Summary
The centromere is defined by the incorporation of the centro-mere-specific histone H3 variant centromere protein A (CENP-A). Like histone H3, CENP-A can form CENP-A-H4 heterotetramers in vitro [1]. However, the in vivo conformation of CENP-A chromatin has been proposed by different studies as hemisomes, canonical, or heterotypic nucleosomes [2–8]. A clear understanding of the in vivo architecture of CENP-A chromatin is important, because it influences the molecular mechanisms of the assembly and maintenance of the centromere and its function in kinetochore nucleation. Akey determinant of this architecture is the number of CENP-A molecules bound to the centromere. Accurate measurement of this number can limit possible centromere architectures. The genetically defined point centromere in the budding yeast Saccharomyces cerevisiae provides a unique opportunity to define this number accurately, as this 120-bp-long centromere can at the most form one nucleosome or hemisome. Using novel live-cell fluorescence microscopy assays, we demonstrate that the budding yeast centromere recruits two Cse4 (ScCENP-A) molecules. These molecules are deposited during S phase and they remain stably bound through late anaphase. Our studies suggest that the budding yeast centromere incorporates a Cse4-H4 tetramer.
Results and Discussion
The Budding Yeast Centromere Binds More Than One Cse4 from S Phase Onward
Previous studies have proposed conflicting numbers of Cse4 molecules per centromere with counts ranging from only one to up to eight [5–7]. A previous study also concluded that while there is only one Cse4 molecule per centromere prior to anaphase, this number doubles in anaphase [5]. To resolve this discrepancy, we wanted to determine whether the budding yeast centromere can incorporate more than one Cse4 prior to anaphase. To this end, we developed an assay based on bimolecular fluorescence complementation (BiFC). In this technique, two proteins of interest are fused to complementing fragments of a fluorescent protein. These fragments generate a fluorophore only if the proteins of interest physically interact [9]. In crystallographic data, the carboxyl termini of either H3 or Cse4 are situated ~ 2 nm apart, suggesting that these termini should facilitate robust BiFC maturation (Figure 1A [8]). Development of BiFC fluorescence can then provide clear, real-time confirmation of close physical proximity of more than one H3 or Cse4 molecule in live cells.
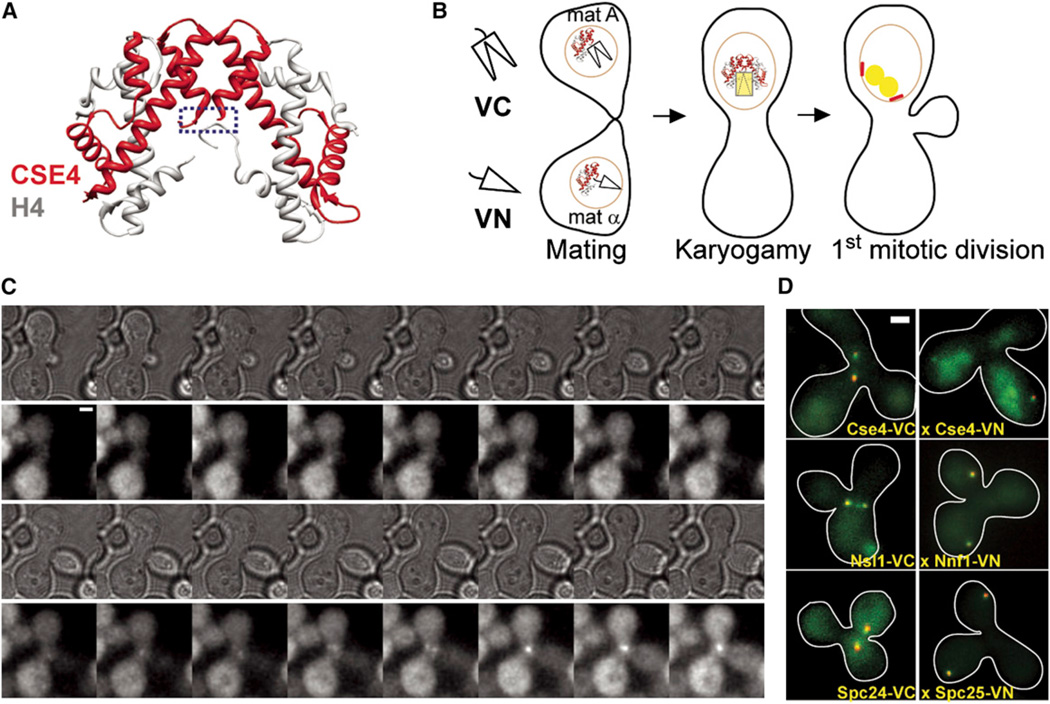
Figure 1. More Than One Cse4 Molecule Is Deposited at the Centromere in S Phase.
(A) Model of the H4-Cse4 heterotetramer shows that the carboxyl termini of the two Cse4 molecules (blue box) are separated by more than 2 nm.
(B) Schematic of approach to monitor the deposition of two or more Cse4 molecules at the centromere. Mating of two haploid strains expressing Cse4-VC (carboxyl fragment of Venus) and Cse4-VN (amino fragment of Venus) is followed by karyogamy, after which the newly diploid cell enters its first mitotic division. The two Cse4-BiFC species can interact with each other only after cell fusion. Therefore, development of Venus fluorescence in the zygote relative to the mitotic spindle (demarcated by red spindle pole bodies) will indicate Cse4 deposition at the centromere.
(C) Time-lapse microscopy showing the development of a single Venus fluorescence focus within the zygote after bud emergence, which coincides with entry into S phase (top panels, transmitted light images; bottom panels, maximum projection YFP images acquired every 5 min, scale bar represents ~ 2 µm).
(D) Colocalization of the BiFC fluorescence with a spindle-pole body marker (either Tub4-mCherry or Spc97-mCherry) confirms that the fluorescence is associated with centromere or kinetochore clusters (scale bar ~ 0.96 µm).
To determine whether more than one Cse4 molecule can bind to the centromere prior to anaphase, we constructed two haploid strains of opposite mating type. Each strain expressed Cse4 (or H3) fused with one BiFC fragment (VC or VN, corresponding to Venus carboxyl or amino fragment, respectively, [Figure 1B]). BiFC is possible only after these strains are allowed to mate and form diploid cells. As expected, we observed robust BiFC signal when the BiFC fragments were fused to H3, after two haploid cells were allowed to mate (Movie S1, available online). Surprisingly, we also detected BiFC signal with Cse4 within the zygote during the first mitotic division, soon after bud emergence (Figure 1C, n = 5/5, Movie S2). This fluorescence was located between two spindle-pole bodies, coincident with centromere distribution during mitosis (Figure 1D). We also applied BiFC to two known heterodimers within the Mtw1 and the Ndc80 complexes of the kinetochore. These heterodimers showed similar kinetics of BiFC fluorescence development in the zygote (Figure 1D). Therefore, the development of Cse4 BiFC fluorescence is consistent with the deposition of more than one Cse4 molecule in close physical proximity to the centromere.
To ensure that BiFC does not lead to nonnative dimerization, we fused VC and VN to the C termini of Nuf2 and Spc24, which are in the same complex but more than 10 nm apart, and histone H3 and Cse4. BiFC fluorescence could not be detected in either case. We also verified the presence of more than one Cse4 per centromere within close physical proximity using Förster resonance energy transfer (FRET, Figure S1).
High BiFC Maturation Efficiency Reveals Uniform Centromere Composition
Next, we investigated whether every centromere associates with more than one Cse4. To this end, we needed to determine the efficiency of Cse4-BiFC formation in live cells. This efficiency can be determined by measuring the ratio of fluorescence from a set number of molecules of Venus and from the same number of molecules of BiFC-Venus.
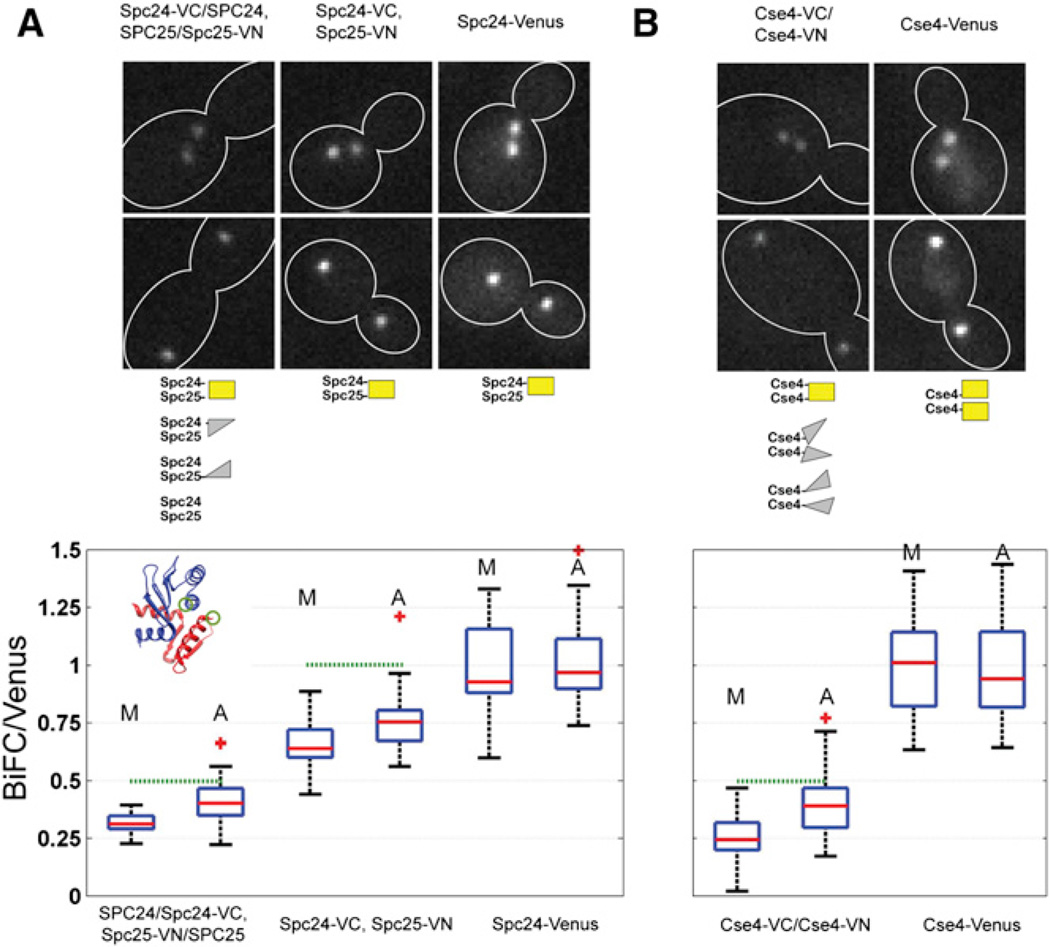
To generate strains with the same number of Venus and BiFC-Venus molecules, we labeled the Spc24/Spc25 heterodimeric subunits of the Ndc80 complex. The C termini of Spc24p and Spc25p are 2.4 nm apart, a separation similar in magnitude to the separation between Cse4 C termini in the Cse4-H4 heterotetramer (inset, Figure 2A). Furthermore, because the number of Spc24/Spc25 molecules per kinetochore is uniform, kinetochores in different strains will have the same number of Spc24/Spc25 molecules [10, 11]. Thus, strains carrying Spc24-Venus should have the same Venus intensity as those expressing both Spc24-VC and Spc25-VN, assuming 100% BiFC maturation efficiency (Figure 2A). We also constructed a heterozygous diploid strain (SPC24/SPC24-VC, SPC25-VN/ SPC25). In this strain, the BiFC/Venus ratio should not exceed 0.5. This is because only 25% of Spc24/Spc25 dimers possess the complementary BiFC fragments to form a Venus molecule, but the diploid contains twice as many kinetochores.
Figure 2. High BiFC Maturation Efficiency Suggests that Every Centromere Associates with More Than One Cse4 Molecule.
(A) Representative images of metaphase and anaphase cells from the Spc24-VC, Spc25-VN, and Spc24-Venus strains. The middle panel displays the possible Spc24/Spc25 dimer species. Box plot displays the relative BiFC and Venus fluorescence intensities (M, metaphase; A, anaphase). Inset shows the structure of the globular domain of the Spc24/25 dimer with the labeled carboxyl termini (green circles). The maximum attainable BiFC to Venus ratio in each case is indicated by the green dotted line.
(B) Representative images of metaphase and anaphase cells from the Cse4-VC/Cse4-VN and Cse4-Venus strains. Comparison of the Cse4-BiFC signal with the signal from a haploid strain expressing Cse4-Venus.
The box plots display the median with a red line, the 25th and 75th percentiles by the edges of the box, and the most extreme data points by the whiskers. Outliers are plotted individually. Note that the integration time and excitation intensity was lowered for Spc24/25 measurements. Hence, intensities should not be compared between the Cse4 and Spc24/25 panels.
The ratio of BiFC fluorescence from the haploid strain to Spc24-Venus fluorescence was 0.65 ± 0.16 in metaphase, and 0.75 ± 0.16 in anaphase (Figure 2A). In the heterozygous diploid strain, the ratio was measured to be 0.32 ± 0.07 in metaphase and 0.41 ± 0.11 in anaphase. Thus, the maximal maturation efficiency for BiFC involving Spc24/Spc25 ranges from 75% to 82%. The increase in the maturation efficiency from metaphase to anaphase represents an ongoing maturation process, because the number of Ndc80 complex molecules does not change [11]. Therefore, BiFC formation efficiency is less than the ideal value of 100% as expected, reaching ~ 80% in this control experiment.
Next, we used the same approach with Cse4 to determine BiFC efficiency. In the heterozygous diploid CSE4-VC/CSE4-VN strain, we measured a ratio of 0.26 ± 0.11 in metaphase, and 0.40 ± 0.16 in anaphase (Figure 2B). These ratios are consistent with the value of 0.5 expected from the relative concentrations of CSE4-VC/CSE4-VN heterodimer and homodimers that do not allow formation of BiFC (either VC/VC or VN/VN, middle panel, Figure 2B). The high BiFC efficiency of 80% in anaphase demonstrates that every centromere carries more than one Cse4 molecule in metaphase and anaphase.
It should be noted that the centromeres and kinetochores in the heterozygous diploid strains can be assembled from either the BiFC heterodimers or the BiFC homodimers (VC/VC or VN/VN), or even from wild-type proteins in the case of Spc24/Spc25. Our measurements show that the amount of BiFC heterodimers recruited at the centromeres/kinetochores conforms to the stoichiometry that can be expected from their intracellular concentration. Therefore, we conclude that the BiFC interaction does not perturb either cellular concentration or centromere/kinetochore incorporation of Cse4 or Spc24/Spc25.
Photobleaching-Assisted Counting Shows that the Centromere Associates with Two Cse4-GFP Molecules
Although the BiFC data demonstrate that the centromere associates with at least two Cse4 molecules, they do not reveal the absolute number. Previous studies compared fluorescence from Cse4 fused with green fluorescent protein (Cse4-GFP) with exogenous or in vitro standards of GFP to determine this number [6, 7]. However, such comparisons are susceptible to effects of the in vivo microenvironment (pH, etc.) on the photophysical properties of GFP. Therefore, to control for such effects, we conducted photobleaching-assisted counting of kinetochore proteins in the nucleus of live yeast cells. We could not use Cse4 or subunits of the Ctf19 or Cbf3 complex for this experiment because these proteins also associate with noncentromeric chromosomal loci and produce a significant and variable nuclear background (see Figure 2B). In contrast, Ndc80 complex subunits produce a low and uniform nuclear background when labeled with GFP. They are stably bound to the kinetochore, and each kinetochore contains a specific and reproducible number of molecules. The Ndc80 complex is thus ideally suited for this experiment.
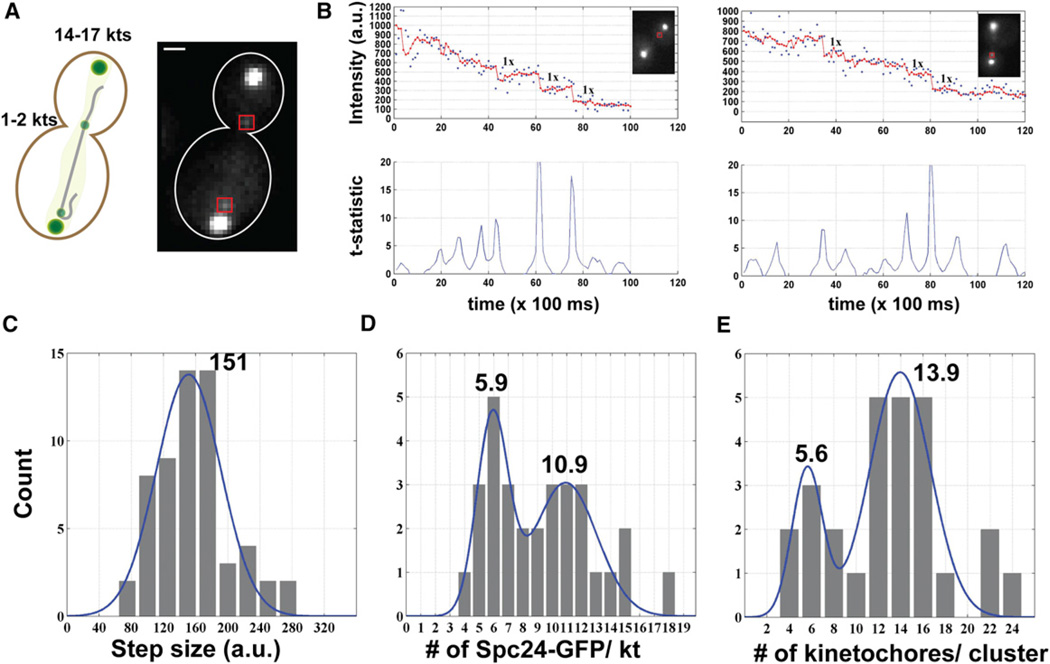
The main challenge in photobleaching-assisted counting of kinetochore proteins is the large number of GFP molecules within the kinetochore cluster. Large fluorophore numbers produce significant shot noise and noise due to GFP blinking. To overcome this challenge, we used a previously described method to generate lagging yeast kinetochores [11]. Fluorescence of such lagging kinetochores labeled with Spc24-GFP was continuously recorded to reveal steps during photo-bleaching (Figure 3A). We used previously described methods to detect step changes in the fluorescence signal [6, 12]. Briefly, the signal was filtered with the Chung-Kennedy algorithm using a five time-point window (100 ms/point) to detect edges in the data. We used the t test over the five time-point window in the filtered data to assign a statistical threshold (p < 0.01), and included visually verifiable steps for further analysis (Figures 3B and S2).
Figure 3. Photobleaching-Assisted Counting of Kinetochore Proteins in Live Yeast Cells.
(A) Conditionally dicentric chromosomes were used to create and image single lagging kinetochores, as described previously.
(B) Signal intensity during continuous photobleaching of one (left) or two (right) kinetochores (blue dots, raw data; red dots, data filtered with the Chung-Kennedy filter and a window size of 5). Bottom panel displays the t-statistic used to ensure a statistical significance of p < 0.01 on all detected steps. Visually verified events were selected for further analysis.
(C) Histogram of detected steps (n = 60 from 31 photobleaching experiments). When fit with a Gaussian (blue line, R2 = 0.90), the photobleaching step was found to be 151 ± 40 a.u.
(D) Histogram depicting the number of Spc24-GFP molecules per photobleached spot obtained by dividing the initial intensity of the spot (average over the first three time points) with the photobleaching step size (n = 30). Multipeak Gaussian fitting of the data, as above, suggests that there are 5.9 ± 1.1 Spc24-GFP molecules per kinetochore (blue curve, n = 30, R2 = 0.87).
(E) Histogram depicting the number of kinetochores in the main kinetochore cluster obtained by dividing the initial intensities of kinetochore clusters by the initial intensity of photobleached spots. Gaussian fit to the data yielded a ratio of 13.9 ± 2.7 verifying that the photobleached spots were either single lagging kinetochores or two kinetochores clustered close together (blue curve, n = 27, R2 = 0.77).
We fitted the histogram of measured step sizes with a Gaussian function to determine the basic step size of 151 ± 40 arbitrary units (a.u.) (Figure 3C). The ratio of this step size with the initial kinetochore fluorescence revealed that each kinetochore contains ~ 5.9 ± 1.1 Spc24-GFP molecules (Figure 3D). We verified that the diffraction-limited spots in these experiments were lagging kinetochores by comparing their initial intensity with the intensity of the nearby kinetochore cluster (Figure 3E). These scale-insensitive fluorescence measurements show that the anaphase kinetochore contains ~ 6 Ndc80 molecules. Using the 3.5:1 stoichiometry between Spc24-GFP and Cse4-GFP, the number of Cse4-GFP molecules per centromere can be calculated as 1.7 [7, 11]. Together with the BiFC measurements discussed above, these measurements suggest that there are only two Cse4-GFP molecules in metaphase and anaphase.
Our results are in partial agreement with a recent study that concluded that there are two Cse4-GFP molecules per centromere in anaphase [5]. However, we find no evidence for only one Cse4 molecule in metaphase. Notably, the study that reported one Cse4 in metaphase used a new method that is sensitive to changes in fluorophore density as well as number [5]. We previously observed that in metaphase, the kinetochore clusters are less compact [11]. Using model convolution to simulate images of kinetochore clusters, we found that the apparent increase in Cse4 copy number in anaphase can be explained by an increase in the kinetochore density from metaphase to anaphase (Figure S3).
Concentration Dependence of the Amount of CENP-A Recruited at Point and Regional Centromeres
Our findings are consistent with sequence-specific incorporation of Cse4 at the budding yeast point centromere. However, this conclusion is inconsistent with a previous study that suggested that the point centromere behaves like a regional centromere by incorporating four to six extracentromeric Cse4 molecules [6, 7]. Furthermore, this small number of molecules must be randomly dispersed within a 35 kb stretch of pericentric DNA independently of the sequence so that they are not detected by high-resolution biochemical methods based on chromatin immunoprecipitation and sequencing [13, 14]. This model of regional centromere-like behavior of the point centromere predicts that the number of Cse4 in the pericentromeric region should increase with Cse4 over-expression because the pericentric region provides significant excess capacity.
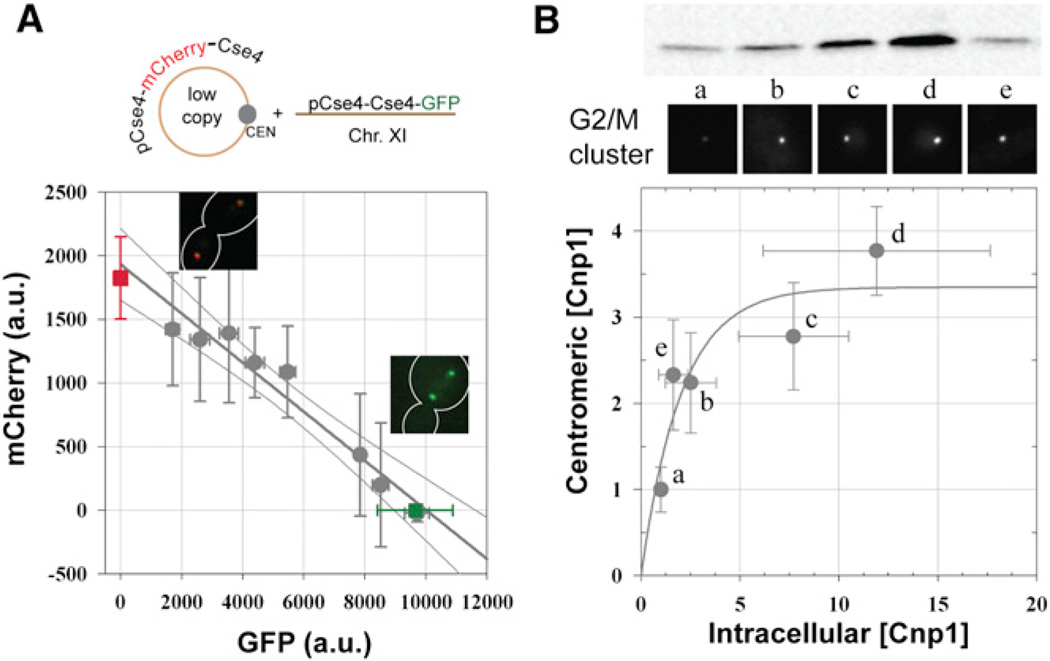
To test whether Cse4 recruitment increases with over-expression, we expressed mCherry-Cse4 ectopically from a low-copy-number plasmid in a strain constitutively expressing Cse4-GFP (Figure 4A). We reasoned that the cumulative fluorescence of Cse4-GFP and mCherry-Cse4 would be higher than the fluorescence of Cse4-GFP (or mCherry) alone, due to the additional recruitment of mCherry-Cse4 in the pericentromeric chromatin. However, we found that the GFP and mCherry signals measured for each centromere cluster were inversely correlated: any increase in the fluorescence of one color was accompanied by an opposite change in the fluorescence of the other (Figure 4A). The absolute value of the slope of the line (0.19) is consistent with the relative brightness of GFP and mCherry under the experimental imaging conditions (~ 0.26 as determined from Ndc80-GFP and Ndc80-mCherry signals). This experiment demonstrates that despite a 2-fold overexpression of Cse4, the number recruited to the centromere remains invariant. Moreover, a strict, negative correlation in the amount of Cse4-GFP and mCherry-Cse4 is observed. These data strongly suggest that the budding yeast centromere recruits a rigidly defined number of Cse4 molecules. Our data are consistent with the model that only the genetically defined point centromere recruits Cse4, as was determined by high-resolution biochemical experiments (Supplemental Information, Note S1 [13–15]).
Figure 4. Dependence of CENP-A Recruited at Point and Regional Centromere on Intracellular CENP-A Concentration.
(A) Relative levels of mCherry-Cse4 and Cse4-GFP recruited in anaphase centromere clusters in a strain expressing chromosomal Cse4-GFP and ectopic mCherry-Cse4 (top panel). Linear regression of the data (n = 79, binned based on the GFP signal) shows inverse correlation between the amount of the two Cse4 species (gray line, R2 = 0.96; data represent mean ± SD). The regression also predicts the signals measured in strains expressing only Cse4-GFP (green square) or Cse4-mCherry (red square, 95% confidence intervals represented by gray curves).
(B) Dependence of Cnp1-GFP recruited at the centromere in G2/M cells on its intracellular concentration. Relative intracellular protein concentration was quantified using western blot analysis (top inset) and normalized relative to the lowest expressing strain. Synchronized culture was not used, because the majority of fission cells in an asynchronous culture are in G2/M due to the long duration of this cell-cycle phase. The centromeric Cnp1-GFP in G2/M cells was quantified using previously described methods from the strains (representative images show clusters of six centromeres in G2/M cells) and normalized to the strain with the lowest centromeric signal (strain key: a: YWY277, b: AJY687, c: AJY686, d: AJY688, e: JW3523). Gray curve displays fit to the data with a single exponential rising to the maximum value (n = 4 for western blots, n > 30 for fluorescence measurements, error bars represent SD. R2 = 0.93).
To confirm that regional centromeres can recruit additional Cse4, we used the fission yeast (Schizosaccharomyces pombe) centromere. This regional centromere recruits Cnp1 (SpCENP-A) over a large centromeric span (~ 30–38 kb over three centromeres) independently of its sequence [16, 17]. We selected five fission yeast strains expressing Cnp1-GFP at different but stable levels, including endogenous expression level, as assessed by western blots (Figure 4B, top). We quantified the amount of centromeric Cnp1-GFP using fluorescence measurements in G2/M fission yeast cells (Figure 4B, top). We found that centromeric Cnp1-GFP amount increases linearly at low Cnp1-GFP expression levels, but levels off to a maximum value at higher expression levels (Figure 4B, gray curve-fit with single exponential rising to the maximum). These data demonstrate that Cnp1 incorporation at the regional centromere is a function of its intracellular concentration. Published reports have similarly found that Cnp1 levels at fission yeast centromeres can be depressed by histone H3 overexpression or by the deletion of Med20, a regulator of centromeric Pol-II activity [18, 19]. This plasticity in Cnp1 number demonstrates that the behavior of regional centromeres is distinct from point centromeres.
This experiment also provides additional support for our conclusion that there are two Cse4 molecules per budding yeast kinetochore. The fluorescence intensity of Cnp1-GFP in wild-type strains is 2.25 to 3-fold higher than the intensity of Cse4-GFP. If we assume that there are two Cse4-GFP molecules per budding yeast centromere and physiochemical conditions of the budding and fission yeast nuclei are similar, this ratio and our finding of two Cse4-GFP molecules per budding yeast centromere together estimate that there are 80–100 Cnp1-GFP molecules in the G2/M centromeric cluster of fission yeast. This value is consistent with ~ 100 Cnp1-GFP molecules reported recently (Supplemental Information, Note S2 [17]).
In conclusion, our data show that the budding yeast centromere contains two Cse4 molecules starting from S phase until late anaphase or telophase. Although these experiments cannot reveal the architecture of the centromeric locus directly, they suggest the presence of a Cse4-H4 tetramer. This tetrameric structure is consistent with the cell-cycle-dependent localization of the Cse4 chaperone Scm3. Scm3-GFP is excluded from the centromeres prior to anaphase and only localizes to the centromeres in anaphase in substoichiometric levels [1, 20]. Because Scm3 does not bind tetramers, it suggests that Cse4 is incorporated into a tetramer prior to anaphase. Additional work is needed to verify whether the Cse4-H4 tetramer exists at the centromere and, if not, the functional significance of two Cse4 molecules.
Our data prescribe the protein count for the budding yeast kinetochore and also for the fission yeast kinetochore nucleated by its regional centromeres [11]. The stoichiometry of proteins within the budding yeast kinetochore is well defined and invariant. Intriguingly, the same stoichiometry is conserved in regional centromeres, despite the ability to incorporate excess Cnp1 molecules. Thus, the count of two Cse4 molecules per centromere provides a key parameter for defining the kinetochore architecture at both point and regional centromeres.
Experimental Procedures
Strains and plasmids used in the study are listed in Table S1. Experimental procedures are described in detail in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
The authors thank Dr. Steve Henikoff and Dr. Xiangwei He for discussion, Dr. Richard Baker for plasmids and guidance with plasmid construction, Dr. Won-Ki Huh for the BiFC plasmids, and Dr. Mara Duncan for helpful comments on the manuscript. A.P.J. is supported by a Career Award at the Scientific Interface from the Burroughs-Wellcome Fund.
Footnotes
Supplemental Information
Supplemental Information includes three figures, two notes, one table, two movies, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2013.03.042.
References
- 1.Cho U-S, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 5.Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leake MC, Wilson D, Gautel M, Simmons RM. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys. J. 2004;87:1112–1135. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff S, Henikoff JG. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics. 2012;190:1575–1577. doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, Klenerman D, Carr AM, Sauer M, Allshire RC, et al. Quantitative single-molecule microscopy reveals that CENPA(Cnp1) deposition occurs during G2 in fission yeast. Open Biol. 2012;2 doi: 10.1098/rsob.120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 18.Carlsten JO, Szilagyi Z, Liu B, Lopez MD, Szászi E, Djupedal I, Nyström T, Ekwall K, Gustafsson CM, Zhu X. Mediator promotes CENP-a incorporation at fission yeast centromeres. Mol. Cell. Biol. 2012;32:4035–4043. doi: 10.1128/MCB.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castillo AG, Mellone BG, Partridge JF, Richardson W, Hamilton GL, Allshire RC, Pidoux AL. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007;3:e121. doi: 10.1371/journal.pgen.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luconi L, Araki Y, Erlemann S, Schiebel E. The CENP-A chaperone Scm3 becomes enriched at kinetochores in anaphase independently of CENP-A incorporation. Cell cycle. 2011;10:3369–3378. doi: 10.4161/cc.10.19.17663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.