Abstract
Cardiovascular malformations and cardiomyopathy are among the most common phenotypes caused by deletions of chromosome 1p36 which affect approximately 1 in 5000 newborns. Although these cardiac-related abnormalities are a significant source of morbidity and mortality associated with 1p36 deletions, most of the individual genes that contribute to these conditions have yet to be identified. In this paper, we use a combination of clinical and molecular cytogenetic data to define five critical regions for cardiovascular malformations and two critical regions for cardiomyopathy on chromosome 1p36. Positional candidate genes which may contribute to the development of cardiovascular malformations associated with 1p36 deletions include DVL1, SKI, RERE, PDPN, SPEN, CLCNKA, ECE1, HSPG2, LUZP1, and WASF2. Similarly, haploinsufficiency of PRDM16–a gene which was recently shown to be sufficient to cause the left ventricular noncompaction–SKI, PRKCZ, RERE, UBE4B and MASP2 may contribute to the development of cardiomyopathy. When treating individuals with 1p36 deletions, or providing prognostic information to their families, physicians should take into account that 1p36 deletions which overlie these cardiac critical regions may portend to cardiovascular complications. Since several of these cardiac critical regions contain more than one positional candidate gene–and large terminal and interstitial 1p36 deletions often overlap more than one cardiac critical region–it is likely that haploinsufficiency of two or more genes contributes to the cardiac phenotypes associated with many 1p36 deletions.
Introduction
Approximately 1 in 5000 newborns has a terminal deletion affecting chromosome 1p36, making it the most common telomeric deletion in humans [1]. Individuals with terminal 1p36 deletions share a common set of phenotypes that constitute the 1p36 deletion syndrome [2], [3]. These phenotypes include typical craniofacial features, cognitive impairment, behavioral problems, seizures, postnatal growth deficiency, eye/vision problems, hearing loss, cleft palate, cardiovascular malformations, cardiomyopathy and renal anomalies.
The distal critical regions for most 1p36 deletion syndrome phenotypes have been determined to reside within approximately 4 Mb from the 1p telomere [4]. However, non-overlapping interstitial deletions involving the proximal region of 1p36, starting approximately 8 Mb from the 1p telomere, have also been shown to cause many of the phenotypes associated with distal 1p36 deletions including cognitive impairment, seizures, postnatal growth deficiency, cardiovascular malformations and cardiomyopathy [5]. Some individuals have deletions of both the distal and proximal regions of 1p36 [6]. In such cases, the additive effects of haploinsufficiency of genes within both of these regions may account for the observation that individuals carrying larger 1p36 deletions are more severely affected and exhibit more of the features typically associated with 1p36 deletions [7], [8].
Cardiovascular malformations and cardiomyopathy are among the most acutely life-threatening conditions associated with both distal and proximal deletions of 1p36. Heterozygous loss-of-function mutations in PRDM16–a gene in the distal portion of 1p36 (2,985,742–3,355,185; hg19)–were recently shown to be sufficient to cause left ventricular noncompaction [9]. Similarly, a heterozygous loss-of-function mutation in ECE1 (21,543,740–21,672,034; hg19) has been identified in an individual with patent ductus arteriosus, a small subaortic ventricular septal defect, and a small atrial septal defect, suggesting that haploinsufficiency of this proximal 1p36 gene may be sufficient to cause cardiovascular malformations in humans [10]. However, the other genomic regions and dosage-sensitive genes that contribute to the cardiac phenotypes caused by 1p36 deletions have not been clearly defined. This paucity of information makes it difficult for physicians to create individualized medical plans and provide accurate prognostic information to patients and families affected by 1p36 deletions. This is true even though detailed information regarding the extent and location of an individual’s 1p36 deletion can be readily obtained on a clinical basis. Defining the cardiac-related regions and genes on 1p36 will not only allow physicians to provide improved medical care, but is also a prerequisite to understanding the molecular mechanisms that underlie the development of 1p36-related cardiovascular malformations and cardiomyopathy. Elucidating these molecular mechanisms may lead to the development of novel preventative and/or therapeutic interventions for cardiovascular disorders.
Using clinical and molecular cytogenetic data from individuals with isolated 1p36 deletions defined by array-based copy number analysis, we have identified five non-overlapping critical regions for cardiovascular malformations and two non-overlapping critical regions for cardiomyopathy on chromosome 1p36. A bioinformatic analysis of each of these cardiac critical regions revealed at least one positional candidate gene whose deletion may contribute to the development of cardiac phenotypes based on human studies and/or animal models. In some cases, haploinsufficiency of two or more such genes may contribute to the cardiac phenotypes associated with a 1p36 deletion. This is particularly likely in the case of large terminal and interstitial deletions that overlap more than one cardiac critical region.
Materials and Methods
Ethics Statement
These studies were performed under research protocols approved by the institutional review board of Baylor College of Medicine. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from parents or guardians on behalf of study participants all of whom were minors/children at the time of enrollment.
Patient Identification and Accrual
Molecular and clinical data from individuals with isolated 1p36 deletions associated with cardiovascular malformation and/or cardiomyopathy were collected from four sources. The first source consisted of data from 33,566 de-identified individuals referred to the Medical Genetics Laboratories at Baylor College of Medicine for array-based copy number analysis. Only data from individuals with isolated 1p36 deletions and an indication for testing that included a cardiovascular malformation or cardiomyopathy were included in this study.
The second source consisted of data from patients with isolated 1p36 deletions confirmed by array-based copy number analyses who were accrued from a group of individuals receiving care at Texas Children’s Hospital in Houston, TX, USA and by self-referral. In these cases, informed consent was obtained under a protocol approved by the institutional review board of Baylor College of Medicine after which clinical and molecular cytogenetic data was obtained from a review of the medical record and correspondence with the individual’s parents/family members.
The third source consisted of data from individuals with isolated 1p36 deletions associated with cardiac phenotypes who were listed in the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER; http://decipher.sanger.ac.uk/). In each case, representatives of the contributing institution–where consent for the submission of clinical and molecular cytogenetic data were obtained–were contacted and given an opportunity to provide further details of the phenotypes present in their patients.
The final source was a literature review conducted to identify additional individuals whose cardiac phenotypes were caused by isolated 1p36 deletions that had been molecularly defined using array-based copy number analysis. Reports of individuals whose 1p36 deletions were not interrogated using such techniques were not included, since traditional cytogenetic techniques may miss copy number changes on other chromosomes, may fail to identify complex rearrangements involving deletion and duplications of various regions of 1p36, and rarely provide robust delineation of the boundaries of the deletion.
Individuals with complex rearrangements of 1p36 that included both deleted and duplicated regions, individuals with 1p36 deletions associated with unbalanced translocations, and individuals with deleterious deletions or duplications on other chromosomes were also excluded from this study.
Defining Cardiac Critical Regions
Individual cardiac critical regions on chromosome 1p36 were defined based on the smallest, non-overlapping deletion present within a single individual with a cardiovascular malformation and/or cardiomyopathy. In cases where both a minimal and a maximal deleted region were defined, the breakpoints of the maximal deleted region were used to define the critical region. This approach minimizes the risk that a cardiac-related gene located between the minimal and maximal deleted region will be erroneously excluded from the critical region.
Identifying Cardiac-related Genes within Critical Regions
Data regarding each gene located completely or partially within a cardiac critical region were downloaded into a searchable spreadsheet using GeneDistiller2 (http://www.genedistiller.org/). This publically-available online program allows the user to access information on genes within a defined interval from a variety of online sources. Information reviewed to identify cardiac-related candidate genes included: Online Mendelian Inheritance in Man (OMIM; http://www.omim.org/) reports, interaction data from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org/) and the Unified Human Interactome (UniHI; http://www.unihi.org/), and phenotype data from the Human Phenotype Ontology website (http://www.human-phenotype-ontology.org/), the Mouse Genome Database (MGD; http://www.informatics.jax.org/phenotypes.shtml) and the Gene Ontology Database (http://www.geneontology.org/). This information was augmented with manually curated data from recently published manuscripts cited in PubMed (http://www.ncbi.nlm.nih.gov/pubmed).
Results
To map and identify dosage-sensitive genes and genomic regions that may contribute to the development of cardiovascular malformations and/or cardiomyopathy, we identified individuals with these phenotypes who had isolated 1p36 deletions defined by array-based copy number detection techniques as described in the Materials and Methods. The location and extent of the 1p36 deletions identified in each individual are provided in Tables 1–6 along with a description of the individual’s cardiovascular malformations (Tables 1–3) or cardiomyopathy (Tables 4–6). These deletions are also represented graphically in Figures 1 and 2.
Table 1. Summary of individuals with isolated 1p36 deletions and cardiovascular malformations referred to the Medical Genetics Laboratory (MGL) at Baylor College of Medicine.
| Identifier | Start-Stop(hg19) | Size(Mb) | Heredity | Cardiovascular Malformations | Other | CriticalRegions |
| Patient 1 | 1–2694017 | 2.7 | De novo | Hypoplastic right heart | 1 | |
| Patient 2 | 1–3581432 | 3.6 | De novo | Bicommissural aortic valve, mild aorticdilatation | Developmental delay, mild unilateralconductive hearing loss, concern for seizures | 1 |
| Patient 3 | 1–6551698 | 6.6 | De novo | Moderate secundum atrial septal defect,dilation of main pulmonary artery | Developmental delay, dysmorphic features,conductive hearing loss | 1 |
| Patient 4 | 1–6804034 | 6.8 | De novo | Moderate patent ductus arteriosus, multiplesmall muscular ventricular septal defects,small secundum atrial septal defect, heavilytrabeculated left ventricle with normalfunction | 1 | |
| Patient 5 | 1–6921434 | 6.9 | Unknown | Patent ductus arteriosus, multiple ventricularseptal defects, secundum atrial septaldefect, aberrant left subclavian artery | Developmental delay, cognitive impairment,dysmorphic features, intractable seizures,cortical blindness, contractures involvingsmall joints of hand | 1 |
| Patient 6 | 8803013–11739523 | 2.9 | De novo | Ventricular septal defect | Microcephaly | 2 |
| Patient 7 | 12726755–20540759 | 7.8 | Unknown | Tetralogy of Fallot,bicommissural aortic valve | Developmental delay, cognitive impairment,seizures, kyphosis of spine | 3* |
| Patient 8 | 27803719–31404471 | 3.6 | De novo | Coarctation of the aorta | Moderate developmental delay, cognitiveimpairment, failure to thrive, gastrointestinalproblems | 5* |
= Deletion defines a cardiac critical region.
Table 6. Summary of individuals with isolated 1p36 deletions and cardiomyopathy identified from the literature.
| Reference and Patient Identifier | Start-Stop (hg19) | Size (Mb) | Heredity | Cardiomyopathy (Co-ExistingCardiovascular Malformations) | Other | CriticalRegions |
| Cremer et al. 2008 [15] | 1–4888723 | 4.9 | De novo | Left ventricular noncompaction(Ventricular septal defect) | Large anterior fontanel, cleft palate,right sided choanal stenosis,hypothyroidism | 1 |
| Saito et al. 2008 [18] | 1–11809959 | 11.8 | De novo | Left ventricular noncompaction(Enlargement of right atrium,narrowing of right ventricle,ventricular septal defects,patent ductus arteriosus,Ebstein anomaly) | Bilateral perisylvian polymicrogyria,periventricular nodular heterotopia,seizures, hypotonia, feeding difficulties,dysmorphic features | 1,2 |
| Arndt et al. 2013, Patient 6 [9] | 564205–10821909 | 10.3 | De novo | Left ventricular noncompaction(Atrial septal defect) | Developmental delay,microcephaly, hypotonia, deepset eyes | 1 |
| Arndt et al. 2013, Patient 13 [9] | 810485–3548853 | 2.7 | De novo | Cardiomyopathy | Developmental delay | 1 |
| Arndt et al. 2013, Patient 4 [9] | 1891455–3465029 | 1.6 | De novo | Left ventricular noncompaction | Developmental delay | 1 |
| Gajeka et al. 2010, Patient 1 [19] | 1916639–3429762 | 1.5 | De novo | Left ventricular noncompaction | Developmental delay, moderatesensorineural hearing loss, mildconductive hearing loss, dysmorphicfeatures, hypoglycemia | 1* |
| Gajeka et al. 2010, Patient 2 [19] | 1916639–3429762 | 1.5 | De novo | Left ventricular noncompaction | Developmental delay, bilateralsensorineural hearing loss,dysmorphic features | 1* |
| Kang et al. 2007, Case 3 [5] | 3768946–18563553 | 14.8 | Unknown | Severe biventricular hypertrophy(Cleft mitral valve, redundanttricuspid valve leaflets,ventricular septal defect,small atrial septal defect,mild pulmonary valvestenosis, patent ductus arteriosus) | Hypotonia, bilateral nasolacrimalduct obstruction, gastroesophagealreflux, moderate sensorineuralhearing loss, bilateral cleft lipand palate, posteriorly rotated ears,digital contractures | 2 |
| Arndt et al. 2013, Patient 16 [9] | 4089259–12054030 | 8.0 | Unknown | Cardiomyopathy (Ventricularseptal defect) | Cognitive impairment,microcephaly, ptosis | 2 |
| Kang et al. 2007, Case 4 [5] | 8395179–11362893 | 3.0 | Unknown | Dilated cardiomyopathy (Perimembranousventricular septal defect,small septum secundum atrial septal defect) | Developmental delay, failureto thrive, truncal hypotonia,dysmorphic features | 2* |
= Deletion defines a cardiac critical region.
Table 3. Summary of individuals with isolated 1p36 deletions and cardiovascular malformations identified from the literature.
| Reference and Patient Identifier | Start-Stop (hg19) | Size (Mb) | Heredity | Cardiovascular Malformations(Co-existing Cardiomyopathy) | Other | CriticalRegions |
| El-Hattab et al. 2010 [11] | 1–2418935 | 2.4 | De novo | Ventricular septal defect, atrial septaldefect, PDA, right-sided aortic arch | Developmental delay, renal malpositionand malrotation, omphalocele, cloacalexstrophy, imperforate anus, multiple sacralsegmentation defects, genital anomalies,diastasis of symphysis pubis,limb deformities | 1* |
| Cremer et al.2008 [15] | 1–4888723 | 4.9 | De novo | Ventricular septal defect (LVNC) | Cleft palate, unilateral choanal stenosis,hypothyroidism | 1 |
| Campeau et al. 2008, Patient 1 [16] | 1–10247416 | 10.2 | De novo | Asymmetric ventricles, muscularventricular septal defect, tortuousaortic arch, PDA | Hypotonia, single febrile seizure, bilateralcolpocephaly, moderate to severenon-obstructive hydrocephalus,sensorineural hearing loss, shortfemurs, unilateral club foot, submucouscleft palate, velopharyngealincompetence, dysmorphic features | 1,2 |
| Bursztejn et al. 2009 [17] | 1–11809959 | 11.8 | De novo | Atrial septal defect, ventricularseptal defect | Infantile spasms, partial seizures,agenesis of the corpus callosum,ventricular dilation,dysmorphic features | 1,2 |
| Saito et al. 2008 [18] | 1–11809959 | 11.8 | De novo | Enlargement of right atrium,narrowing of right ventricle,ventricular septal defects, PDA,Ebstein anomaly (LVNC) | Bilateral perisylvian polymicrogyria,periventricular nodular heterotopia,seizures, hypotonia, feeding difficulties,dysmorphic features | 1,2 |
| Nicoulaz et al. 2011 [6] | 1–16177338 | 15.6 | De novo | Tetralogy of Fallot | Joint contractures, ventriculomegaly,marked pachygyria, absent septumpellucidum, thinned corpus callosum,dysmorphic features | 1,2,3 |
| Arndt et al. 2013, Patient 6 [9] | 564205–10821909 | 10.3 | De novo | Atrial septal defect (LVNC) | Developmental delay, microcephaly,hypotonia, deep set eyes | 1,2 |
| Kang et al. 2007, Case 3 [5] | 3768946–18563553 | 14.8 | Unknown | Cleft mitral valve, redundanttricuspid valve leaflets,ventricular septal defect, small atrial septal defect,mild pulmonary valvestenosis, PDA | Hypotonia, bilateral nasolacrimal ductobstruction, gastroesophageal reflux,severe biventricular hypertrophy,moderate sensorineural hearing loss,bilateral cleft lip and palate,posteriorly rotated ears,digital contractures | 2,3 |
| Arndt et al. 2013, Patient 16 [9] | 4089259–12054030 | 8.0 | Unknown | Ventricular septal defect(Cardiomyopathy) | Cognitive impairment,microcephaly, ptosis | 2 |
| Kang et al. 2007, Case 5 [5] | 4317448–13867316 | 9.5 | Unknown | Partial anomalous pulmonaryvenous return with the leftpulmonary veins draininginto the innominate vein | Developmental delay, seizures,failure to thrive, hemivertebra, Wolff-Parkinson-White syndrome, ataxia,unsteady gait,appendicular hypertonia | 2,3 |
| Kang et al. 2007, Case 2 [5] | 4843323–16397974 | 11.6 | Unknown | Two small right coronary arteryfistulae terminating in theleft atrium andright ventricle | Developmental delay, seizures,peripheral hypertonia, gastroesophagealreflux, dysmorphic features | 2,3 |
| Kang et al. 2007, Case 4 [5] | 8395179–11362893 | 3.0 | Unknown | Perimembranous ventricular septaldefect, small septum secundumatrial septal defect | Developmental delay,failure to thrive, truncalhypotonia, dysmorphicfeatures | 2* |
| Kang et al. 2007, Case 1 [5] | 9124551–21782714 | 12.7 | Unknown | High and mid muscularventricular septal defect,bicuspid aortic valve,patent foramen ovale, PDA | Developmental delay, seizures,dysmorphic features | 2, 3,4 |
= Deletion defines a cardiac critical region, LVNC = left ventricular noncompaction, PDA = patent ductus arteriosus.
Table 4. Summary of individuals with isolated 1p36 deletions and cardiomyopathy referred to the Medical Genetics Laboratory (MGL) or recruited into a 1p36 research study at Baylor College of Medicine.
| Identifier | Start-Stop (hg19) | Size (Mb) | Heredity | Cardiomyopathy | Other | Critical Regions |
| Patient 9 | 1–4470448 | 4.5 | Unknown | Dilated cardiomyopathy | 1 | |
| Patient 10 | 1–4330413 | 4.3 | De novo | Dilated cardiomyopathy | Developmental delay, infantile spasms, hypotonia | 1 |
| Patient 11 | 1–4078518 | 4.1 | De novo | Bilateral dilatedcardiomyopathy | Seizures, cleft lip, intellectual disability, sensorineural hearing loss | 1 |
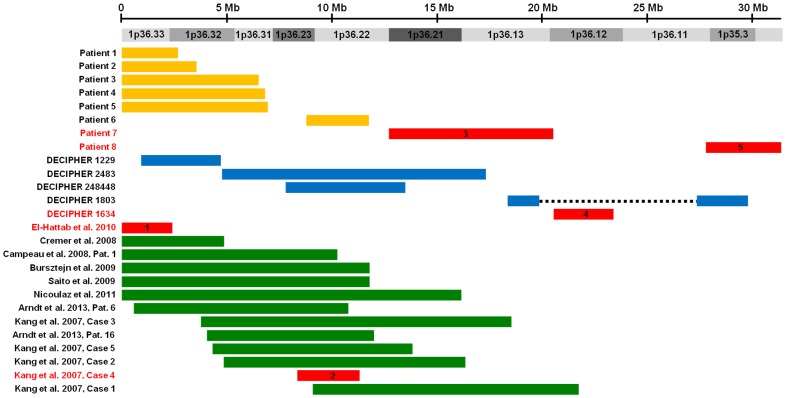
Figure 1. Delineation of five critical regions for cardiovascular malformations on 1p36.
Solid bars represent isolated 1p36 deletions identified in individuals with cardiovascular malformations. These deletions are grouped based on the source from which data was obtained: 1) patients referred to the Medical Genetics Laboratories at Baylor College of Medicine for copy number analysis and 2) patients recruited into a research study on 1p36 deletions at Baylor College of Medicine [orange], 3) the DECIPHER database [blue], and 4) a review of the literature [green]. Deletions that define non-overlapping critical region for cardiovascular malformation are shown in red and are numbered sequentially from the telomere.
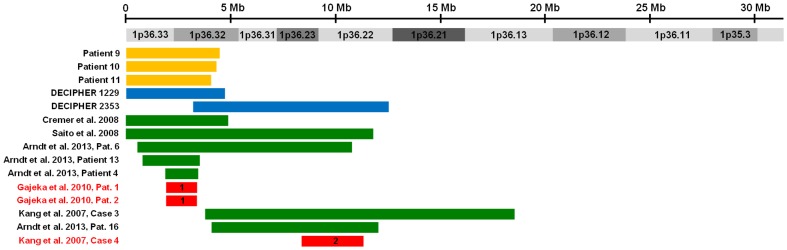
Figure 2. Delineation of two critical regions for cardiomyopathy on 1p36.
Solid bars represent isolated 1p36 deletions identified in individuals with cardiomyopathy. These deletions are grouped based on the source from which data was obtained: 1) patients referred to the Medical Genetics Laboratories at Baylor College of Medicine for copy number analysis and 2) patients recruited into a research study on 1p36 deletions at Baylor College of Medicine [orange], 3) the DECIPHER database [blue], and 4) a review of the literature [green]. Deletions that define non-overlapping critical region for cardiomyopathy are shown in red and are numbered sequentially from the telomere.
Table 2. Summary of individuals with isolated 1p36 deletions and cardiovascular malformations identified from the DECIPHER database.
| DECIPHERIdentifier | Start-Stop(hg19) | Size(Mb) | Heredity | CardiovascularMalformations (Co-existing Cardiomyopathy) | Other | CriticalRegions |
| 1229 | 928301–4708254 | 3.8 | De novo | Ventricular septal defect(Cardiomyopathy) | Intellectual disability, epileptic spasms,delayed cranial suture closure, stenosisof external auditory canal, dysmorphic features | 1 |
| 2483 | 4795388–17364849 | 12.6 | Not maternal,fatherunavailable | Secundum atrialseptal defect | Intellectual disability, delayed speech andlanguage development, feeding difficultiesin infancy, microcephaly, submucous cleft hard palate,prenatal short stature, scoliosis, dysmorphic features | 2, 3 |
| 248448 | 7812397–13488491 | 5.7 | De novo | Mild pulmonary valvestenosis | Intellectual disability, feeding difficulties ininfancy, recurrent infections, sensorineuralhearing impairment, proportionate shortstature, cryptorchidism, short phalanges,broad thumbs, dysmorphic features | 2, 3 |
| 1803 Distal | 18382579–19879460 | 1.5 | Unknown | Atrial septal defect | Intellectual disability, feeding difficultiesin infancy, high palate, dysmorphicfeatures | 3 |
| 1803 Proximal | 27358936–29807278 | 2.4 | De novo | Atrial septal defect | See above | 5 |
| 1634 | 20555776–23438888 | 2.9 | De novo | Patent ductus arteriosus,coarctation of the aorta,small ventricular septaldefect | Intellectual disability, behavioralproblems with aggression and mood swings,dysmorphic features, non-cleft velopharyngealdysfunction, hypospadias,bifid thumb, stiffnessand progressive joint contractures withfixed kyphosis, fusion of 1st and 2ndcervical vertebrae | 4* |
= Deletion defines a cardiac critical region.
Table 5. Summary of individuals with isolated 1p36 deletions and cardiomyopathy identified from the DECIPHER database.
| DECIPHER Identifier | Start-Stop (hg19) | Size (Mb) | Heredity | Cardiomyopathy (Co-existing Cardiovascular Malformations) | Other | CriticalRegions |
| 1229 | 1–4708254 | 4.7 | De novo | Cardiomyopathy (Ventricular septal defect) | Intellectual disability, development delay, infantile spasms, narrow/atretic auditory canal, dysmorphic features | 1 |
| 2353 | 3224674–12540397 | 9.3 | Unknown | Dilated cardiomyopathy | Intellectual disability, development delay, seizures, myopia, sensorineural hearing loss, dysmorphic features | 1,2 |
Using this data, we defined five non-overlapping critical regions for cardiovascular malformations (Table 7, Figure 1). For clarity, each critical region has been numbered sequentially starting with the most distal region on chromosome 1p36. The first critical region is defined by a de novo ∼2.4 Mb terminal deletion reported by El-Hattab and colleagues in a patient with a ventricular septal defect, an atrial septal defect, patent ductus arteriosus and a right-sided aortic arch [11].
Table 7. Cardiac-related genes within cardiovascular malformation critical regions.
| Cardiovascular Malformation Region 1: Chr1∶1–2418935 (2.4 Mb), 111 genes | |||||
| Gene | Start (hg19) | Stop (hg19) | Related Cardiovascular Phenotypes | References | |
| DVL1 | 1270658 | 1284492 | No cardiovascular phenotypes have been documented in Dvl1-null mice.However, an extra copy of Dvl1 was able to rescue the lethal conotruncalheart defects seen in Dvl3-null mice suggesting that Dvl1has redundant functions in cardiac development. | [20], [21] | |
| SKI | 2160134 | 2241652 | Mutations in SKI with putative dominant-negative potential havebeen shown to cause Shprintzen-Goldberg syndrome whose featuresinclude mitral valve prolapse, aortic root dilatation, vascular tortuosityand aortic aneurysms. Knockdown of the 2 paralogs of mammalianSKI in zebrafish (skia and skib) results in severe cardiac anomaliescharacterized by partial to complete failure in cardiac looping andmalformations of the outflow tract. | [22] | |
| Cardiovascular Malformation Region 2: Chr1∶8395179–11362893 (3.0 Mb), 55 genes | |||||
| RERE | 8412464 | 8877699 | Rere-null mouse embryos die of cardiac failure aroundE9.5 with unlooped hearts. RERE-deficient mouse embryoshave aortic arch anomalies, double outlet right ventricle,transposition of the great arteries, andperimembranous ventricular septal defects. | [23], [24] | |
| Cardiovascular Malformation Region 3: Chr1∶12726755–20540759 (7.8 Mb), 175 genes | |||||
| PDPN | 13910252 | 13944452 | Pdpn-null mouse embryos have hypoplastic and perforatedcompact and septal myocardium, hypoplasticatrioventricular cushions resulting in atrioventricularvalve abnormalities, and coronary arteryabnormalities, hypoplasia of the sinoatrial node, andthin, perforated cardinal and pulmonary vein walls. | [25]–[27] | |
| SPEN | 16174359 | 16266950 | Spen-null mouse embryos die in utero and havedefects of the cardiac septum and muscles. | [28] | |
| CLCNKA | 16348486 | 16360545 | A loss-of-function variant in the human CLCNKAgene is a risk factor for heart failure in Caucasians. | [29] | |
| Cardiovascular Malformation Region 4: Chr1∶20555776–23438888 (2.9 Mb), 50 genes | |||||
| ECE1 | 21543740 | 21672034 | A loss-of-function mutation in ECE1 was identified in anindividual with patent ductus arteriosus, a small subaorticventricular septal defect, and a small atrial-septal defect,Hirschsprung disease, and autonomic dysfunction. Ece1-nullmice have interrupted aortic arch, absent right subclavianartery, poorly developed endocardial cushions, doubleoutlet right ventricle, truncus arteriosus, double aortic arch,overriding aorta and ventricular septal defects. | [10], [30], [31] | |
| HSPG2 | 22148737 | 22263750 | HSPG2-deficient mouse embryos have hyperplasticconotruncal endocardial cushions, transposition of the greatarteries, and malformations of the semilunar valves. However,recessive mutations in HSPG2 have been shown to causeSchwartz-Jampel syndrome, type 1 and dyssegmental dysplasia,Silverman-Handmaker type, neither of which are commonlyassociated with cardiac defects. | [32]–[34] | |
| LUZP1 | 23410516 | 23495351 | Luzp1-null mice have double outlet right ventricle,transposition of the great arteries, and ventricularseptal defects. | [35] | |
| Cardiovascular Malformation Region 5: Chr1∶27803719–31404471 (3.6 Mb), 55 genes | |||||
| WASF2 | 27730734 | 27816678 | Wasf2-null mouse embryos die before E11.5 withabnormalities in vasculogenesis. These embryos also havesmall dorsal aortas and anterior cardinal veins at the 22somite stage, and display incomplete cardiac looping andsmall hearts at E10.5. | [36] | |
The second critical region is defined by an ∼3.0 Mb interstitial deletion identified in Case 4 reported by Kang and colleagues in an individual with a perimembranous ventricular septal defect and a small septum secundum atrial septal defect [5]. In using Kang et al. Case 4 to define the second critical region, we note that the ∼2.9 Mb interstitial deletion in Patient 6, who had a ventricular septal defect, begins slightly centromeric to the deletion in Kang et al. Case 4 and, therefore, does not involve the SCL45A1 gene. However, the deletion in Patient 6 extends farther into the proximal region of 1p36 than the deletion in Kang et al. Case 4 and includes UBE2V2P3, PTCHD2, FBXO2, FBXO44, FBXO6 and MAD2L2.
The third critical region is defined by an ∼7.8 Mb interstitial deletion in Patient 7 who was referred to the Medical Genetics Laboratories at Baylor College of Medicine for copy number analysis with an indication for testing that included tetralogy of Fallot and a bicommissural aortic valve. The fourth critical region is defined by DECIPHER patient #1634 who had a patent ductus arteriosus, coarctation of the aorta and a small ventricular septal defect caused by a de novo ∼2.9 Mb interstitial deletion. The fifth critical region is defined by a de novo ∼3.6 Mb interstitial deletion in Patient 8 who was referred to the Medical Genetics Laboratories at Baylor College of Medicine for copy number analysis with an indication for testing that included coarctation of the aorta.
Using this same dataset, we defined two critical regions for cardiomyopathy (Table 8, Figure 2). The first critical region contains the PRDM16 gene and is defined by an ∼1.5 Mb interstitial deletion in two siblings with left ventricular noncompaction described by Gajeka and colleagues. The second critical region is defined by an ∼3.0 Mb interstitial deletion in an individual reported by Kang and colleagues (Case 4) who had dilated cardiomyopathy which coexisted with a perimembranous ventricular septal defect and a small septum secundum atrial septal defect [5]. Kang and colleagues indicated that both cardiac malformations spontaneously closed but the dilated cardiomyopathy remained and was still being treated with medications when the child was 5 years old. While this history suggests that he had a primary cardiomyopathy, we cannot rule out the possibility that his cardiovascular malformations contributed to his dilated cardiomyopathy. However, further evidence of the role of this region in the development of primary cardiomyopathy comes from Patient 16 described by Arndt and colleagues who had an overlapping ∼8 Mb deletion associated with early-onset cardiomyopathy with transient heart failure whose only structural heart defect was a clinically insignificant ventricular septal defect [9].
Table 8. Cardiac-related genes within cardiomyopathy critical regions.
| Cardiomyopathy Region 1: Chr1∶1916639–3429762 (1.5 Mb), 28 genes | |||||
| Gene | Start (hg19) | Stop (hg19) | Related Cardiovascular Phenotypes | References | |
| PRKCZ | 1981909 | 2116834 | PRKCZ regulates the phosphorylation and de-phosphorylation ofcardiac sarcomeric proteins including cardiac troponin T. Mutationsin the TNNT2 gene, which encodes cardiac troponinT, have been found in individuals with leftventricular noncompaction, and dilated, restrictive,and hypertrophic cardiomyopathy. | [37]–[42] | |
| SKI | 2160134 | 2241652 | In zebrafish, co-injection of subthreshold doses ofskia and prdm16 morpholinos reduced cardiac outputsuggesting that these genes act synergistically toaffect cardiac contractility. | [22] | |
| PRDM16 | 2985742 | 3355185 | PRDM16-deficient mice have ventricular hypoplasiaand abnormal ventricular morphology with a cleftbetween ventricles. Morpholino knockdown ofprdm16 in zebrafish causes contractile dysfunction, partialuncoupling of cardiomyocytes and impairedcardiomyocyte proliferative capacity. Deleteriousmutations in PRDM16 were identified in individualswith left ventricular noncompaction and novel,putatively deleterious sequence variants wereidentified in individuals with dilated cardiomyopathy. | [43], [44] | |
| Cardiomyopathy Region 2: Chr1∶8395179–11362893 (3.0 Mb), 55 genes | |||||
| RERE | 8412464 | 8877699 | In the absence of cardiovascular malformations, asubset of RERE-deficient mice spontaneouslydevelops cardiac fibrosis and cardiomegaly. | [24] | |
| UBE4B | 10093041 | 10241297 | Ube4b-null mice have underdeveloped and compactmyocardial layers, defective assembly of myosin incardiac muscle cells, reduced cardiac trabeculationand cardiac restricted apoptosis that affects mostcardiomyocytes at E13.5. | [45] | |
| MASP2 | 11086580 | 11107296 | MASP2 genotypes that generate low MASP2 levelsare associated with a high risk of cardiomyopathy in thesetting of Trypanosoma cruzi infection (Chagas disease).However, in a model of transient myocardialischemia/reperfusion injury, Masp2-null mice hadsignificantly smaller infarct volumes than their wild-type littermates. | [46], [47] | |
Each of the cardiac-related critical regions we identified contains one or more positional candidate genes whose haploinsufficiency may contribute to the development of cardiovascular phenotypes. Genes which may contribute to the development of cardiovascular malformations associated with 1p36 deletions include DVL1, SKI, RERE, PDPN, SPEN, CLCNKA, ECE1, HSPG2, LUZP1, and WASPF2. Genes which may contribute to the development of cardiomyopathy associated with 1p36 deletions include SKI, PRKCZ, PRDM16, RERE, UBE4B and MASP2. The cardiac-related phenotypes associated with each of these genes in humans, mice and zebrafish are summarized in Tables 7 and 8.
Discussion
Terminal deletions of chromosome 1p36 are the most common telomeric deletions in humans and carry a high risk of cardiac-related medical problems. In one large cohort, 71% of individuals with terminal 1p36 deletions had cardiovascular malformations and 27% had cardiomyopathy [2]. The most common cardiovascular malformations seen were atrial and ventricular septal defects, patent ductus arteriosus, valvular anomalies, tetralogy of Fallot and coarctation of the aorta [2]. Among individuals with cardiomyopathy, 85% had noncompaction defects and the remaining 15% had dilated cardiomyopathy. Similar patterns of cardiac anomalies were seen in the patients with terminal 1p36 deletion described in this report. Individuals with interstitial deletions of 1p36 also have high rates of cardiac abnormalities, although their exact incidences likely vary from one region to another and are more difficult to estimate based on the relatively low number of patients that have been identified [5].
Due to the high risk of cardiac-related problems, most children with 1p36 deletions are screened for cardiovascular anomalies at baseline and followed over time for the development of cardiomyopathy–often having several echocardiograms even if they are asymptomatic. Although this type of careful monitoring can help identify potentially treatable problems, it also places a burden on families and the health care system which could be avoided or reduced if an individual’s risk of having these problems could be estimated based on the location and extent of their individual 1p36 deletion. This type of risk stratification will require a better understanding of the 1p36 genes and genomic regions associated with the development of cardiovascular anomalies and cardiomyopathy.
In this report, we take the first step towards addressing this issue by defining five non-overlapping critical regions for cardiovascular malformations and two non-overlapping critical regions for cardiomyopathy. Each of these cardiac critical regions is defined by an isolated 1p36 deletion identified in a single individual. Hence, they represent genomic regions whose deletions are sufficient to cause cardiovascular malformation or cardiomyopathy. The size of these regions vary with the smallest being 1.5 Mb (28 genes) and the largest being 7.8 Mb (175 genes). As more 1p36 deletions are described, it is likely that these regions will be refined and some may be subdivided into more than one region. It is also possible that data from additional deletions will reveal novel critical regions on 1p36 that are associated with cardiac problems.
It is reasonable to assume that each of the cardiac critical regions we have delineated on chromosome 1p36 contains at least one dosage-sensitive, cardiac-related gene or regulatory region. Although one or more positional candidate genes from each interval can be identified based on their known function and/or their phenotypes in animal models, haploinsufficiency of only two genes have been shown to cause cardiac phenotypes in humans. Heterozygous loss-of-function mutations in PRDM16 have been shown to be sufficient to cause left ventricular noncompaction and a loss-of-function mutation in ECE1 was identified in an individual with patent ductus arteriosus, a small subaortic ventricular septal defect and a small atrial septal defect, Hirschsprung disease, and autonomic dysfunction [9], [10].
Loss-of-function mutations in the remainder of the positional candidate genes identified in 1p36 cardiac critical regions–DVL1, SKI, RERE, PDPN, SPEN, CLCNKA, HSPG2, LUZP1, WASPF2, PRKCZ, UBE4B and MASP2–have yet to be identified in individuals with cardiovascular phenotypes. The identification of de novo loss-of-function mutations in these positional candidate genes, or dominantly inherited loss-of-function mutations segregating with a cardiac phenotype in multiple family members, would serve to confirm their pathogenic roles. Experience with other chromosomal regions suggests that these types of mutations are most likely to be identified in genes with particularly high impacts on cardiac development and function like GATA4 on 8p23.1, ZFPM2 on 8q23.1, and TBX1 on 22q11.2 [12]–[14].
De novo loss-of-function mutations in cardiac genes whose haploinsufficiency makes a more modest contribution to cardiac risk may be particularly difficult to identify. Loss-of-function mutations in such genes are more likely to be inherited from an asymptomatic parent and may combine with other genetic, environmental or stochastic factors to cause cardiac-related problems in sporadic cases following a multifactorial inheritance pattern. In such cases, animal models may provide the first evidence that a gene within one of the critical regions plays a role in cardiac development and/or function. Not only can animal models be used to effectively identify low penetrance phenotypes caused by haploinsufficiency of a candidate gene, but they can also be used to explore cardiac-related phenotypes–including defects that lead to embryonic lethality–that only become apparent when the expression of an individual gene is reduced by more than 50%.
Deleterious changes in genes that make a modest contribution to cardiac risk may also be identified in asymptomatic individuals from the general population. One source of information on potentially deleterious changes in various populations is the Database of Genomic Variants (http://dgv.tcag.ca/) which catalogues genomic variation among “normal controls” and population-based cohorts. It is interesting to note that exon-containing deletions in each of the positional candidate genes identified in this study have been reported in this database (Table S1). This suggests that the cardiac-related phenotypes caused by haploinsufficiency of each of these genes alone are likely to be incompletely penetrant or may go undetected in some individuals. This is consistent with the incomplete penetrance for cardiovascular malformations and cardiomyopathy seen among patients with overlapping terminal and interstitial deletions of 1p36 [2], [5].
Several of the cardiac critical regions we have identified on chromosome 1p36 contain more than one positional candidate gene. We also note that large terminal and interstitial deletions often overlap more than one cardiac critical region. This suggests that haploinsufficiency of two or more genes may contribute to the cardiac phenotypes associated with many 1p36 deletions. Future studies aimed at understanding how adjacent 1p36 genes work together to impact cardiac development and function may provide information which can be used to design effective therapeutic and/or preventative measures which can minimize the impact of cardiovascular malformations and/or cardiomyopathy in such cases.
Supporting Information
Exon-containing deletions in 1p36 cardiac-related genes found in various control and population-based cohorts catalogued in the Database of Genomic Variants.
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Shaffer LG, Lupski JR (2000) Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34: 297–329. [DOI] [PubMed] [Google Scholar]
- 2. Battaglia A, Hoyme HE, Dallapiccola B, Zackai E, Hudgins L, et al. (2008) Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 121: 404–410. [DOI] [PubMed] [Google Scholar]
- 3. Slavotinek A, Shaffer LG, Shapira SK (1999) Monosomy 1p36. J Med Genet 36: 657–663. [PMC free article] [PubMed] [Google Scholar]
- 4. Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, et al. (2003) Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 72: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang SH, Scheffer A, Ou Z, Li J, Scaglia F, et al. (2007) Identification of proximal 1p36 deletions using array-CGH: a possible new syndrome. Clin Genet 72: 329–338. [DOI] [PubMed] [Google Scholar]
- 6. Nicoulaz A, Rubi F, Lieder L, Wolf R, Goeggel-Simonetti B, et al. (2011) Contiguous approximately 16 Mb 1p36 deletion: Dominant features of classical distal 1p36 monosomy with haplo-lethality. Am J Med Genet A 155A: 1964–1968. [DOI] [PubMed] [Google Scholar]
- 7. Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FF, et al. (1997) Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet 61: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu YQ, Heilstedt HA, Bedell JA, May KM, Starkey DE, et al. (1999) Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Hum Mol Genet 8: 313–321. [DOI] [PubMed] [Google Scholar]
- 9. Arndt AK, Schafer S, Drenckhahn JD, Sabeh MK, Plovie ER, et al. (2013) Fine Mapping of the 1p36 Deletion Syndrome Identifies Mutation of PRDM16 as a Cause of Cardiomyopathy. Am J Hum Genet 93: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hofstra RM, Valdenaire O, Arch E, Osinga J, Kroes H, et al. (1999) A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet 64: 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. El-Hattab AW, Skorupski JC, Hsieh MH, Breman AM, Patel A, et al. (2010) OEIS complex associated with chromosome 1p36 deletion: a case report and review. Am J Med Genet A 152A: 504–511. [DOI] [PubMed] [Google Scholar]
- 12. Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, et al. (2003) Role of TBX1 in human del22q11.2 syndrome. Lancet 362: 1366–1373. [DOI] [PubMed] [Google Scholar]
- 13. Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, et al. (2009) Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A 149A: 1661–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pizzuti A, Sarkozy A, Newton AL, Conti E, Flex E, et al. (2003) Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum Mutat 22: 372–377. [DOI] [PubMed] [Google Scholar]
- 15. Cremer K, Ludecke HJ, Ruhr F, Wieczorek D (2008) Left-ventricular non-compaction (LVNC): a clinical feature more often observed in terminal deletion 1p36 than previously expected. Eur J Med Genet 51: 685–688. [DOI] [PubMed] [Google Scholar]
- 16. Campeau PM, Ah Mew N, Cartier L, Mackay KL, Shaffer LG, et al. (2008) Prenatal diagnosis of monosomy 1p36: a focus on brain abnormalities and a review of the literature. Am J Med Genet A 146A: 3062–3069. [DOI] [PubMed] [Google Scholar]
- 17. Bursztejn AC, Bronner M, Peudenier S, Gregoire MJ, Jonveaux P, et al. (2009) Molecular characterization of a monosomy 1p36 presenting as an Aicardi syndrome phenocopy. Am J Med Genet A 149A: 2493–2500. [DOI] [PubMed] [Google Scholar]
- 18. Saito S, Kawamura R, Kosho T, Shimizu T, Aoyama K, et al. (2008) Bilateral perisylvian polymicrogyria, periventricular nodular heterotopia, and left ventricular noncompaction in a girl with 10.5–11.1 Mb terminal deletion of 1p36. Am J Med Genet A 146A: 2891–2897. [DOI] [PubMed] [Google Scholar]
- 19. Gajecka M, Saitta SC, Gentles AJ, Campbell L, Ciprero K, et al. (2010) Recurrent interstitial 1p36 deletions: Evidence for germline mosaicism and complex rearrangement breakpoints. Am J Med Genet A 152A: 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lijam N, Paylor R, McDonald MP, Crawley JN, Deng CX, et al. (1997) Social interaction and sensorimotor gating abnormalities in mice lacking Dvl1. Cell 90: 895–905. [DOI] [PubMed] [Google Scholar]
- 21. Etheridge SL, Ray S, Li S, Hamblet NS, Lijam N, et al. (2008) Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet 4: e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doyle AJ, Doyle JJ, Bessling SL, Maragh S, Lindsay ME, et al. (2012) Mutations in the TGF-beta repressor SKI cause Shprintzen-Goldberg syndrome with aortic aneurysm. Nat Genet 44: 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zoltewicz JS, Stewart NJ, Leung R, Peterson AS (2004) Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development 131: 3–14. [DOI] [PubMed] [Google Scholar]
- 24. Kim BJ, Zaveri HP, Shchelochkov OA, Yu Z, Hernandez-Garcia A, et al. (2013) An Allelic Series of Mice Reveals a Role for RERE in the Development of Multiple Organs Affected in Chromosome 1p36 Deletions. PLoS One 8: e57460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahtab EA, Wijffels MC, Van Den Akker NM, Hahurij ND, Lie-Venema H, et al. (2008) Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: Correlation with abnormal epicardial development. Dev Dyn 237: 847–857. [DOI] [PubMed] [Google Scholar]
- 26. Douglas YL, Mahtab EA, Jongbloed MR, Uhrin P, Zaujec J, et al. (2009) Pulmonary vein, dorsal atrial wall and atrial septum abnormalities in podoplanin knockout mice with disturbed posterior heart field contribution. Pediatr Res 65: 27–32. [DOI] [PubMed] [Google Scholar]
- 27. Mahtab EA, Vicente-Steijn R, Hahurij ND, Jongbloed MR, Wisse LJ, et al. (2009) Podoplanin deficient mice show a RhoA-related hypoplasia of the sinus venosus myocardium including the sinoatrial node. Dev Dyn 238: 183–193. [DOI] [PubMed] [Google Scholar]
- 28. Kuroda K, Han H, Tani S, Tanigaki K, Tun T, et al. (2003) Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18: 301–312. [DOI] [PubMed] [Google Scholar]
- 29. Cappola TP, Matkovich SJ, Wang W, van Booven D, Li M, et al. (2011) Loss-of-function DNA sequence variant in the CLCNKA chloride channel implicates the cardio-renal axis in interindividual heart failure risk variation. Proc Natl Acad Sci U S A 108: 2456–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, et al. (1998) Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J Clin Invest 102: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, et al. (1998) Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development 125: 825–836. [DOI] [PubMed] [Google Scholar]
- 32. Costell M, Carmona R, Gustafsson E, Gonzalez-Iriarte M, Fassler R, et al. (2002) Hyperplastic conotruncal endocardial cushions and transposition of great arteries in perlecan-null mice. Circ Res 91: 158–164. [DOI] [PubMed] [Google Scholar]
- 33. Arikawa-Hirasawa E, Wilcox WR, Le AH, Silverman N, Govindraj P, et al. (2001) Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat Genet 27: 431–434. [DOI] [PubMed] [Google Scholar]
- 34. Nicole S, Davoine CS, Topaloglu H, Cattolico L, Barral D, et al. (2000) Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia). Nat Genet 26: 480–483. [DOI] [PubMed] [Google Scholar]
- 35. Hsu CY, Chang NC, Lee MW, Lee KH, Sun DS, et al. (2008) LUZP deficiency affects neural tube closure during brain development. Biochem Biophys Res Commun 376: 466–471. [DOI] [PubMed] [Google Scholar]
- 36. Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, et al. (2003) WAVE2 is required for directed cell migration and cardiovascular development. Nature 424: 452–456. [DOI] [PubMed] [Google Scholar]
- 37. Wu SC, Solaro RJ (2007) Protein kinase C zeta. A novel regulator of both phosphorylation and de-phosphorylation of cardiac sarcomeric proteins. J Biol Chem 282: 30691–30698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, et al. (2008) Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation 117: 2893–2901. [DOI] [PubMed] [Google Scholar]
- 39. Luedde M, Ehlermann P, Weichenhan D, Will R, Zeller R, et al. (2010) Severe familial left ventricular non-compaction cardiomyopathy due to a novel troponin T (TNNT2) mutation. Cardiovasc Res 86: 452–460. [DOI] [PubMed] [Google Scholar]
- 40. Hershberger RE, Pinto JR, Parks SB, Kushner JD, Li D, et al. (2009) Clinical and functional characterization of TNNT2 mutations identified in patients with dilated cardiomyopathy. Circ Cardiovasc Genet 2: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peddy SB, Vricella LA, Crosson JE, Oswald GL, Cohn RD, et al. (2006) Infantile restrictive cardiomyopathy resulting from a mutation in the cardiac troponin T gene. Pediatrics 117: 1830–1833. [DOI] [PubMed] [Google Scholar]
- 42. Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, et al. (1994) Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77: 701–712. [DOI] [PubMed] [Google Scholar]
- 43. Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR (2010) Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet 19: 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herron BJ, Lu W, Rao C, Liu S, Peters H, et al. (2002) Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet 30: 185–189. [DOI] [PubMed] [Google Scholar]
- 45. Kaneko-Oshikawa C, Nakagawa T, Yamada M, Yoshikawa H, Matsumoto M, et al. (2005) Mammalian E4 is required for cardiac development and maintenance of the nervous system. Mol Cell Biol 25: 10953–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boldt AB, Luz PR, Messias-Reason IJ (2011) MASP2 haplotypes are associated with high risk of cardiomyopathy in chronic Chagas disease. Clin Immunol 140: 63–70. [DOI] [PubMed] [Google Scholar]
- 47. Schwaeble WJ, Lynch NJ, Clark JE, Marber M, Samani NJ, et al. (2011) Targeting of mannan-binding lectin-associated serine protease-2 confers protection from myocardial and gastrointestinal ischemia/reperfusion injury. Proc Natl Acad Sci U S A 108: 7523–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exon-containing deletions in 1p36 cardiac-related genes found in various control and population-based cohorts catalogued in the Database of Genomic Variants.
(DOC)