression to birth weight, by showing that IGF2 in term placenta is decreased in SGA pregnancies compared to controls [23], [24]. However, the first trimester IGF2 expression data reported in this study, provides the first evidence for the role of this paternally expressed imprinted gene as an in utero fetal growth enhancer this early in human pregnancy. This is compatible with results previously described in animal studies [9], [32]. For example, in mouse experiments, paternally expressed Igf2 was reported to control 40% of fetal growth, with the maternally expressed gene Igf2r limiting fetal growth [10], [33]. In humans, IGF2 is consistently maternally imprinted and therefore a paternal-expression driven growth promoter. However, in humans, IGF2R is polymorphic for its imprinting status with 90% of individuals showing biallelic expression [31]. This indicates a possible shift in the mechanism of its regulation of expression away from that used in the mouse. Interestingly, here, the samples showing monallelic expression of IGF2R had similar levels to those showing biallelic expression.
As fetal size is subject to both positive and negative regulation, IGF2 and IGF2R genes can exert opposite forces on the fetus, achieving a fine degree of control over the growth process. As levels of IGF2 increase in the fetus, the levels of IGF2R can also increase, resulting in clearance of IGF2 from plasma and tissue fluids, correcting and maintaining levels of IGF2 in the circulation. This acts as a fine-tuning rheostat designed to prevent babies from overgrowth in order to maintain a normal growth trajectory. This is also supported by the fact that no association was observed between the IGF2/IGF2R expression levels and birth weight, as the ligand and the receptor work tightly together to regulate growth. The same applies for IGF1 and its regulating receptor IGF1R.
In this study, expression of IGF1 in chorionic villi was not associated with birth weight outcome. These results directly contradict findings from studies involving mouse models which highlighted IGF1 role in the regulation of both pre- and postnatal growth [34]. There also remains a strong possibility that IGF1 is indeed relevant to intrauterine growth in the later stages of pregnancy as it is preparing the baby for a postnatal life. While fetal chorionic fetal samples in the present cohort proved an opportunity to assess correlation between gene expression and fetal growth in early gestation (11–13 weeks), this narrow window might not be the best to investigate IGF1 expression, as IGF1 does not appear in the fetal circulation until later [35].
Although previous studies have shown that placental PHLDA2 expression is negatively associated with birth weight [16], [17], in our study no statistically significant association was observed between PHLDA2 expression levels in CVS and birth weight. This suggests that maternally expressed PHLDA2 is suppressing the baby's growth later in pregnancy rather than early on. Previous studies investigating term placental IGF2 expression have shown inconclusive results. However, in our study a statistically significant association was observed between IGF2 expression levels in CVS and birth weight, suggesting that the paternal genome is promoting the baby's growth earlier in pregnancy. Therefore, the two parental genomes appear to be acting at different times during pregnancy to control the fetal weight. This supports the idea that whilst increased fetal growth is important early on, it must still require careful regulation by the mother to ensure a successful birth.
In our study, SGA neonates had significantly lower IGF2 expression levels compared to AGA neonates, but no differences were observed between the levels of the receptors IGF2R and IGF1R. This highlights the importance of the IGF2 ligand rather than the receptor level in determining size of the neonate in SGA pregnancies. This is in agreement with previous studies investigating IGF2 mRNA levels in the placentas from growth-restricted pregnancies [23], [24].
To better understand the causes and consequences of fetal growth restriction as a human pregnancy complication, we focused on available tissue from the first trimester, as this time window is more likely to accurately reflect the in utero growth potential. We used a larger sample size and we investigated the whole growth spectrum including not only SGA but also LGA neonates. We reasoned that extending the investigation of these genes to samples displaying macrosomic birth weight would also reveal further correlations between growth parameters and gene expression. In this study, LGA neonates had higher IGF2R expression levels compared to AGA neonates and lower IGF1R levels compared to SGA babies. Similar results have been reported in placenta tissue for IGF1R mRNA levels alone [36], and this suggests that fetuses with high birth weight are producing more IGF2R and correspondingly less IGF1R, which could remove IGF2. This balance may represent another important compensatory mechanism in response to fetal overgrowth. This is also supported by the fact that a significant association was observed between the ratio of IGF2 to IGF1R and birth weight, suggesting that the levels of IGF1R in LGA neonates decrease as their IGF2 levels increase, to avoid any further increase in size.
The “small baby syndrome hypothesis” proposed by Barker et al., (1993) suggests that there is an inverse linear relation between birth weight and T2D [37]. This is also supported by a large meta-analysis [38]. The mechanisms by which birth weight is related to T2D is still under debate, Barker et al. claim that this relationship reflects long-term consequences of under-nutrition in utero [37], whereas others do not see under-nutrition as playing a significant role [39]. Interestingly, specific fetal IGF2 paternal haplotypes are linked to higher maternal glucose levels that increase the risk of gestational diabetes [40]. Nevertheless the amount of IGF2 available from the placenta early in the first trimester is likely to be a major factor in promoting growth.
An important aim for antenatal care is the prediction, detection and treatment of anomalous fetal growth. We know that variation in size at birth results from interaction between fetal genetic factors and the maternal genetic and uterine environment. In this study we also implicate the important role of the father's genes in fetal growth in utero. Understanding the developmental role, function and parent-of-origin effect of critical imprinted genes during human fetal growth will make an important contribution to effective clinical evaluation of growth disorders and their association with longer term, metabolically related, health risks such as T2D. Determination of paternal IGF2 expression in the first trimester, potentially in maternal blood, may act as a predictor of fetal growth trajectory for later in the pregnancy. An early growth biomarker could be invaluable to alert Obstetricians and Neonatologists towards closer ‘at risk’ pregnancy surveillance.
Supporting Information
Confounding factors. Correlation of birth weight with gestational age at term (p = 4.8×10−15; Fig. S1A), with maternal BMI (p = 0.0012; Fig. S1B), with gender (p = 1.4×10−7; Fig. S1C), with parity (p = 7.5×10−5; Fig. S1D), and maternal smoking status (p = 0.33; Fig S1E).
(TIF)
Funding Statement
CD is funded by Save the Baby Unit (http://www.savethebabyunit.org/). PS is funded by Great Ormond Street Hospital Children's Charity (http://www.gosh.org/gen/). GEM Fetal growth and development research team is funded by the MRC (http://www.mrc.ac.uk/index.htm), Wellbeing of Women (http://www.wellbeingofwomen.org.uk/), Sparks (http://www.sparks.org.uk/) and the Wellcome Trust (http://www.wellcome.ac.uk/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pollack RN, Divon MY (1992) Intrauterine growth retardation: definition, classification, and etiology. Clin Obstet Gynecol 35: 99–107. [DOI] [PubMed] [Google Scholar]
- 2. Brodsky D, Christou H (2004) Current concepts in intrauterine growth restriction. J Intensive Care Med 19: 307–319. [DOI] [PubMed] [Google Scholar]
- 3. Albertsson-Wikland K, Wennergren G, Wennergren M, Vilbergsson G, Rosberg S (1993) Longitudinal follow-up of growth in children born small for gestational age. Acta Paediatr 82: 438–443. [DOI] [PubMed] [Google Scholar]
- 4. Barker DJ (1992) Fetal growth and adult disease. Br J Obstet Gynaecol 99: 275–276. [DOI] [PubMed] [Google Scholar]
- 5. Hales CN, Barker DJ, Clark PM, Cox LJ, Fall C, et al. (1991) Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 303: 1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harder T, Rodekamp E, Schellong K, Dudenhausen JM, Plagemann A (2007) Birth weight and subsequent risk of type 2 diabetes: A meta-analysis. Am J Epidemiol 165: 849–857. [DOI] [PubMed] [Google Scholar]
- 7. Moore T, Haig D (1991) Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49. [DOI] [PubMed] [Google Scholar]
- 8. Willison K (1991) Opposite imprinting of the mouse Igf2 and Igf2r genes. Trends Genet 7: 107–109. [DOI] [PubMed] [Google Scholar]
- 9. DeChiara TM, Efstratiadis A, Robertson EJ (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345: 78–80. [DOI] [PubMed] [Google Scholar]
- 10. Wang ZQ, Fung MR, Barlow DP, Wagner EF (1994) Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372: 464–467. [DOI] [PubMed] [Google Scholar]
- 11. Weksberg R, Shen DR, Fei YL, Song QL, Squire J (1993) Disruption of insulin-like growth factor-2 in Beckwith Wiedemann syndrome. Nat Genet 5: 143–149. [DOI] [PubMed] [Google Scholar]
- 12. Eggermann T, Meyer E, Obermann C, Heil I, Schuler H, et al. (2005) Is maternal duplication of 11p15 associated with Silver-Russell syndrome? J Med Genet 42: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qian N, Frank D, O'Keefe D, Dao D, Zhao L, et al. (1997) The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum Mol Genet 6: 2021–2029. [DOI] [PubMed] [Google Scholar]
- 14. Frank D, Fortino W, Clark L, Musalo R, Wang W, et al. (2002) Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci USA 99: 7490–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, et al. (2006) Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27: 540–549. [DOI] [PubMed] [Google Scholar]
- 16. Apostolidou S, Abu-Amero S, O'Donoghue K, Frost J, Olafsdottir O, et al. (2007) Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med 85: 379–387. [DOI] [PubMed] [Google Scholar]
- 17. Ishida M, Monk D, Duncan AJ, Abu-Amero S, Chong J, et al. (2012) Maternal inheritance of a promoter variant in the imprinted PHLDA2 gene significantly increases birth weight. Am J Hum Genet 90: 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klammt J, Pfaffle R, Werner H, Kiess W (2008) IGF signaling defects as causes of growth failure and IUGR. Trends Endocrinol Metab 19: 197–205. [DOI] [PubMed] [Google Scholar]
- 19. Le Roith D, Scavo L, Butler A (2001) What is the role of circulating IGF-I? Trends Endocrinol Metab 12: 48–52. [DOI] [PubMed] [Google Scholar]
- 20. Koutsaki M, Sifakis S, Zaravinos A, Koutroulakis D, Koukoura O, et al. (2011) Decreased placental expression of hPGH, IGF-I and IGFBP-1 in pregnancies complicated by fetal growth restriction. Growth Horm IGF Res 21: 31–36. [DOI] [PubMed] [Google Scholar]
- 21. Abuzzahab MJ, Schneider A, Goddard A, Grigorescu F, Lautier C, et al. (2003) IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N Engl J Med 349: 2211–22. [DOI] [PubMed] [Google Scholar]
- 22. Abu-Amero SN, Ali Z, Bennett P, Vaughan JI, Moore GE (1998) Expression of the insulin-like growth factors and their receptors in term placentas: a comparison between normal and IUGR births. Mol Reprod Dev 49: 229–235. [DOI] [PubMed] [Google Scholar]
- 23. Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, et al. (2011) Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med 28: 481–487. [DOI] [PubMed] [Google Scholar]
- 24. Guo L, Choufani S, Ferreire J, Smith A, Chitayat D, et al. (2008) Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Developmental Biology 320: 79–91. [DOI] [PubMed] [Google Scholar]
- 25. Street ME, Seghini P, Fieni S, Ziveri MA, Volta C, et al. (2006) Changes in interleukin-6 and IGF system and their relationships in placenta and cord blood in newborns with fetal growth restriction compared with controls. Eur J Endocrinol 155: 567–574. [DOI] [PubMed] [Google Scholar]
- 26. Laviola L, Perrini S, Belsanti G, Natalicchio A, Montrone C, et al. (2005) Intrauterine growth restriction in humans is associated with abnormalities in placental insulin-like growth factor signaling. Endocrinology 146: 1498–1505. [DOI] [PubMed] [Google Scholar]
- 27. Klauwer D, Blum WF, Hanitsch S, Rascher W, Lee PD, et al. (1997) IGF-I, IGF-II, free IGF-I and IGFBP-1, -2 and -3 levels in venous cord blood: relationship to birthweight, length and gestational age in healthy newborns. Acta Paediatr 86: 826–833. [DOI] [PubMed] [Google Scholar]
- 28. Smerieri A, Petraroli M, Ziveri MA, Volta C, Bernasconi S, et al. (2011) Effects of cord serum insulin, IGF-II, IGFBP-2, IL-6 and cortisol concentrations on human birth weight and length: pilot study. PLoS One 6: e29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ong K, Kratzsch J, Kiess W, Costello M, Scott C, et al. (2000) Size at birth and cord blood levels of insulin, insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-1 (IGFBP-1), IGFBP-3, and the soluble IGF-II/mannose-6-phosphate receptor in term human infants. The ALSPAC Study Team. Avon Longitudinal Study of Pregnancy and Childhood. J Clin Endocrinol Metab 85: 4266–4269. [DOI] [PubMed] [Google Scholar]
- 30. Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C (1993) Parental genomic imprinting of the human IGF2 gene. Nat Genet 4: 98–101. [DOI] [PubMed] [Google Scholar]
- 31. Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, et al. (2006) Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA 103: 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Constância M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, et al. (2002) Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417: 945–948. [DOI] [PubMed] [Google Scholar]
- 33. Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, et al. (1994) Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev 8: 2953–2963. [DOI] [PubMed] [Google Scholar]
- 34. Le Bouc Y, Gircquel C, Holzenberger M (2003) Physiology of somatotropic axis: interest of gene inactivation experiments. Bull Acad Natl Med 187: 1225–43. [PubMed] [Google Scholar]
- 35. Demendi C, Börzsönyi B, Nagy ZB, Rigó J Jr, Pajor A, et al. (2011) Gene expression patterns of insulin-like growth factor 1, 2 (IGF-1, IGF-2) and insulin-like growth factor binding protein 3 (IGFBP-3) in human placenta from preterm deliveries: influence of additional factors. Eur J Obstet Gynecol 160: 40–44. [DOI] [PubMed] [Google Scholar]
- 36. Iniguez G, Gonzalez CA, Argandona F, Kakarieka E, Johnson MC, et al. (2010) Expression and protein content of IGF-I and IGF-I receptor in placentas from small, adequate and large for gestational age newborns. Horm Res Paediatr 73: 320–327. [DOI] [PubMed] [Google Scholar]
- 37. Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, et al. (1993) Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 36: 62–67. [DOI] [PubMed] [Google Scholar]
- 38. Whincup PH, Kaye SJ, Owen CG, Huxley R, Cook DG, et al. (2008) Birth weight and risk of type 2 diabetes A systematic review. JAMA 300: 2886–2897. [DOI] [PubMed] [Google Scholar]
- 39. Hofman PL, Regan F, Jackson WE, Jefferies C, Knight DB, et al. (2004) Premature birth and later insulin resistance. N Engl J Med 351: 2179–2186. [DOI] [PubMed] [Google Scholar]
- 40. Petry CJ, Seear RV, Wingate DL, Manico L, Acerini CL, et al. (2011) Associations between paternally transmitted fetal IGF2 variants and maternal circulating glucose concentartions in pregancy. Diabetes 60: 3090–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confounding factors. Correlation of birth weight with gestational age at term (p = 4.8×10−15; Fig. S1A), with maternal BMI (p = 0.0012; Fig. S1B), with gender (p = 1.4×10−7; Fig. S1C), with parity (p = 7.5×10−5; Fig. S1D), and maternal smoking status (p = 0.33; Fig S1E).
(TIF)