Abstract
The Auxin/Indole-3-Acetic Acid (Aux/IAA) and Auxin Response Factor (ARF) are two important families that play key roles in auxin signal transduction. Both of the families contain a similar carboxyl-terminal domain (Domain III/IV) that facilitates interactions between these two families. In spite of the importance of protein-protein interactions among these transcription factors, the mechanisms involved in these interactions are largely unknown. In this study, we isolated six intragenic suppressors of an auxin insensitive mutant, Osiaa23. Among these suppressors, Osiaa23-R5 successfully rescued all the defects of the mutant. Sequence analysis revealed that an amino acid substitution occurred in the Tryptophan (W) residue in Domain IV of Osiaa23. Yeast two-hybrid experiments showed that the mutation in Domain IV prevents the protein-protein interactions between Osiaa23 and OsARFs. Phylogenetic analysis revealed that the W residue is conserved in both OsIAAs and OsARFs. Next, we performed site-specific amino acid substitutions within Domain IV of OsARFs, and the conserved W in Domain IV was exchanged by Serine (S). The mutated OsARF(WS)s can be released from the inhibition of Osiaa23 and maintain the transcriptional activities. Expression of OsARF(WS)s in Osiaa23 mutant rescued different defects of the mutant. Our results suggest a previously unknown importance of Domain IV in both families and provide an indirect way to investigate functions of OsARFs.
Introduction
The phytohormone auxin is critical for plant growth and development, including lateral root development, embryonic development, tropic responses, apical dominance and vascular development [1]. Auxin is also involved in the crown root initiation and quiescent center maintenance in rice [2], [3], [4].
The transmission of auxin signaling is controlled by the interaction between Aux/IAA and ARF proteins [5]. Limited interaction studies suggested that, in the absence of auxin, the Aux/IAA repressors interact with ARFs and recruit co-repressors of TOPLESS (TPL) family, preventing the ARFs from regulating auxin response genes [6]. In the presence of auxin, the Aux/IAA proteins are degraded by the ubiquitin-proteasome pathway. In this process, auxin promotes the interaction between Aux/IAA proteins and TIR1 (Transport Inhibitor Response1) F-box (or its homologues) of the SCF (Skp1-Cullin-F-box) complex that acts as an auxin co-receptor [7], [8], [9], [10]. Destruction of the Aux/IAA repressors would then release the ARFs to regulate the transcription of auxin response genes [11]. Aux/IAA and ARF proteins contain a similar carboxyl-terminal domain (Domain III/IV), and Domain III/IV serve for dimerization with Aux/IAA and ARF proteins [12], [13], [14]. Most ARF proteins consist of a conserved amino-terminal DNA-binding domain that recognize TGTCTC auxin response elements in promoters of genes responding to auxin, followed by a middle region that functions as an activation domain or repression domain, and ending with Domain III/IV similar to those in the Aux/IAAs in the carboxyl-terminal domain [15].
The Aux/IAAs are short-lived nuclear proteins and generally contain four conserved domains (i.e., referred to as Domain I, II, III and IV) [16]. The rapid degradation of Aux/IAA proteins require the core sequence GWPPV at positions 4–8 in the 13-amino acid consensus sequence in Domain II [17]. The Domain II interacts with the SCF complex in an auxin-dependent manner and confers instability to the Aux/IAA proteins [14], [18]. Mutations in the core sequence of Domain II block the degradation of Aux/IAAs and interrupt the transmission of the auxin signaling pathway by constitutively suppressing ARF activity. Many auxin-insensitive mutants containing gain-of-function mutant alleles of iaa have been reported in rice [4], [19], [20]. Of all the iaa mutants, Osiaa23 was the most interesting mutant reported in rice. Osiaa23 exhibits pleiotropic defects in both root and shoot [4]. This implies that a number of OsARFs have been suppressed by the stabilized Osiaa23. However, the mechanisms in protein-protein interactions between Osiaa23 and OsARFs and the functions of these OsARFs are still unknown.
In this research, we isolated six intragenic suppressors of Osiaa23. One of these suppressors rescued all the defects of Osiaa23. Sequence analysis revealed that an amino acid substitution occurred in a conserved W residue in Domain IV of Osiaa23. Yeast two-hybrid experiments and analysis of transgenic plants expressing mutated OsARF(WS)s in the background of Osiaa23 revealed a previously unknown importance of Domain IV in both families and provide an indirect way to investigate functions of OsARFs.
Materials and Methods
Plant growth conditions
Rice was grown in culture solution in growth room at temperature regimes of 28/22°C (day/night) and 70% humidity under a 12-h photoperiod. The hydroponic solution contained 3.0 mM NH4NO3, 1.0 mM CaCl2, 0.32 mM NaH2PO4, 0.51 mM K2SO4, 1.65 mM MgSO4, 3.13 mM MnCl2, 1.52 mM (NH4)6Mo7O24, 1.5 mM H3BO3, 1.5 mM ZnSO4, 1.6 mM CuSO4, 35 mM FeCl3, and 70 mM citric acid. The pH of the solution was adjusted to 5.5.
Isolation of suppressors of Osiaa23-3
Osiaa23-3 is a weak allele of Osiaa23 in the genetic background of indica cultivar ‘Kasalath’. The suppressors of Osiaa23-3 were screened from M2 population of EMS treated Osiaa23-3 seeds. Osiaa23-3 has no lateral roots, so 7-day-old seedlings with lateral roots were isolated as suppressors. The OsIAA23 genes of all the suppressors were cloned and sequenced for checking the intragenic mutations. We screened 20,000 M2 plants and isolated six suppressors of Osiaa23-3. Sequence analysis showed that they were all intragenic suppressors.
Yeast two-hybrid analysis
Yeast two-hybrid analysis was performed according to the instructions for the MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech). The coding sequences of Osiaa23-3 and Osiaa23-R5 were amplified by primers OsIAA23-U and OsIAA23-L, cloned into pGBKT7 and transformed into yeast Y187. The coding sequences of OsARFs and mutated OsARF(WS)s were amplified by primers OsARF6-U and OsARF6-L for OsARF6 and OsARF6(WS), OsARF12-U, OsARF12-L for OsARF12 and OsARF12(WS), OsARF16-U, OsARF16-L for OsARF16 and OsARF16(WS), OsARF17-U, OsARF17-L for OsARF17 and OsARF17(WS), OsARF25-U, OsARF25-L for OsARF25 and OsARF25(WS). These coding sequences were cloned into pGADT7 and transformed into yeast AH109. Yeast Y187 containing Osiaa23-3 or Osiaa23-R5 were mated with AH109 containing OsARFs or OsARF(WS)s, according to the manufacturer's protocol. Mated strains were spread on low stringency SD–Leu/–Trp and high stringency SD–Ade/–His/–Leu/–Trp. The sequences of primers are listed in Table S2. Interaction of pGBKT7-53 and pGADT7-T was used as a positive control, and non-interaction of pGBKT7-Lam and pGADT7-T was used as a negative control.
Self-activation test in the yeast system
The MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech) was used to detect the autonomous activation of OsARF16 and OsARF16(WS). The coding sequences of OsARF16 and OsARF16(WS) were amplified by primers OsARF16-SU and OsARF16-SU, and inserted into pGBKT7 in-frame fused with the GAL4 DNA-BD. The fusion constructs were transformed into yeast strain AH109 and selected on the minimal medium SD/-Trp and SD/-Trp-His-Ade to examine the reporter gene expression. The sequences of primers are listed in Table S2.
PCR site-directed mutagenesis of OsARFs
PCR Site-Directed Mutagenesis of OsARFs were performed according to the instructions for the Fast Mutagenesis System (FM111, TransGenBiotech). The primers were OsARF6-MU, OsARF6-ML for OsARF6, OsARF12-MU, OsARF12-ML for OsARF12, OsARF16-MU, OsARF1-ML for OsARF16, OsARF17-MU, OsARF17-ML for OsARF17 and OsARF25-MU, OsARF25-ML for OsARF25. The sequences of primers are listed in Table S2.
Construction of vectors and transgenic plants development
The coding sequences of OsARF(WS)s were amplified by OsARF6-PU, OsARF6-PL for OsARF6(WS), OsARF12-PU, OsARF12-PL for OsARF12(WS), OsARF16-PU, OsARF16-PL for OsARF16(WS), OsARF17-PU, OsARF17-PL for OsARF17(WS) and OsARF25-PU, OsARF25-PL for OsARF25(WS). These coding sequences were cloned into a binary vector pHB, which had 35S promoter to drive these coding sequences. The sequences of primers are listed in Table S2.
These constructs were transformed into callus initiated from mature Osiaa23-3 seeds by Agrobacterium tumefaciens (strain EHA105)-mediated transformation [21].
RT-PCR analysis
For RT-PCR experiments, 5 µg of total RNA was denatured at 65°C for 5 min followed by quick chill on ice in a 14-μl reaction containing 1 µl oligo (dT)12–18(500 µg ml−1) primer, and 1 µl of 10 mM dNTP mixture (10 mM each dATP, dGTP, dCTP, and dTTP at neutral pH). After addition of 4 µl 5×reaction buffer (Promega), the reaction was incubated at 37°C for 2 min, 1 µl (200 units) of M-MLV RTa (Promega) was added to the reaction and incubated at 42°C for another 50 min. After terminating, the reaction was heated at 70°C for 15 min for inactivating. The primers used were as follows: OsARF6-RTU, OsARF6-RTL for OsARF6, OsARF12-RTU, OsARF12-RTL for OsARF12, OsARF16-RTU, OsARF16-RTL for OsARF16, OsARF17-RTU, OsARF17-RTL for OsARF17, OsARF25-RTU, OsARF25-RTL for OsARF25 and OsACTIN-RTU, OsACTIN-RTL for OsACTIN. The sequences of primers are listed in Table S2.
Results
Intragenic mutations rescued the defects of Osiaa23-3 mutant
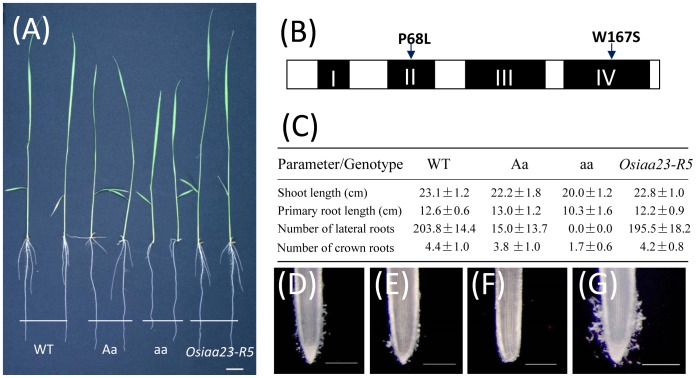
In the previous research, we reported an auxin insensitive mutant designated as Osiaa23, which had multiple defects in both root and shoot development [4]. The Osiaa23-3 is a weak allele of Osiaa23. The defects of Osiaa23-3 are similar to that of Osiaa23 reported before, except that Osiaa23-3 can produce a few crown roots and can complete the life cycle (Figure 1). This weak allele was used in this research. In order to investigate different auxin signaling pathways corresponding to different defects, homozygous seeds of Osiaa23-3 were mutagenized with ethyl methane sulfonate (EMS), and the M2 plants were screened for suppressors of Osiaa23-3. After the morphological screen of about 20,000 M2 plants, we successfully isolated six suppressors with different extent of recovery (Figure S1). Sequence analysis showed that all the suppressors had second site mutations in the Osiaa23 gene. Furthermore, all the mutations changed the amino acids between Domain III and Domain IV of Osiaa23 (Figure S2), which is considered to be essential for the interaction between IAAs and ARFs [13].
Figure 1. Intragenic mutation in Domain IV fully rescued the defects of Osiaa23-3 mutant.
(A) Phenotypes of 7-day-old seedlings of wild type (WT), heterozygous mutant of Osiaa23-3 (Aa), homozygous mutant of Osiaa23-3 (aa) and the suppressor of Osiaa23-3, Osiaa23-R5. Bar = 2 cm. (B) Diagram of conserved domains of OsIAA23. Black rectangles marked by Roman numerals indicate four domains of OsIAA23, and two mutated sites in Domain II and Domain IV are marked by arrows. (C) Growth parameters of 7-day-old seedlings of WT, Aa, aa and Osiaa23-R5. (D-G) Root tips of 7-day-old seedlings of WT (D), Aa (E), aa (F) and Osiaa23-R5 (G). Bars = 250 µm.
Amino acid substitution in Domain IV of Osiaa23 prevents the protein-protein interactions between Osiaa23 and OsARFs
Of all the suppressors isolated, Osiaa23-R5 fully rescued all the defects in Osiaa23-3 (Figure 1; Figure S3). This indicated that all the functions of OsARFs blocked by Osiaa23 have been released in Osiaa23-R5. Sequence analysis of Osiaa23-R5 showed that a second point mutation occurred in Osiaa23 resulting in an amino acid substitution from W to S in Domain IV (Figure 1B; Figure S2). To confirm that the amino acid substitution in Domain IV prevents the protein-protein interactions between Osiaa23 and OsARFs, we selected the appropriate OsARF candidates for yeast two-hybrid assay.
We chose OsARFs which had high expression in the root tip (Based on our previous microarray results of gene expression profile in rice root tip, Table S1) and the full-length cDNAs of OsARF6, OsARF12, OsARF17 and OsARF25 were cloned. In addition, we also cloned the OsARF16, the homolog of ARF7 and ARF19, which play key roles in lateral root initiation in Arabidopsis [22], [23]. To confirm the microarray results and explore their expression profiles in other tissues in rice, RT-PCRs were performed with total RNAs isolated from root, shoot base in young seedlings and stem, leaf, young panicle in adult plants. The analysis showed that all the five OsARFs are expressed in the selected tissues and none of the OsARFs have the tissue specific expression pattern (Figure S4).
The coding sequences of Osiaa23, Osiaa23-R5 and five OsARFs (OsARF6, OsARF12, OsARF16, OsARF17 and OsARF25) were inserted into the yeast expression vectors pGADT7 and pGBKT7 respectively. All the transformed yeast cells formed colonies on the medium with histidine and ade (SD -Leu/ -Trp), indicating a successful transformation of these vectors (Figure 2A), while on the medium without histidine and ade (SD -Leu/ -Trp/ -His/ -Ade), only the transformed yeast cells expressing both Osiaa23 and any of the OsARFs formed colonies, none of the transformed yeast cells formed colonies when Osiaa23-R5 was used instead of Osiaa23 (Figure 2B). These results indicated that Osiaa23 can interact with selected OsARFs and the W residue in Domain IV of Osiaa23 is crucial for the interactions.
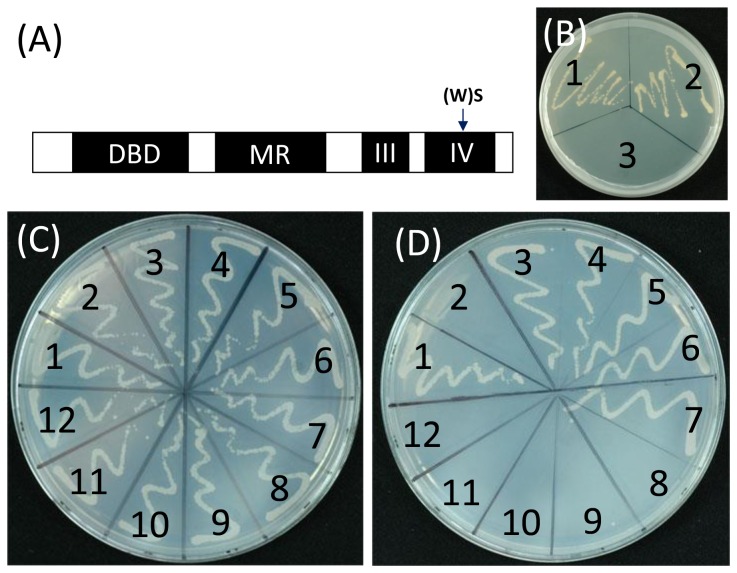
Figure 2. Amino acid substitution in Domain IV of Osiaa23 prevents the protein-protein interactions between Osiaa23 and OsARFs.
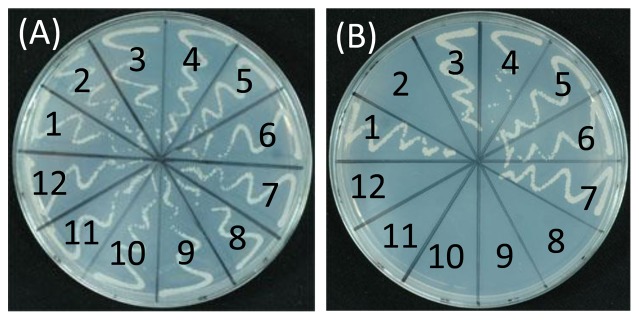
Interactions between Osiaa23 and OsARFs, Osiaa23-R5 and OsARFs in the yeast two-hybrid system. 1, positive control; 2, negative control; 3, Osiaa23 + OsARF6; 4, Osiaa23 + OsARF12; 5, Osiss23 + OsARF16; 6, Osiaa23 + OsARF17; 7, Osiaa23 + OsARF25; 8, Osiaa23-R5 + OsARF6; 9, Osiaa23-R5 + OsARF12; 10, Osiaa23-R5 + OsARF16; 11, Osiaa23-R5 + OsARF17; 12, Osiaa23-R5 + OsARF25. Yeast was grown on medium without leucine and tryptophan (SD -Leu/ -Trp) as a contral (A) and medium without leucine, tryptophan, histidine and ade (SD -Trp/ -Leu/ -Ade/ -His) to test the protein-protein interactions (B).
The W residue in Domain IV is conserved in both OsIAAs and OsARFs
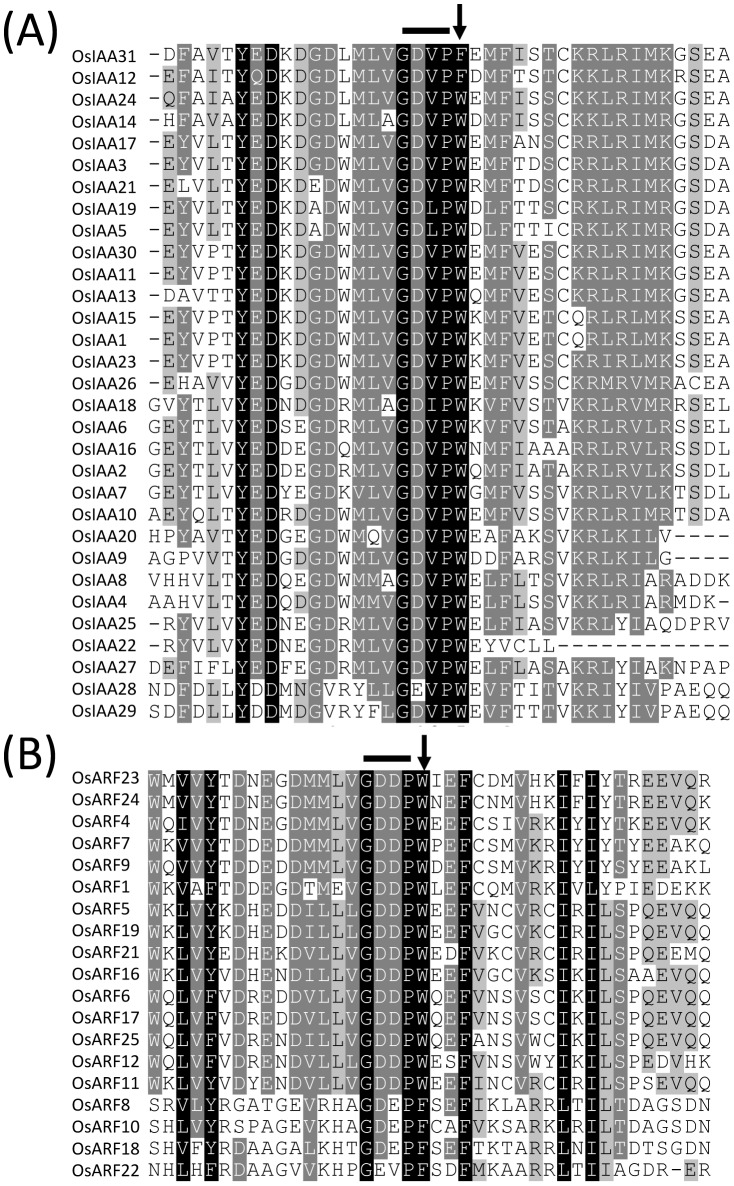
Because of the importance of W residue in Domain IV of OsIAA23, the alignment of Domain IV in all the 31 rice Aux/IAA proteins was performed using the ClustalX program. The result showed that the W residue was in the middle of Domain IV, and near the conserved motif GDVP [24]. Further analysis showed that the W residue is conserved among 29 OsIAAs. Although OsIAA12 and OsIAA31 have the Phenylalanine (F) residue instead of W, both F and W are aromatic amino acids and may have the similar properties (Figure 3A).
Figure 3. The W residue in Domain IV is conserved in both OsIAAs and OsARFs.
(A) The alignment of Domain IV of OsAux/IAAs in rice, the conserved W is marked by the arrow. Conserved GDVP motif is indicated by thick line above the alignment. (B) The alignment of Domain IV of OsARFs in rice, the conserved W is marked by the arrow. Conserved GDDP motif is indicated by thick line above the alignment.
The protein-protein interactions between OsIAAs and OsARFs are mediated by the similar Domain III/IV in both protein families. So it is interesting to investigate whether OsARFs have the same conserved W residue in Domain IV. Of all the 25 OsARF proteins, 19 OsARFs have the conserved Domain IV [23]. The alignment of Domain IV in the 19 OsARFs was performed, and we found that the W residue also existed near the conserved motif GDDP in the Domain IV of OsARFs [24]. Although OsARF8, 10, 18 and 22 have F instead of W, they may have similar properties as mentioned above (Figure 3B).
The W residue in Domain IV of OsARFs is crucial for the protein-protein interactions between Osiaa23 and OsARFs
To examine whether the conserved W residue in Domain IV of OsARFs affects the protein-protein interactions between Osiaa23 and OsARFs, the W residue in the Domain IV of OsARF6, 12, 16, 17 and 25 was exchanged with S respectively, as the same change occurred in Osiaa23-R5 (Figure 4A). The coding sequences of mutated OsARF6(WS), OsARF12(WS), OsARF16(WS), OsARF17(WS) and OsARF25(WS) were inserted into yeast expression vector pGBKT7 and transformed into yeast cells along with Osiaa23. The results showed that none of the transformed yeast cells expressing OsARF6(WS)-Osiaa23, OsARF12(WS)-Osiaa23, OsARF16(WS)-Osiaa23, OsARF17(WS)-Osiaa23, OsARF25(WS)-Osiaa23 formed colonies (Figure 4C, D). These results showed that the W residue in Domain IV of OsARFs is crucial for the protein-protein interactions between Osiaa23 and OsARFs.
Figure 4. The W residue in Domain IV of OsARFs is crucial for the protein-protein interactions between Osiaa23 and OsARFs.
(A) Diagram of conserved domains of OsARF protein. Black rectangles indicate four domains of OsARF, DBD and MR represent DNA binding Domain and middle region respectively, III and IV represent Domain III and Domain IV, which are similar to that of OsIAA. The substitution from W to S in Domain IV is indicated by arrow. (B) Self-activation test of OsARF16 and OsARF16(WS), which has an amino acid substitution from W to S in Domain IV. The transformed yeast was grown on medium without tryptophan, histidine and ade (SD -Trp/ -His/ -Ade). 1, OsARF16; 2, OsARF16(WS); 3, negative control. (C-D) Interactions between Osiaa23 and OsARFs, Osiaa23 and OsARF(WS)s in the yeast two-hybrid system. 1, positive control; 2, negative control; 3, Osiaa23 + OsARF6; 4, Osiaa23 + OsARF12; 5, Osiss23 + OsARF16; 6, Osiaa23 + OsARF17; 7, Osiaa23 + OsARF25; 8, Osiaa23 + OsARF6(WS); 9, Osiaa23 + OsARF12(WS); 10, Osiaa23 + OsARF16(WS); 11, Osiaa23 + OsARF17(WS); 12, Osiaa23 + OsARF25(WS). Yeast was grown on medium without leucine and tryptophan (SD –Leu/ -Trp) as a contral (C) and medium without leucine, tryptophan, histidine and ade (SD -Trp/ -Leu/ -Ade/ -His) to test the protein-protein interactions (D).
Mutated OsARF(WS)s rescued different defects of Osiaa23-3
To examine the transcriptional activities of the OsARF(WS)s mutated in Domain IV, the autonomous gene activation test was performed in the yeast system. The full length cDNA of OsARF16 and OsARF16(WS) were fused to the DNA-BD of the yeast transcription factor GAL4 and transformed into yeast strain AH109. Both strains with OsARF16 and OsARF16(WS) could grow well on SD -Trp/ -His/ -Ade (Figure 4B). This result indicates that the mutation in Domain IV does not affect the transcriptional activities of OsARFs.
In order to investigate the functions of OsARFs suppressed in Osiaa23, the coding sequences of mutated OsARF(WS)s were driven by the constitutive promoter (35S) and transformed into Osiaa23-3. All the transgenic lines were confirmed by RT-PCR to insure their enhanced expressions (Figure S5). The phenotypes of transgenic rice were compared with that of Osiaa23-3.
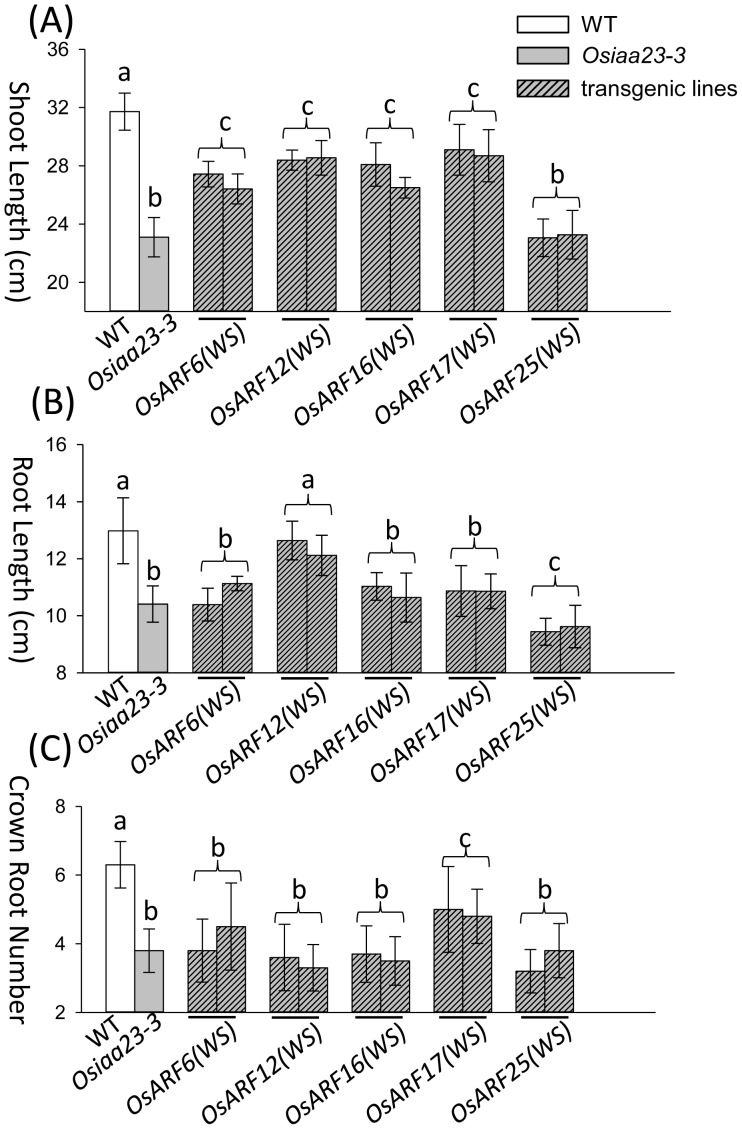
None of the transgenic rice rescued the root cap or lateral root defects of Osiaa23-3 (Figure S5; Figure S6). Further analysis revealed that over expression of OsARF6(WS), OsARF12(WS), OsARF16(WS), and OsARF17(WS) in Osiaa23-3 partially rescued the shoot length of the mutant, while over expression of OsARF25(WS) had no effect to the shoot length of Osiaa23-3 (Figure 5A; Figure S5). In the aspect of root length, over expression of OsARF12(WS) in Osiaa23-3 fully rescued the root length, while over expression of OsARF25(WS) reduced root growth in Osiaa23-3 (Figure 5B; Figure S5). Over expression of OsARF17(WS) partially rescued the number of crown roots as compared with Osiaa23-3 (Figure 5C).
Figure 5. Mutated OsARF(WS)s rescued different defects of Osiaa23-3.
The phenotypes of transgenic rice, over expressing OsARF(WS)s in the Osiaa23-3 background. Two independent lines of transgenic rice over expressing OsARF(WS)s are compared with WT and Osiaa23-3 in the aspects of shoot length (A), root length (B) and crown root number (C). Statistically distinct groups are marked by a, b and c (n = 10).
Discussion
Intragenic suppressor Osiaa23-R5 fully rescued all the defects of Osiaa23-3
These is no information on the 3D structures of Domain III/IV in Aux/IAA or ARF proteins, and little is known about which amino acid residues are important for protein-protein interactions. Current knowledge comes from intragenic suppressors of gain-of-function iaa mutants that specific amino acid substitutions in Domain III/IV revert the mutant phenotypes to wild type phenotypes, presumably by suppressing protein-protein interactions [24]. Although intragenic suppressors of iaa mutants have been reported in Arabidopsis, none of them fully rescued the defects of iaa mutants, this indicated that these amino acid residues in Domain III/IV may not vital for protein-protein interactions [25], [26]. In this study, we described an intragenic suppressor of Osiaa23 mutant, Osiaa23-R5, which fully rescued all the defects of Osiaa23-3. Sequence analysis revealed a second site mutation in Osiaa23-R5, resulting in an amino acid substitution in Domain IV. Yeast two-hybrid experiments showed that Osiaa23 can interact with selected OsARFs, while the Osiaa23-R5, which has an amino acid substitution in Domain IV, cannot interact with any of these OsARFs. These results partially explained the reasons why the amino acid substitution of W in Domain IV of Osiaa23-R5 fully rescued the defects of Osiaa23-3, and indicated that W residue in Domain IV of OsIAA23 may crucial for protein-protein interactions.
It was originally proposed that the Domain III/IV of Aux/IAA and ARF families contain a secondary structure consisting of a beta sheet (β) followed by two alpha helices (α). It was suggested that the predicted amphipathic βαα motif might function in dimerization [24]. Interestingly, the W residue is at the beginning of α2 motif. This implied that α2 motif may play an important role in protein-protein interactions between Aux/IAA and ARF families. Studies of other suppressors showed that although βαα motif has an important role in dimerization, residues outside of βαα motif may also involve in protein-protein interactions. One suppressor, Osiaa23-R3, has an amino acid substitution between Domain III and Domain IV, which is outside of βαα motif, shows weak extent of recovery (Figure S1).
The verified functions of ARFs
Most of knowledge about the functions of ARFs has been revealed by forward genetic approaches. Examination of phenotypic defects in knock-out arf mutants is a direct way to find out the functions of ARFs.
arf2/hss has defects in apical hook formation and has increased seed size [27], [28], arf3/ett lost the abaxial identity in the gynoecium [29]; arf5/mp has defects in embryo development and vascular tissue formation [30], arf7/nph4 has defects in hypocotyl tropisms and resistance to auxin and ethylene [31]; arf8 uncouples fruit development from fertilization [32], arf19 shows insensitivity to auxin and ethylene [33]. Identification and characterization of T-DNA insertion lines for 18 of the ARFs showed that most of the lines fail to show an obvious growth phenotype except for the previously identified arf2/hss, arf3/ett, arf5/mp, and arf7/nph4 mutants, suggesting that there are functional redundancies among the ARF proteins [34].
Double mutants of closely related ARFs generally show much stronger phenotypes than single mutants. arf1 arf2 double mutant enhance the defects of arf2 [27], [35]. arf3 arf4 double mutant has reduced abaxial identity in all organs compared with defects only in the gynoecium [36]. arf5 arf7 double mutant enhances the defects in the embryo patterning and vasculature of arf5 [37]. arf6 arf8 double mutant shows flower arrested development and is completely infertile, while both arf6 and arf8 single mutants only show delayed flower maturation and reduced fertility [38]. arf7 arf19 double mutant shows a strong auxin-related phenotype and severely impaired lateral root formation and abnormal gravitropism in both hypocotyl and root. These defects were not observed in the arf7 and arf19 single mutants [34], [39]. arf10 and arf16 single mutants have no defects, while double mutant of arf10 arf16 shows root cap defects and abnormal root gravitropism [40].
A family of OsARFs has been described in rice with 25 OsARFs compared with 23 ARFs in Arabidopsis [23], [41]. The phylogenetic relationship analysis showed that the organization of rice OsARFs were very similar to that of Arabidopsis ARFs, implying that rice and Arabidopsis ARFs were derived from a common ancestor, and they existed before the divergence of monocots and dicots [23].
Limited information has been obtained in the functions of OsARFs in rice. OsARF1 (or OsARF23) is the first OsARF gene described in rice, and it is closely related to ARF1 and ARF2 in Arabidopsis. Knock-down of OsARF1 has defects in vegetative and reproductive development, which is similar to the double mutant of arf1 arf2 in Arabidopsis [35], [42], [43]. OsARF12 has been proved to regulate root elongation and affect iron accumulation in rice [44]. On the other hand, OsARF11 was the orthologue of ARF5, and OsARF16 was the orthologue of ARF7 and ARF19. However, transposon insertions in OsARF11 and OsARF16 do not show similar defects as arf5 and arf7 arf19 double mutant in Arabidopsis [23]. Interestingly, OsARF16 is required for both auxin and phosphate starvation response in rice [45]. These results showed similarities and differences of ARF functions between rice and Arabidopsis.
Recent studies showed that four OsARFs (OsARF6, OsARF12, OsARF17 and OsARF25) are negatively controlled by miR167. The transgenic rice over expressing miR167 showed a substantial decrease in the expression of these four OsARF genes. Moreover, the transgenic rice were small in stature with remarkably reduced tiller number [46]. These results showed that these four OsARFs are important to the normal growth and development, while the functions of different OsARFs are still to be characterized.
An indirect way to investigate functions of OsARFs
Osiaa23 exhibits pleiotropic defects in both shoot and root development, this means that the stabilized Osiaa23 restricted many OsARFs, which was supported by our yeast two-hybrid experiments (Figure 2B, C). In this research, the mutated OsARF(WS)s can be released from the inhibition of Osiaa23 and maintain the transcriptional activities. These results provide an indirect way to investigate functions of OsARFs.
The mutated OsARF(WS)s were transformed into Osiaa23-3 mutant, and the phenotypes of transgenic rice were compared with that of Osiaa23-3. A large-scale analysis of the Aux/IAA-ARF interactome predicted a strong buffering capacity of the Aux/IAA-ARF network in the shoot apex of Arabidopsis [47]. In our research, over expression of OsARF6(WS), OsARF12(WS), OsARF16(WS) and OsARF17(WS) partially rescued the shoot length of Osiaa23-3, this implies that these OsARFs may be redundantly involved in the shoot development. Over expression of OsARF12(WS) rescued the root length of Osiaa23-3, this implies that OsARF12 may be involved in the root development. This is in agreement with the recent finding that OsARF12 regulates root elongation in rice [44]. Over expression of OsARF17(WS) partially rescued the crown root number of Osiaa23-3, this implies that OsARF17 may be involved in the crown root initiation in rice. Interestingly, over expression of OsARF25(WS) didn’t rescue any defects in Osiaa23-3, hence, the root length was even shorter. This implies that OsARF25 may function as a negative regulator in rice root development.
Over expression of site-specific mutated OsARF(WS)s rescued several defects of Osiaa23-3, while none of the transgenic rice rescued the root cap or lateral root defects. Considering that auxin gradient is needed in both the initiation of lateral root and maintenance of root apical meristem [48], [49], native promoter of OsARFs may be required to rescue the root development of Osiaa23-3. Alternatively, considering the possibility of false positives in yeast two-hybrid experiments [24], the selected five OsARFs may not interact with Osiaa23 in vivo, and the rest of OsARFs may be involved in these process or more than one OsARFs should work together to regulate these developments.
Supporting Information
Suppressors of Osiaa23-3 with different extent of recovery. (A) Root phenotypes of 7-day-old suppressors of Osiaa23-3. 1, wild type; 2, Osiaa23-3; 3, Osiaa23-R5, which fully rescued all the defects of Osiaa23-3; 4-8, the rest of the suppressors, which partially rescued defects of Osiaa23-3. 4, Osiaa23-R1; 5, Osiaa23-R2; 6, Osiaa23-R3; 7, Osiaa23-R4; 8, Osiaa23-R6. Bar = 2 cm. (B) Lateral root numbers of revertant mutants of Osiaa23. 1, wild type; 2, Osiaa23, which has no lateral root; 3, Osiaa23-R5; 4-8, the rest of the suppressors. 4, Osiaa23-R1; 5, Osiaa23-R2; 6, Osiaa23-R3; 7, Osiaa23-R4; 8, Osiaa23-R6.
(TIF)
The mutation sites of intragenic suppressors of Osiaa23-3 . The amino acid sequence of OsIAA23, four domains of OsIAA23 are underlined. Red arrow in Domain II represents the mutation site of Osiaa23-3, the other 6 arrows represent mutation sites of six intragenic suppressors, these sites are distributed between Domain III and Domain IV. The substitutions of K to M, V to E, A to G, M to T, W to S and R to Q result in the phenotypes of Osiaa23-1, Osiaa23-2, Osiaa23-3, Osiaa23-4, Osiaa23-5 and Osiaa23-6 respectively.
(TIF)
The expression patterns of selected OsARFs . Semi-quantitative RT-PCR analysis of OsARF6, OsARF12, OsARF16, OsARF17 and OsARF25 expressions in root (R), stem-base (SB) of 7-d-old wild-type seedlings, and in stem (S), leaf (L) and panicle (P) of adult plants.
(TIF)
Phenotypes of transgenic rice. Phenotypes of transgenic rice over expressing OsARF6(WS) (A), OsARF12(WS) (B), OsARF16(WS) (C), OsARF17(WS) (D) and OsARF25(WS) (E) in the Osiaa23-3 background. From left to right are wild type, Osiaa23-3 and two independent transgenic lines in the Osiaa23-3 background. Bars = 2 cm. The lowers are RT-PCR results of these transgenic lines.
(TIF)
Root tips of transgenic rice over expressing OsARF(WS)s . From left to right are wild type, Osiaa23-3 mutant and five transgenic rice over expressing different OsARF(WS)s in the background of Osiaa23-3. None of the transgenic rice recovered the root tip defect. Bars = 0.5 mm.
(TIF)
The microarray results of gene expression profile in rice root tip. Total RNA was extracted from root tips (1 cm) of 7-day-old rice. The experiment included two biological replicates (Signal 1 and Signal 2). Microarray analysis was carried out using an Affymetrix technology platform and Affymetrix GeneChip rice genome array.
(ZIP)
The sequences of primers used in this paper.
(XLSX)
Acknowledgments
We thank Professor James N. Siedow (Duke University) for critical reading of this manuscript. We also thank Dr. Keke Yi (Zhejiang Academy of Agricultural Sciences) and Dr. Feihua Wu (Hangzhou Normal University) for their helpful comments.
Funding Statement
This work was funded by Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ13C020005, and Qianjiang talents project of Science Technology Department of Zhejiang Province (2012R10059). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Annals of Botany 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, et al. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell 17: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu H, Wang S, Yu X, Yu J, He X, et al. (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. The Plant Journal 43: 47–56. [DOI] [PubMed] [Google Scholar]
- 4. Ni J, Wang GH, Zhu ZX, Zhang HH, Wu YR, et al. (2011) OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. The Plant Journal 68: 433–442. [DOI] [PubMed] [Google Scholar]
- 5. Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49: 387–400. [PubMed] [Google Scholar]
- 6. Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- 7. Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- 8. Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Developmental Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- 9. Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- 10. Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- 11. Tromas A, Perrot-Rechenmann C (2010) Recent progress in auxin biology. Comptes Rendus Biologies 333: 297–306. [DOI] [PubMed] [Google Scholar]
- 12. Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Sciences of the United States of America 94: 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. The Plant Cell 13: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guilfoyle TJ, Hagen G (2007) Auxin response factors. Current Opinion in Plant Biology 10: 453–460. [DOI] [PubMed] [Google Scholar]
- 16. Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Molecular Biology 49: 373–385. [PubMed] [Google Scholar]
- 17. Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. The Plant Cell 13: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, et al. (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. The Plant Journal 21: 553–562. [DOI] [PubMed] [Google Scholar]
- 19. Kitomi Y, Inahashi H, Takehisa H, Sato Y, Inukai Y (2012) OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Science 190: 116–122. [DOI] [PubMed] [Google Scholar]
- 20. Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, et al. (2012) A Gain-of-Function Mutation in OsIAA11 Affects Lateral Root Development in Rice. Molecular Plant 5: 154–161. [DOI] [PubMed] [Google Scholar]
- 21. Chen S, Jin W, Wang M, Zhang F, Zhou J, et al. (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. The Plant Journal 36: 105–113. [DOI] [PubMed] [Google Scholar]
- 22. Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . The Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang DK, Pei KM, Fu YP, Sun ZX, Li SJ, et al. (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24. [DOI] [PubMed] [Google Scholar]
- 24. Guilfoyle TJ, Hagen G (2012) Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Science 190: 82–88. [DOI] [PubMed] [Google Scholar]
- 25. Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. [DOI] [PubMed] [Google Scholar]
- 26. Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, et al. (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiology 123: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis . Developmental Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- 28. Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, et al. (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261. [DOI] [PubMed] [Google Scholar]
- 29. Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, et al. (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- 30. Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. The EMBO Journal 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, et al. (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell 12: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis . The Plant Cell 18: 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li JS, Dai XH, Zhao YD (2006) A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiology 140: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19 . The Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, et al. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana . Development 132: 4563–4574. [DOI] [PubMed] [Google Scholar]
- 36. Pekker I, Alvarez JP, Eshed Y (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant Cell 17: 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, et al. (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4 . Development 131: 1089–1100. [DOI] [PubMed] [Google Scholar]
- 38. Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118. [DOI] [PubMed] [Google Scholar]
- 39. Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, et al. (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal 43: 118–130. [DOI] [PubMed] [Google Scholar]
- 40. Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, et al. (2005) Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. The Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sato Y, Nishimura A, Ito M, Ashikari M, Hirano HY, et al. (2001) Auxin response factor family in rice. Genes & Genetic Systems 76: 373–380. [DOI] [PubMed] [Google Scholar]
- 42. Waller F, Furuya M, Nick P (2002) OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Molecular Biology 50: 415–425. [DOI] [PubMed] [Google Scholar]
- 43. Attia KA, Abdelkhalik AF, Ammar MH, Wei C, Yang J, et al. (2009) Antisense Phenotypes Reveal a Functional Expression of OsARF1, an Auxin Response Factor, in Transgenic Rice. Current Issues in Molecular Biology 11: I29–i34. [PubMed] [Google Scholar]
- 44. Qi YH, Wang SK, Shen CJ, Zhang SN, Chen Y, et al. (2012) OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytologist 193: 109–120. [DOI] [PubMed] [Google Scholar]
- 45. Shen CJ, Wang SK, Zhang SN, Xu YX, Qian Q, et al. (2013) OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell and Environment 36: 607–620. [DOI] [PubMed] [Google Scholar]
- 46. Liu H, Jia SH, Shen DF, Liu J, Li J, et al. (2012) Four AUXIN RESPONSE FACTOR genes downregulated by microRNA167 are associated with growth and development in Oryza sativa . Functional Plant Biology 39: 736–744. [DOI] [PubMed] [Google Scholar]
- 47. Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- 49. Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, et al. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppressors of Osiaa23-3 with different extent of recovery. (A) Root phenotypes of 7-day-old suppressors of Osiaa23-3. 1, wild type; 2, Osiaa23-3; 3, Osiaa23-R5, which fully rescued all the defects of Osiaa23-3; 4-8, the rest of the suppressors, which partially rescued defects of Osiaa23-3. 4, Osiaa23-R1; 5, Osiaa23-R2; 6, Osiaa23-R3; 7, Osiaa23-R4; 8, Osiaa23-R6. Bar = 2 cm. (B) Lateral root numbers of revertant mutants of Osiaa23. 1, wild type; 2, Osiaa23, which has no lateral root; 3, Osiaa23-R5; 4-8, the rest of the suppressors. 4, Osiaa23-R1; 5, Osiaa23-R2; 6, Osiaa23-R3; 7, Osiaa23-R4; 8, Osiaa23-R6.
(TIF)
The mutation sites of intragenic suppressors of Osiaa23-3 . The amino acid sequence of OsIAA23, four domains of OsIAA23 are underlined. Red arrow in Domain II represents the mutation site of Osiaa23-3, the other 6 arrows represent mutation sites of six intragenic suppressors, these sites are distributed between Domain III and Domain IV. The substitutions of K to M, V to E, A to G, M to T, W to S and R to Q result in the phenotypes of Osiaa23-1, Osiaa23-2, Osiaa23-3, Osiaa23-4, Osiaa23-5 and Osiaa23-6 respectively.
(TIF)
The expression patterns of selected OsARFs . Semi-quantitative RT-PCR analysis of OsARF6, OsARF12, OsARF16, OsARF17 and OsARF25 expressions in root (R), stem-base (SB) of 7-d-old wild-type seedlings, and in stem (S), leaf (L) and panicle (P) of adult plants.
(TIF)
Phenotypes of transgenic rice. Phenotypes of transgenic rice over expressing OsARF6(WS) (A), OsARF12(WS) (B), OsARF16(WS) (C), OsARF17(WS) (D) and OsARF25(WS) (E) in the Osiaa23-3 background. From left to right are wild type, Osiaa23-3 and two independent transgenic lines in the Osiaa23-3 background. Bars = 2 cm. The lowers are RT-PCR results of these transgenic lines.
(TIF)
Root tips of transgenic rice over expressing OsARF(WS)s . From left to right are wild type, Osiaa23-3 mutant and five transgenic rice over expressing different OsARF(WS)s in the background of Osiaa23-3. None of the transgenic rice recovered the root tip defect. Bars = 0.5 mm.
(TIF)
The microarray results of gene expression profile in rice root tip. Total RNA was extracted from root tips (1 cm) of 7-day-old rice. The experiment included two biological replicates (Signal 1 and Signal 2). Microarray analysis was carried out using an Affymetrix technology platform and Affymetrix GeneChip rice genome array.
(ZIP)
The sequences of primers used in this paper.
(XLSX)