Abstract
Background
Besides gene duplication and de novo gene generation, horizontal gene transfer (HGT) is another important way of acquiring new genes. HGT may endow the recipients with novel phenotypic traits that are important for species evolution and adaption to new ecological niches. Parasitic systems expectedly allow the occurrence of HGT at relatively high frequencies due to their long-term physical contact. In plants, a number of HGT events have been reported between the organelles of parasites and the hosts, but HGT between host and parasite nuclear genomes has rarely been found.
Results
A thorough transcriptome screening revealed that a strictosidine synthase-like (SSL) gene in the root parasitic plant Orobanche aegyptiaca and the shoot parasitic plant Cuscuta australis showed much higher sequence similarities with those in Brassicaceae than with those in their close relatives, suggesting independent gene horizontal transfer events from Brassicaceae to these parasites. These findings were strongly supported by phylogenetic analysis and their identical unique amino acid residues and deletions. Intriguingly, the nucleus-located SSL genes in Brassicaceae belonged to a new member of SSL gene family, which were originated from gene duplication. The presence of introns indicated that the transfer occurred directly by DNA integration in both parasites. Furthermore, positive selection was detected in the foreign SSL gene in O. aegyptiaca but not in C. australis. The expression of the foreign SSL genes in these two parasitic plants was detected in multiple development stages and tissues, and the foreign SSL gene was induced after wounding treatment in C. australis stems. These data imply that the foreign genes may still retain certain functions in the recipient species.
Conclusions
Our study strongly supports that parasitic plants can gain novel nuclear genes from distantly related host species by HGT and the foreign genes may execute certain functions in the new hosts.
Keywords: Cuscuta, Horizontal gene transfer, New genes, Orobanche, Parasitic plants, Strictosidine Synthase-Like Genes
Background
New genes provide novel traits to organisms and thus contribute to the adaption of species to new ecological niches [1,2]. The mechanisms by which organisms acquire new genes have been intensively studied and the rapidly accumulating genomic data have further supported the idea that most genes were originated from duplication events [3,4]. Besides gene duplication, retroposition, exon shuffling, trans-splicing, and de novo gene evolution, horizontal gene transfer (HGT) represents another critical source of gaining novel genes by directly introducing new genes to distantly related recipient species [1].
A large number of HGT events have been recognized in prokaryotes [2] and unicellular eukaryotes [5-7]. Although in multicellular animals, the separation of germ line cells from the soma expectedly leads to low HGT frequencies, genes of prokaryotic, fungal or plant origins have been discovered in diverse animals including bdelloid rotifer [8,9], tunicates [10], jelly fishes [11], starlet sea anemones [12], nematodes [13], aphids [14,15] and other insects [16,17]. The evolutionary significance of HGT has been well illustrated in some animals. For instance, acquiring two carotenoid biosynthetic genes from fungi endowed the pea aphids with red body color, based on which the variation between green and red aphids is further driven by the predators and parasites [18].
HGT is also involved in the adaptation and genome evolution of plants [19,20]. In nonvascular plants, Yue et al. identified 57 gene families in a moss nuclear genome with prokaryotic, fungal, and viral origins [21]. Most of these genes were most likely transferred to the ancestor of green plants and may have played significant roles during the transition of plants from being aquatic to terrestrial [21]. Extensive HGT has occurred between the organellar genomes of higher plants [19,22-24]. However, gene transfer between plant nuclear genomes was rarely reported [19,22]. Some exceptions are that a Mu-like element (MULE) in higher plants was found to be transferred between rice and Setaria nuclear genomes [25], and similarly, four independent genes in the C4 photosynthesis pathway were recurrently transferred from C4 to C3 plants in Panicoideae, and such HGT events may explain the origin of C4 plants in different species [26]. These HGTs were all between nuclear genomes and it is still unclear how these HGT events happened, since there are no direct physical interactions between donors and recipients.
Physical contact between the donors and the recipients theoretically facilitate HGT occurrence, since it increases the chance of genetic material transfer [7,27]. Parasites and hosts can form long-lasting intimate physical contact and thus parasitism may result in relatively high frequencies of HGT [5,7]. Consistent with this scenario, the majority of HGT events in higher plants found so far are from plant parasitic systems. In plants, hundreds of host organelle genes have been reported to be transferred from hosts to parasites [23,28,29], or vice versa [30-32], with most being targeted to the recipient mitochondria. In contrast, only few cases of HGT between the nuclear genomes of parasitic plants and hosts were reported [19,22]. The first confirmed HGT in parasitic systems involving nuclear genomes was that a gene with unknown function was transferred from a monocot host (likely Sorghum) to the eudicot parasite plant Striga hermonthica[33], and this was detected by analyzing the expressed sequence tags of S. hermonthica. With the rapid development of the next-generation sequencing technologies, increasing amount of transcriptome data become available. Rafflesia cantleyi (Rafflesiaceae) is an obligate and holoparasitic plant, and by screening the transcriptome data of this parasite and its obligate host Tetrastigma rafflesiae, Xi et al. proposed that several dozen genes were transferred from the host to the parasite, with most probably encoded by the nuclear genomes [34]. Likewise, Zhang et al. found that an albumin gene, encoding a seed storage and insect toxin protein, was transferred from legumes to the root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta pentagona and probably retained the same function after HGT [35].
Plants in the genus Orobanche (Orobanchaceae) are root parasites and form one of the largest groups of holoparasitic plants. Most Orobanche species have a narrow host range, while O. aegyptiaca is one of the exceptions, as it parasitizes many plants including Brassicaceae, Leguminosae, Solanaceae, Apiaceae, Asteraceae, and Cucurbitaceae [35,36]. O. aegyptiaca is mainly distributed in Mediterranean region and western Asia and can also been found sporadically in Africa, Australia, America, and eastern Asia [37]. Cuscuta (Convolvulaceae) plants are shoot parasites and represent another group of obligate parasitic plants, and most members of this genus have broad host ranges, which greatly overlap with the plant families infected by O. aegyptiaca[38], and have a wide geographic distribution that is similar to that of O. aegyptiaca[37].
The inconsistence between a species tree and a gene tree is an important indication of the occurrence of HGT. The genome data of Mimulus guttatus and Solanum (S. tuberosum and S. lycopersicum), the ordinal relatives of Orobanche and Cuscuta respectively, have been released [39]. M. guttatus (Phrymaceae) and O. aegyptiaca belong to the order Lamiales, and Solanum spp. (Solanaceae) and C. australis (Convolvulaceae) belong to the order Solanales. The available genomic information from M. guttatus and Solanum has provided an excellent opportunity for identifying HGT in Orobanche and Cuscuta. Here, we report that strictosidine synthase-like (SSL) genes from Brassicaceae were co-opted by the two parasitic plants, O. aegyptiaca and C. australis. Furthermore, these SSL genes belong to a new member of the SSL gene family and were originated by gene duplication uniquely in Brassicaceae. The presence of introns in the foreign SSLs of parasitic plants strongly suggests that host DNA, but not mRNA, was directly integrated into O. aegyptiaca and C. australis respectively. Furthermore, the expression levels of the SSL genes in O. aegyptiaca and C. australis varied in different developmental stages and organs and the SSL gene in C. australis was inducible after wounding. These results support the scenario that during parasitization nuclear genes can be transferred from hosts to parasitic plants, and the foreign genes may provide their new hosts with novel traits, which might be beneficial for adaptation.
Results
Identification of foreign SSL genes in Orobanche aegyptiaca and Cuscuta australis
O. aegyptiaca is a root holoparasitic plant that parasitizes many plant species. Recently, a large set of transcriptome data was released (Parasitic Plant Genome Project; [40]). Taking advantage of these published databases, we downloaded the assembled transcriptomes of O. aegyptiaca and screened for foreign genes (see Additional file 1 for the procedure). More than 2100 sequences were obtained initially which were predicted as HGT candidates by AlienG [41]. Among these, a 691-bp transcript fragment showed 88 and 89% identity at the nucleotide and amino acid level, respectively, to a strictosidine synthase-like (SSL) gene in Arabidopsis thaliana (hereafter, Arabidopsis) (AT2G41300), while it was highly divergent from the homologs in Orobanche relative Mimulus guttatus (49% identity at the amino acid level and no significant similarities at the nucleotide level). To obtain the complete SSL transcript sequence, we re-assembled the RNA-seq datasets of O. aegyptiaca using Trinity [42] and obtained a 1597-bp cDNA sequence with a putatively complete open reading frame (ORF) encoding 369 amino acids (hereafter OaSSL, NCBI accession number: KF817594).
Considering that the shoot parasitic plant C. australis also parasitizes Brassicaceous plants, we investigated whether C. australis also gained the SSL gene. A homemade transcriptome assembly of C. australis was searched using OaSSL as the query, and 2 highly similar transcripts were found, which were very likely derived from one gene by alternative splicing (see below). This C. australis SSL (hereafter CaSSL) also showed high similarities with the same Arabidopsis SSL at the nucleotide and amino acid level (84 and 88%, respectively), but exhibited much lower similarities with its Solanales homologs in S. tuberosum and S. lycopersicum (49% and 50% at the amino acid level, no significant similarities at the nucleotide level). These data strongly suggested that these two parasitic plants acquired a SSL gene from Arabidopsis or certain other Brassicaceous species.
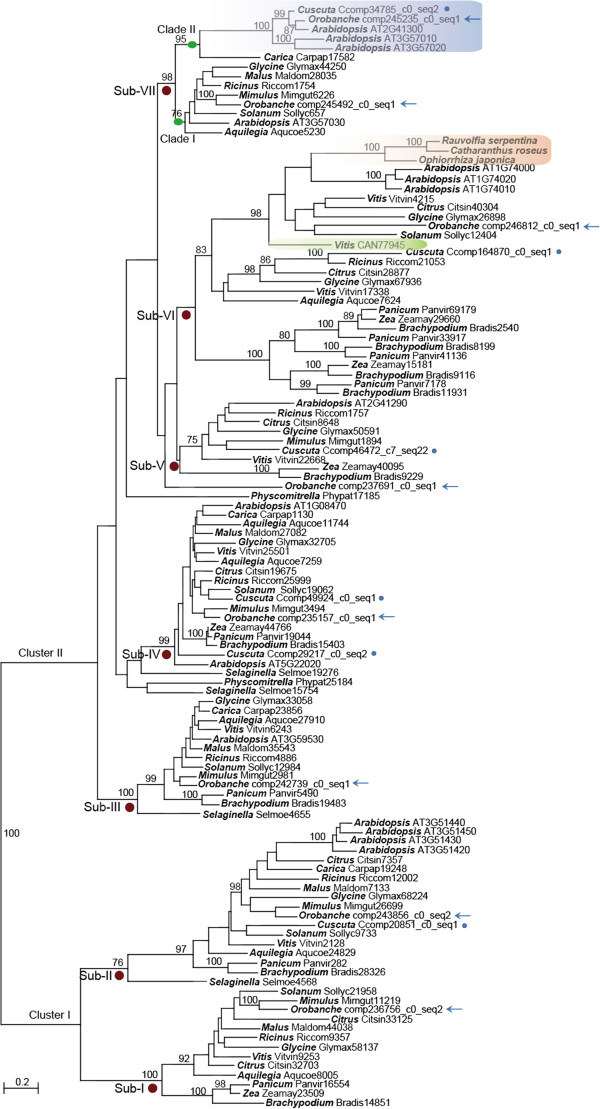
To gain insight into the evolution of SSL genes and rule out the possibility that OaSSL and CaSSL were originated from non-Brassicaceous plants, SSL sequences from 15 representative plant genomes and the two parasitic plants were used for constructing a gene tree (Figure 1). Phylogenetic analysis using all SSL members in Arabidopsis and the representative homolog sequences from other species (the sequences that clustered as sister branches in the phylogenetic tree were removed) revealed that SSL gene family contained two highly divergent clusters, Cluster I and Cluster II, and multiple subfamilies. Cluster I comprised 2 subfamilies with each containing homologs from the major clades of land plants, including eudicots and monocots; Cluster II contained 5 subfamilies, among which sub-family VII (Sub-VII) was only composed of the homologs from eudicots but the other 4 subfamilies included homologs from both eudicots and monocots. Transcriptome analysis revealed that both parasitic plants contain multiple SSL copies, 7 copies in O. aegyptiaca belonging to 6 subfamilies and 5 copies in C. australis from 5 subfamilies.
Figure 1.
Molecular phylogeny of the strictosidine synthase-like (SSL) protein family in plants. To reduce the size of the tree, only one sequence among multiple highly similar copies in each plant was used, except that all the SSLs of Arabidopsis thaliana were used. Genus names and sequence IDs are shown in the nodes. Numbers above branches indicate bootstrap support values of maximum likelihood when they are more than 75%. Red circles below the branches indicate Sub-families I to VII (Sub-I to Sub-VII). Green circles indicate the branches of Clade I and (Brassicales-specific genes). The sequences from parasitic plants O. aegyptiaca and C. australis are indicated by arrows and dots, respectively. The interested SSL genes in O. aegyptiaca and C. australis and their Brassicaceae donor are indicated in blue. Three sequences from Rauvolfia serpenti, Catharanthus roseus, and Ophiorrhiza japonica with biochemical evidence for the activity of strictosidine synthase are indicated in orange. SSL in Vitis vinifera, which has no strictosidine synthase activity detected, is indicated in green.
Sub-VII contained 2 clades, Clade I and Clade II. Although Clade I contained homologs from diverse species of eudicots, Clade II only included SSL sequences from Brassicales and the 2 parasitic plants (Figure 1), and the OaSSL and CaSSL clustered with 3 Arabidopsis SSL genes with 100% bootstrap support. Thus, Clade II may be originated in Brassicales by gene duplication from Clade I before the divergence of Brassicales (the SSL genes in Clade I are hence called the original copies of the SSL genes in Clade II), and OaSSL and CaSSL were very likely derived from Brassicaceae by HGT.
To gain more insight into the potential gene donors of the SSL genes in Orobanche and Cuscuta, we collected all SSL sequences from species with available genome sequences and also from the transcriptome assemblies of 3 shoot parasitic plants, Cuscuta pentagona (Convolvulaceae), Cassytha filiformis (Lauraceae), and Pilostyles thurberi (Apodanthaceae) from the 1KP Project (http://www.onekp.com/project.html), and 2 root parasitic plants, Triphysaria versicolor and Striga hermonthica from the Parasitic Plant Genome Project [40]. Phylogenetic analysis showed that only certain Brassicaceae SSL genes clustered closely with OaSSL and CaSSL. All these sequences clustered were used to infer the specific origins of foreign SSLs. The tree showed 3 groups, Group I to III, and the SSL genes from the two parasitic plants were assigned to Group II with 100% bootstrap support (Figure 2). Importantly, both OaSSL and CaSSL shared a single amino acid deletion and multiple unique amino acids that are uniquely conserved in Group II SSL proteins (Figure 2). These results indicated that OaSSL and CaSSL were originated from Brassicaceae, although we were not able to determine the exact donor species, since OaSSL and CaSSL showed ambiguous relationships with other species in Group II (Figure 2).
Figure 2.
Molecular phylogeny and partial alignment of the Brassicaceae-specific SSL proteins and the foreign SSLs in O. aegyptiaca and C. australis. Sequence IDs are shown after the species names. Numbers above or below the branches indicate bootstrap support values of maximum likelihood no less than 75%. The sequences from parasitic plants O. aegyptiaca and C. australis are highlighted in boxes. The respective partial protein sequences are shown on the right side of the species names and sequence IDs. The amino acids and the single amino acid deletion uniquely conserved in Group II are shaded.
The structures of Brassicaceae-specific SSL genes, OaSSL in O. aegyptiaca, and CaSSL in C. australis
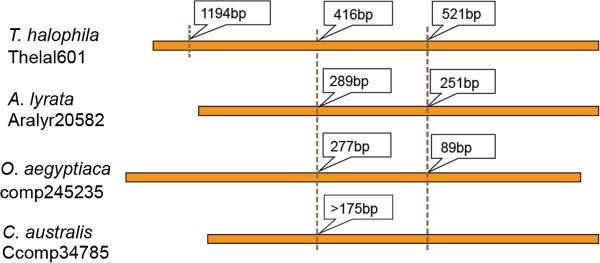
Comparison of 23 SSL mRNA sequences with their respective genomic DNA sequences in 5 Brassicaceae species indicated that Brassicaceae SSL genes contained maximally 3 introns and among them 18 genes lacked the first intron. The positions of intron 2 and intron 3 were well conserved but the intron lengths varied within and between species (Additional file 2).
Only one copy of OaSSL transcript was identified in the transcriptome of O. aegyptiaca. Using genomic DNA as the template, we cloned its genomic sequence (NCBI accession number: KF817597): it contained 2 introns, whose positions were identical to the intron 2 and 3 of the SSL genes in Brassicaceae (Figure 3). Analysis of C. australis transcriptome assembly revealed 2 isoforms of CaSSL, which were named Seq1 (NCBI accession number: KF817596) and Seq2 (NCBI accession number: KF817595). Seq1 shared 100% identity with Seq2 except that Seq1 had a 21-bp insertion (Additional file 3). The insertion was located at the same position as intron 2 in Brassicaceae-specific SSL genes, thus Seq1 was very likely originated from partial retention of intron 2. The putative protein sequences of Seq1 and Seq2 aligned well with SSLs from Brassicaceae without any frame shifts, suggesting that Seq1 and Seq2 encode functional proteins (Additional file 3). Genomic PCR was used to amplify the sequence of CaSSL gene, but various primers spanning the intron region failed to produce any products. We speculate that the intron in CaSSL may have complex secondary structures or may be very long. Intron 3 was obviously absent in CaSSL (NCBI accession number: KF817598, Figure 3).
Figure 3.
Gene structures of representative Brassicaceae-specific SSLs and the foreign SSLs in O. aegyptiaca and C. australis. The orange bars indicate the exons. The vertical dotted lines show the intron positions, with intron lengths marked in boxes. The intron length and position in C. australis were inferred from the difference in the two CaSSL mRNA isoforms.
By HGT, DNA can be transferred into nuclear, plastidal, or mitochondrial genomes of plants. All currently known Brassicaceae-specific SSL genes are encoded by the nuclear genomes. Nuclear or mitochondrial genomes from Orobanche or Cuscuta genera are not available, but a BLAST search of multiple chloroplast genomes of Cuscuta species [43] (chloroplast genome of C. australis is yet unavailable) indicated that none of the plastidal genomes contained homologs of CaSSL gene. We speculate that the CaSSL gene is likely in the nuclear genome of C. australis, although the possibilities of being in chloroplast or mitochondrial genome could not be completely ruled out. This might be also true for the OaSSL in O. aegyptiaca, since the current plastid genomes from Orobanche do not possess the SSL genes [44].
Expression analysis of AtSSL1 in A. thaliana, OaSSL in O. aegyptiaca, and CaSSL in C. australis
Because no functional studies have been reported for the Brassicaceae-specific SSL genes, we chose the AtSSL1 gene in Arabidopsis (AT2G41300, in Clade II, Figure 1), which showed the highest similarities to OaSSL in O. aegyptiaca and CaSSL in C. australis, as a representative and investigated its expression profile in various developmental stages, and after diverse stress stimuli and treatments using GENEVESTIGATOR [45]. AtSSL1 was expressed in all developmental stages at rather similar levels (Additional file 4). Strong up-regulation of AtSSL1 expression was observed in inflorescence stem upon addition of 1-naphthaleneacetic acid and in roots inoculated with the nematode Heterodera schachtii; a large down-regulation was found in root after low oxygen treatment (Additional file 5). Therefore, we speculated that AtSSL1 might be involved in plant development and stress responses.
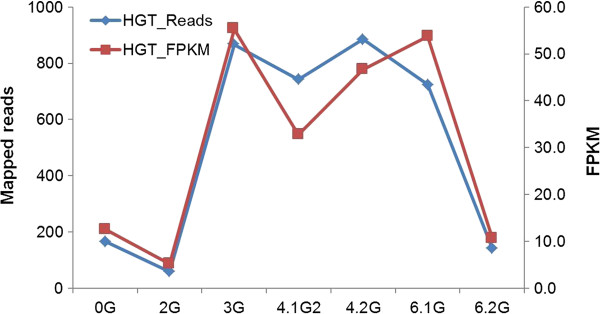
To gain insight into the potential function of OaSSL gene in O. aegyptiaca, we examined the expression profiles of OaSSL using the paired-end RNA-seq datasets in PPGP generated from different developmental stages of O. aegyptiaca[40] (Figure 4). OaSSL shows 3- to 5-fold increased transcript abundance during haustorial formation and vegetable growth stages (Figure 4), suggesting that OaSSL gene might perform functions in haustorial formation and vegetable growth.
Figure 4.
Expression levels of the OaSSL gene at different developmental stages. The numbers of mapped reads mapped to the cDNA were shown in the left Y-axis. The normalized expression levels were estimated by the fragments per kilo base of exon per million fragments mapped (FPKM, one fragment refers to a pair of reads from the paired-end data) in Illumina RNA-seq datasets that are from different developmental stages (PPGP). 0G, seeds imbibed, pre-germination; 2G, seedling after exposure to haustorial induction factors; 3G, haustoria attached to host root; early penetration stages, pre-vascular connection; 4.1G, early established parasite; parasite vegetative growth after vascular connection; 4.2G, spider stage; 5.1G, pre-emergence from soil - shoots; 5.2G, pre-emergence from soil - roots; 6.1G, post emergence from soil - vegetative structures, leaves/stems; 6.2G, post emergence from soil - reproductive structures, floral buds.
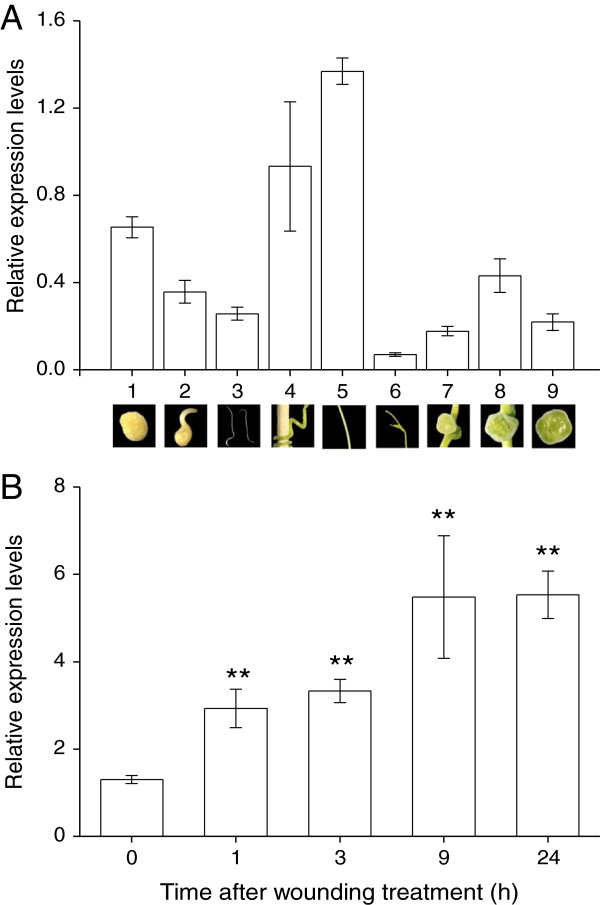
Given that there are no public transcriptome databases available for Cuscuta spp., quantitative real time-PCR (qPCR) was done to measure the expression levels of CaSSL in C. australis in different developmental stages and after wound treatment. Because selection of suitable reference genes is important for qPCR assay, four commonly used reference genes, actin, EF-1α, EF-TU, and PP2A, were evaluated with geNorm [46], DeltaCT [47], BestKeeper [48], Normfinder [49], and RefFinder [50]. The result showed that EF-1α was stably expressed under all conditions (Additional file 6). Thus, EF-1α was used as the reference gene. CaSSL was expressed at all 9 different developmental stages or organs (Figure 5A), and the highest expression level was found in mature stems and the lowest was in shoot tips (the difference was 19.7-fold) (Figure 5A). Additionally the transcriptional levels of CaSSL before and 1, 3, 9, and 24 h after wounding were determined. The expression level of the CaSSL gene was up-regulated 1.3-fold 1 h after treatment and increased over time (increased 3.2-fold 24 h after treatment) (Figure 5B). These results imply that CaSSL might be involved in development and abiotic stresses, such as wounding.
Figure 5.
Expression level analyses of the CaSSL gene in C. australis during different developmental stages and after wounding treatment by qRT-PCR. A. Expression levels (±SE, n = 5) of the CaSSL gene at different developmental stages and in different organs. 1, seeds; 2, just germinated seeds; 3, seedlings; 4, pre-haustoria; 5, stems; 6, shoot tips; 7, floral buds; 8, flowers; 9, capsules. A picture of each developmental stage was shown below its expression labeling. B. Expression levels (±SE, n = 5) of the CaSSL gene after wounding treatment in stems. Asterisks indicate significant differences between treatment and 0-h time point (**P < 0.01; Student’s t-test).
Functional divergence and selection analysis
Compared with its original gene copies (Clade I, Figure 1), the Brassicaceae-specific genes (Clade II, Figure 1) present as a relatively long branch in the phylogenetic tree, which suggests that functional divergence between the two subfamilies probably had occurred after the gene duplication event. To evaluate this scenario, two types of functional divergence (type I and type II) between these two subfamilies were estimated by posterior analysis with DIVERGE 2 [51]. Type I functional divergence (altered functional constrains) represents the conserved amino acid sites in one clade but highly variable in the other; type II functional divergence detected the conserved amino acid sites in both clades but with changed biochemical properties [52]. The coefficient of type I functional divergence was significantly greater than 0 (θ1=0.21040 ± 0.0343, LRT = 37.5692, p < 0.01) and 7 amino acid sites possessed a posterior probability higher than 0.7, which suggested that functional constrains on these 7 sites had shifted for most members between the two clades. No type II-related radical changes happened between the two clades after gene duplication. These results suggested that type I functional divergence was the dominant patter for the genes in Clade I and Clade II and altered site-specific selective constrains probable had led to group-specific functional changes between them.
We further investigated whether strong selection was involved in the evolution of the SSL genes in Clade I and II (Figures 1 and 2) using likelihood ratio tests developed by Yang et al. [53]. We firstly used the site models to detect whether positive selection occurred in some sites among the Brassicaceae-specific SSL genes, their original copies, and the foreign SSL genes in the two parasitic plants. No positive selection was detected in these genes (Additional file 7). Notably, the maximum likelihood estimates of the value under M0 model approached 0, 0.1777 in the large dataset (the Brassicaceae-specific genes and their original copies) and 0.1903 in the small dataset (the Brassicaceae-specific gene copies and two foreign SSL genes in the parasitic plants). The log-likelihood differences between models M0 and M3 were statically different for both the large dataset (LRT = 630.293, p < 0.01) and the small dataset (LRT = 387.605, p < 0.01) (Additional file 7). These suggested that relaxed purifying selection was the main driving force in the evolution of these genes.
Under the branch-site models, the null hypothesis and alternative hypothesis were examined by setting the Brassicaceae-specific SSL genes and each of the two parasitic genes separately as foreground. No evidence indicated that positive selection acted on the Brassicaceae-specific SSL genes and the CaSSL gene in C. australis (Additional file 8). However, two codon sites in the OaSSL gene of O. aegyptiaca have probably undergone positive selection (LRT = 5.754, p < 0.05) with more than 75% posterior probability (Additional file 8). Codon 181 encoded a glycine (G) or arginine (R) in the Brassicaceae-specific SSL genes, whereas it encoded an asparagine (N) in the OaSSL gene of O. aegyptiaca; site 267 was a serine (S) in the OaSSL gene, which showed a 0.975 posterior probability of positive selection, while it encoded an asparagine (N) or glutamic acid (E) in the Brassicaceae-specific genes (Additional file 9).
Discussion
The Brassicaceae-specific SSL genes and the alien origins of SSL genes in the parasitic plants O. aegyptiaca and C. australis
The SSL genes in plants constitute a large gene family that shows high levels of divergence among different subfamilies. Among them, Sub-VII is relatively special since it includes one clade (Clade I) containing SSL genes from different species of eudicots and the other clade (Clade II) only has species from Brassicales and the two parasitic plants studied in present work. It is very likely that SSL genes in Clade II were formed only in Brassicales via gene duplication of an ancestral SSL gene in Clade I.
Intriguingly, in this work we found that two SSL genes of Brassicaceae have been introduced to the two parasitic plants by horizontal gene transfer. This is strongly supported by the following evidence: 1) high identity values in the multiple sequence alignment, 2) shared indels and amino acids, and 3) robust bootstrap values in the phylogeny analyses. From these lines of evidence, the possibility of convergent evolution can be ruled out. Because O. aegyptiaca and C. australis belong to different taxa, the gene transfer events leading to the two foreign genes in the two distantly related parasitic plants should occur independently.
The possible transfer mechanism
The existence of introns in OaSSL and CaSSL gene sequences indicates that both gene transfer events occurred not at the mRNA level but at the DNA level. Similarly, it was reported that the foreign albumin gene in O. aegyptiaca also contains introns and thus should be transferred as DNA [35]. Parasitic plants are known to transport thousands of mRNAs from their hosts [54,55]; however, it is likely that DNA, but not mRNA, transferred from a plant to another is the major form of HGT in plants. Probably this is because mRNA has to be reversed transcribed to DNA before they could be integrated into a plant genome and the activity of this type of reverse transcription is extremely low.
The occurrence of any HGT event at DNA level in plants requires multiple steps: 1) entering the cytoplasm after crossing the cell wall and cell membrane, 2) entering the nucleus through the nuclear membrane, 3) integration into the genome, 4) fixation and transfer to the next generation, 5) spread inside the population. The proposed mechanisms of HGT in plants include illegitimate pollination, vector-mediated, and plant-plant contact hypothesis [5,19]. Illegitimate pollination hypothesis presumes that the pollen grains of the gene donors can germinate on the stigma of the reproductive isolated species, after pollination, recombination may allow some foreign genes to be integrated into the other set of chromosomes, and hybrids with the same species and natural selection may lead to the retain of the target chromosome set with some foreign genes within the population. This HGT mechanism is considered to occur between closely related species, such as the HGT cases between Setaria and Oryza[25], between Poa and Festuca[56], or between C4 and C3 species in Alloteropsis[26]. Because both O. aegyptiaca and C. australis are distantly related with Brassicaceae, the illegitimate pollination hypothesis is very unlikely.
The vector-mediated transfer hypothesis assumes that viruses, pathogens, endophytes, or other media act as vectors to transfer the foreign DNA into the recipients. Because most vectors have limited host ranges, vector-mediated HGT from Brassicaceae (or from Fabaceae, in the case of albumin 1[35]) to Orobanchaceae and Convolvulaceae appears to be unlikely. Only vectors with a wide host range, such as apple latent spherical virus [57], may be able to mediate these transfer events. However, no plant viruses carrying the foreign genes have been discovered so far. This is possibly due to fast replication of virus which may cause loss of foreign genes during amplification. Evidence of virus carrying foreign genes is needed to confirm the vector-mediated HGT as a plausible mechanism.
Plant-Plant contact hypothesis suggests that the foreign DNA is imported into the recipients via the direct contact between plants, such as the long-term intimate contact in parasitic systems. Parasitism establishes a functional symplastic pathway between parasitic plant and host, allowing transport of nutrients, RNAs [54,58], proteins [59-61], viruses [62], and phytoplasma [63] from the hosts to the parasites. Currently, no direct evidence indicates that host DNA molecules can be transported into parasitic plants, but movement of large DNA fragments and even whole chloroplasts has been observed in the graft regions between the same species [64] or between sexually incompatible species [65]. The connections between parasitic plants and their hosts are highly similar to the artificial graft junctions; thus, regardless of whether the transfer of DNA is limited to cell-to-cell in a short range or can occur in relatively long distances, the fusion of host and parasite vascular systems provides possibilities for foreign DNA to travel to the recipients. Because the concentration of imported macromolecules also decreases along the stems [55,61], the probability for the integration of foreign genetic materials decreases with the distance from the haustoria. We hypothesize that Brassicaceae SSL genes were firstly integrated to the cells of O. aegyptiaca and C. australis haustoria or nearby tissues, and certain cells somehow developed into floral meristems, allowing the host genes to pass to the next generations.
Functional implication of the foreign SSL genes in O. aegyptiaca and C. australis
The initial biochemical evidence for the function of a SSL gene was from Rauvolfia Serpentina, in which the activity of a strictosidine synthase (STR) that catalyzes the stereospecific condensation of tryptamine and secologanin to form strictosidine (Pictet-Spengler reaction) was detected [66], and later, the coding gene was cloned [67]. Because strictosidine is the key intermediate in indole alkaloid biosynthesis, which is the precursor of several clinically useful anti-cancer alkaloids, the STR genes were successively elucidated in Catharanthus roseus[68], Rauwolfia verticillata[69], and Ophiorrhiza japonica[70]. However, compared with the catalytic feature and the active site information in the 3D-structure of STRs [71,72], Hickes et al. suggested that the great majority of sequences annotated as STR do not catalyze the Pictet-Spengler reaction [73].
We added all sequences encoding proteins with confirmed STR activity in our comprehensive phylogenetic analysis, and they clustered as a highly supported clade (Figure 1). One sequence from Vitis vinifera (CAN77945) is related to those with verified STR activity in our phylogenetic tree. No STR activities but hydrolytic activities were detected in this SSL gene of V. vinifera[73]. Given the low sequence similarities and distant relationships of the genes in Clade II to the real STRs, the Brassicaceae-specific and the foreign SSL genes in the parasitic plants probably do not catalyze the Pictet-Spengler reaction. We found that the SSL genes in Arabidopsis and the parasitic plants are actively transcribed in multiple developmental stages and following different treatments, but their biological functions need to be further studied.
Parasitic plants face at least two challenges. First, they need to conquer host defenses while at the same time absorbing nutrients from the hosts; second, they have to resist herbivores, bacteria, fungi, or abiotic environment stresses, such as wounding. Previous work had indicated that transferred genes in nematodes from microorganism may have enabled the parasites to modulate the host defense systems [74]. Parasitic oomycetes probably acquired certain secretory proteins from fungi, which inhabit the same niche with the oomycetes, and thus are able to suppress certain host defense responses [75]. Data-mining in the public microarray database revealed that the most related Arabidopsis AtSSL1 also responds to the treatments of hormones, abiotic and biotic stresses. OaSSL in O. aegyptiaca showed different expression levels at different developmental stages and had undergone positive selection. Similarly, in C. australis, CaSSL also exhibited developmental stage- and organ-specific expression levels; furthermore, wounding transcriptionally activated CaSSL. Therefore, the foreign SSL genes in the two parasitic plants might be involved in plant development and secondary metabolism, which is normally associated with herbivore and pathogen resistance [76,77] or other environmental stresses [78].
Conclusion
HGT from the host nuclear genome to parasitic plants has been rarely reported so far, here we provide solid evidence indicating that two nuclear-encoded SSL genes from Brassicaceae species were co-opted by two distantly related holoparasitic plants, the root parasitic plant O. aegyptiaca and the shoot parasitic plants C. australis. Thus, the physical connection with host plant and the parasitic lifestyle including the transport of host nutrients and macromolecules may give parasites the opportunity to obtain host genes by HGT, and HGT between host and parasitic plant nuclear genomes may not be very rare. Our analysis also suggests a potential role of the horizontally transferred genes in the evolution and adaptation to parasitic lifestyle or environment.
Methods
Data sources
All the 12 Illumina datasets from O. aegyptiaca and the original transcriptome assembly OrAeBC4 were retrieved from the Root Parasitic Plant Genome Project (PPGP) website (http://ppgp.huck.psu.edu/). The transcriptome assemblies of other parasitic plants from PPGP, Striga hermonthica (StHeBC2), Triphysaria versicolor (TrPuRnBC1), and Triphysaria pusilla (TrVeBC2), were also downloaded. The protein database needed in AlienG [41] for BLAST search included NCBI non-redundant (nr) database (Sep. 2012) and all predicted proteins of 22 plant genomes which were available at Phytozome (version 9.0) [39] but absent from nr database. The nucleotide database included NCBI nucleotide collection (nt) and all the predicted transcripts from 22 plant genomes from Phytozome. The databases in the 1 KP project (http://www.onekp.com/project.html; which includes the transcriptome assemblies from Cuscuta pentagona, Cassytha filiformis, and Pilostyles thurberi), PlantGDB (http://www.plantgdb.org), and SOL Genomics Network (http://solgenomics.net) were searched online.
Transcriptome screening of horizontally transferred genes in O. aegyptiaca
Because parasitic plants transport thousands of mRNAs from their hosts, the dataset used for the original transcriptome assembly includes samples attached to the hosts, and therefore the assembly contains contamination from the host mRNAs. To remove the contaminated sequences, the reads from two samples that have not attached to the hosts, seed germination (OrAe0G) and germinated seed, radicle emerged, and pre-haustorial growth (OrAe1G), were mapped to the unique sequences using RSEM [79] and the assembled sequences with mapped reads were kept. As for the assembled sequences with length no less than 300 nt, the potential ORFs were predicted locally by OrfPredictor [80] using default settings, and the amino acid sequences whose lengths were not less than 100 were screened for genes with potential alien origins using AlienG [41]. The alien origin of a gene was predicted if the score ratio of the first non-Lamiales hit to the first Lamiales hit was more than 1.2. The obtained candidates were further filtered to exclude those with M. guttatus affiliation in their corresponding cDNA sequences by BLAST search, since some non-coding genes were found to be rigidly translated into proteins.
Total RNA extraction, cDNA library construction and transcriptome sequencing of C. australis
Seeds of C. australis were treated with sulfuric acid for 20 min to loosen the seed coat, and then the sulfuric acid was removed by extensive rinsing with water. The seeds were kept at 25°C for 3–4 days on wet filter paper until the seedlings were ~ 4 cm long. The seedlings were gently fastened to young soybean (Glycine max) plants (~ 20–25 cm tall; cultured at 26 ± 2°C,12 h light) using cotton threads and water was sprayed to these dodder seedlings 2–3 times/day to prevent them from drying out until parasitization was established. Total RNA was extracted from C. australis stem using the RNAeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. Potential DNA contamination was removed by DNase treatment (Qiagen). The cDNA library was constructed using the Truseq RNA and DNA Sample Prep Kit following the user manual except that the insert fragments after adaptor ligation were set to 300 bp (the cDNA insertion length is about 180 bp). The paired short reads (2 × 101 bp) were generated on an Illumina Hiseq™ 2Clade I instrument. Image deconvolution and quality calculation were conducted by using the Illumina GA pipeline 1.6.
Identification of SSLs in the transcriptomes of the two parasitic plants
The potential adaptor tags and their following sequences were removed from all raw reads in the Illumina datasets from O. aegyptiaca and C. australis using cutadapt [81]. The clean reads were obtained by further filtering out bases with low quality values (below 25) using Btrim [82]. De novo assembly of RNA-seq data was carried out using the Trinity software package [42] with the maximum length expected between fragment pairs set to 300. The foreign SSL cDNA fragment from the assembly of O. aegyptiaca in PPGP was used to search against the home made Trinity assembly of this species. The obtained putative full-length cDNA was used to search against the transcriptome assemblies of C. australis and other parasitic plants, including Striga hermonthica, Triphysaria versicolor, Triphysaria pusilla, Cuscuta pentagona, Cassytha filiformis, and Pilostyles thurberi.
Expression level estimation of OaSSL gene by transcriptome analysis, visualization of mapping of RNA-seq Reads to the assembled sequences, and microarray expression analysis of Arabidopsis AtSSL1 gene
The expression levels of the foreign SSL gene in O. aegyptiaca in different tissues and developmental stages were estimated using RSEM [79] by mapping all the clean reads obtained above to our Trinity assemblies in O. aegyptiaca. To check the possible assembly errors, mapping of all the clean reads from all these datasets to the foreign SSL cDNAs was visualized with Tablet [83]. Web-based expression analysis of A. thaliana AtSSL1 gene (AT2G41300) in different tissues and multiple development stages and under various treatments was performed using GENEVESTIGATOR (https://www.genevestigator.com/gv/, [45].
Cloning of SSL genes in parasitic plants
To obtain the genomic sequences of the foreign SSL genes, genomic DNA was extracted from 3 mg dried seeds of O. aegyptiaca and 0.1 g stem of C. australis using a modified cetyltrimethylammonium bromide (CTAB) method [84]. Specific primer pairs oaeF2/R1370 (5′-GCCACGTAGTGTGAAGCTT-3′/5′-CATCTCTTTCTTGAACCTCAC-3′) and cusF52/ R1470 (5′-AGGAACGAAGGGAGTATTT-3′/5′-TATTCATGAACTTCCGATATGG-3′) were designed according to the SSL cDNA sequences in O. aegyptiaca and C. australis, respectively. Amplification products of expected sizes were extracted using AxyPrep DNA Gel Extraction Kit (Axygen Bioscienes), cloned into the pMD18-T vector (Takara), and sequenced.
qRT-PCR of the CaSSL gene in C. australis
Fresh tissues were collected from 9 different development stages: dry seeds, germinated seeds with 2 mm hypocotyl and 3-cm hypocotyl, pre-haustoria (2 d after attaching to pseudo-hosts, bamboo sticks), and mature stems (at least 4 cm far from shoot tips), shoot tips, floral buds, flowers and capsules. For wounding treatment, 2 cm long dodder stems that were 4 cm far from shoot tips were wounded by four rows of thin wires (diameter 0.1 mm) from an iron wire gauze. Samples were harvested 0, 1, 3, 9, and 24 h after treatment. Five biological replicates were taken for each time point. Total RNA from each sample was extracted with Fruit-mate™ and RNAiso Plus (TaKaRa) following the manufacturer’s instructions. RNA concentrations were quantified and 500 ng of each RNA sample was reverse-transcribed using oligo_(dT)18 and RevertAid™ H Minus Reverse Transcriptase (Fermentas) in a total volume of 10 μL. The obtained cDNA samples were diluted to 25 μL. Specific primer pair RT-cus34-F2/RT-cus34-R1 (5′-CTGCGACGGTTACCTTGGAA-3′/5′-CCACCATGTCCACCACTTTCT-3′) was designed according to a shared region by the 2 mRNA isoforms. q-PCR was used to characterize the expression of the foreign SSL gene in C. australis from different development stages and wounding treated stems on an CFX Connect™ Real-Time System (BIO-RAD) using iTaq™ Universal SYBR Green Supermix (BIO-RAD) following the manufacturer’s instructions. The reference gene for normalizing cDNA concentration variations was selected from four candidates, actin, elongation factor 1α (EF-1α), elongation factor Tu (EF-Tu), and serine/threonine-protein phosphatase 2A (PP2A). Their Ct values in all the samplings were recorded using the same amount of total RNA. The most stably expressed gene was selected by comparing their Ct values of samples from different developmental stages and wounding treatment using the online server RefFinder (http://www.leonxie.com/referencegene.php). Changes in expression levels of the foreign SSL gene were assessed using the comparative CT method. Differences in expression levels of SSL induced by wounding treatment were determined by Student’s t test.
Phylogenetic analyses
Protein sequences were sampled from representative plants in Phytozome by BLAST search using the OaSSL gene in O. aegyptiaca as the query. Because OaSSL homolog genes have multiple copy numbers in each species, we constructed a bootstrap NJ tree using ClustalX2 [85] and only kept a representative sequence for each species in each branch. The homologs in O. aegyptiaca and C. australis were obtained by tblastn search against the RNA-seq assembly constructed in-house with the horizontal transferred gene in O. aegyptiaca as the query.
All these collected protein sequences were aligned using ClustalX2 [86]. We visually inspected the alignments and performed manual refinement. Gaps and ambiguously sites were removed from the alignment. The most optimal model of protein substitution matrix and rate heterogeneity was determined by ModelGenerator (v_851) [87]. Phylogenetic trees were reconstructed with a maximum likelihood method using PHYML 3.0 [88]. Bootstrap analyses used 100 pseudo-replicates. The topology structures of trees were viewed and edited with NJplot [89].
Analysis of functional divergence
The coefficients of type I and type II functional divergence between the Brassicales-specific genes and its original copies were estimated by DIVERGE 2.0 [51]. Type I functional divergence (θ1) alters functional constraints on some sites after gene duplication, and type II functional divergence (θ11) results in radical change in amino acid properties between the two duplicated copies. A likelihood ratio test was conducted using distribution with 1 degree of freedom (DF).
Detection of positive selection
The corresponding cDNAs were obtained by BLAST search against all the transcripts from Phytozome and the transcriptome assemblies from the parasitic plants. The selection pressure acting on the SSL coding regions was analyzed by calculating the rate ratio of nonsynonymous to synonymous substitution (or) with the program codeml implemented in PAML v4.7 [53]. We created large and small two datasets. The large dataset included the Brassicales-specific gene copies and their original gene copies; the small dataset included the Brassicales-specific gene copies and the foreign SSL genes in the two parasitic plants. The online server PAL2NAL [90] was used to convert the protein sequence alignments into the corresponding codon alignments, which were used as the input files of codeml in PAML. Unrooted NJ trees with branch lengths were produced using ClustalX 2.1 [85] based on the protein alignments and were fed to codeml in PAML.
We analyzed the site models and branch-site models on the large and small datasets with likelihood ratio tests (LRT), respectively. As for the site models, the LRT of M0-M3 comparison was used to test variable among sites with DF set to 3. The LRTs of M1a-M2a and M7-M8 comparison were used to test positive selection with DF set to 2. As for the branch-site models, the null hypothesis was compared with the alternative hypothesis to test whether positive selection acted on the Brassicales-specific SSLs and the foreign SSLs in the two parasitic plants, which were used as foreground in their own analysis. We chose the Bayes Empirical Bayes (BEB) analysis in codeml to calculate the posterior probability of sites undergone positive selection in the interested lineages.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the study: GS and JQW. Generated the data and performed the analysis: GS DZ JQ JY TS ZY JSW LW. Contributed reagents/materials/analysis tools: JLH SL JFW. Wrote the paper: GS JQW. All authors read and approved the final manuscript.
Supplementary Material
Schematic outline of the identification of horizontally transferred genes in O. aegyptiaca transcriptomes.
Gene loci and the structures of the Brassicaceae-specific SSL genes.
The gene Structure and its two mRNA isoforms in CaSSL .
The AtSSL1 expression levels of A. thaliana in different developmental stages calculated by GENEVESTIGATOR.
The AtSSL1 expression changes of A. thaliana upon different treatments calculated by GENEVESTIGATOR.
Primer sequences of four candidate reference genes and their ranking orders of table expression evaluated by RefFinder.
Results of positive selection using site models.
Results of positive selection using branch-site models.
Sites undergone positive selection in the OaSSL gene of O. aegyptiaca.
Contributor Information
Dale Zhang, Email: zhang521.77@henu.edu.cn.
Jinfeng Qi, Email: qijinfeng@mail.kib.ac.cn.
Jipei Yue, Email: yuej@ecu.edu.
Jinling Huang, Email: huangj@ecu.edu.
Ting Sun, Email: suntinglily@163.com.
Suoping Li, Email: lisuoping@henu.edu.cn.
Jian-Fan Wen, Email: wenjf@mail.kiz.ac.cn.
Christian Hettenhausen, Email: hettenhausen@mail.kib.ac.cn.
Jinsong Wu, Email: jinsongwu@mail.kib.ac.cn.
Lei Wang, Email: leiwang@mail.kib.ac.cn.
Huifu Zhuang, Email: zhuanghuifu@mail.kib.ac.cn.
Jianqiang Wu, Email: wujianqiang@mail.kib.ac.cn.
Guiling Sun, Email: sung@mail.kib.ac.cn.
Acknowledgments
We thank Prof. Yongqing Ma (College of Resources and Environment, Northwest A&F University, Yangling, Shaanxi Province, China) for kindly providing O. aegyptiaca seeds, the PPGP project (Virginia Tech, Penn State University, University of Virginia, and University of California, Davis) for the generation of transcriptome data from Orobanche, Triphysaria, and Striga, the 1 KP project (Gane Ka-Shu Wong, University of Alberta) for generating the transcriptome data from Cuscuta, Cassytha, and Pilostyles. Most computation work was performed in the HPC Center, Kunming Institute of Botany, CAS, China. This work is supported by a grant from the Natural Science Foundation of China (No. 31301037), a grant from the Natural Science Foundation of Yunnan Province of China (No. 2013FB068), a project sponsored by SRF for ROCS, SEM, a Startup grant from Kunming Institute of Botany, CAS, a grant from the Yunnan Recruitment Program of Experts in Sciences (No. 2012HA016), a grant from the Thousand Youth Talents Program, an Open Foundation of the State Key Laboratory of Genetics Resources and Evolution, CAS, and a grant for the Max Planck Partner Group.
References
- Ding Y, Zhou Q, Wang W. Origins of new genes and evolution of their novel functions. Ann Rev Ecol Evol. 2012;43:345–363. doi: 10.1146/annurev-ecolsys-110411-160513. [DOI] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19(12):2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. P Roy Soc B-Biol Sci. 2012;279(1749):5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Syvanen M. Evolutionary Implications of Horizontal Gene Transfer. Annu Rev Genet. 2012;46:341–358. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- Wijayawardena BK, Minchella DJ, Dewoody JA. Hosts, parasites, and horizontal gene transfer. Trends Parasitol. 2013;29(7):329–338. doi: 10.1016/j.pt.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Boschetti C, Carr A, Crisp A, Eyres I, Wang-Koh Y, Lubzens E, Barraclough TG, Micklem G, Tunnacliffe A. Biochemical diversification through foreign gene expression in Bdelloid rotifers. PLoS Genet. 2012;8(11):e1003035. doi: 10.1371/journal.pgen.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320(5880):1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Ni T, Yue J, Sun G, Zou Y, Wen J, Huang J. Ancient gene transfer from algae to animals: mechanisms and evolutionary significance. BMC Evol Biol. 2012;12(1):83. doi: 10.1186/1471-2148-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker E, Bapteste E, Le Guyader H, Manuel M, Rabet N. Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr Biol. 2008;18(18):R858–R859. doi: 10.1016/j.cub.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Starcevic A, Akthar S, Dunlap WC, Shick JM, Hranueli D, Cullum J, Long PF. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc Natl Acad Sci U S A. 2008;105(7):2533–2537. doi: 10.1073/pnas.0707388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EGJ, Rosso MN. Lateral gene transfers have polished animal genomes: lessons from nematodes. Front Cell Infect Microbiol. 2012;2:27. doi: 10.3389/fcimb.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328(5978):624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna R, Padilla BE, Florez-Ramos CP, Rubio JD, Herrera JC, Benavides P, Lee SJ, Yeats TH, Egan AN, Doyle JJ. et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc Natl Acad Sci USA. 2012;109(11):4197–4202. doi: 10.1073/pnas.1121190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Yuen MM, Liao NY, Roderick Docking T, Chan SK, Taylor GA, Palmquist DL, Jackman SD, Nguyen A, Li M. et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14(3):R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T. A fungal past to insect color. Science. 2010;328(5978):574–575. doi: 10.1126/science.1190417. [DOI] [PubMed] [Google Scholar]
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15(1):11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Yue J, Hu X, Huang J. Horizontal gene transfer in the innovation and adaptation of land plants. Plant Signal Behav. 2013;8(5):e24130. doi: 10.4161/psb.24130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Hu X, Sun H, Yang Y, Huang J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun. 2012;3:1152. doi: 10.1038/ncomms2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58(1):1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley RK, Sugumaran M, Marx CJ, Rest JS, Davis CC. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9(2):e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Bellot S. Genomics of Chloroplasts and Mitochondria. Netherlands: Springer; 2012. pp. 223–235. (Horizontal Gene Transfer in Eukaryotes: Fungi-to-Plant and Plant-to-Plant Transfers of Organellar DNA). [Google Scholar]
- Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4(1):e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP. Adaptive Evolution of C-4 Photosynthesis through Recurrent Lateral Gene Transfer. Curr Biol. 2012;22(5):445–449. doi: 10.1016/j.cub.2012.01.054. [DOI] [PubMed] [Google Scholar]
- Huang J. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays. 2013;35(10):868–875. doi: 10.1002/bies.201300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science. 2004;305(5684):676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang TC, Qiao Q, Ren Z, Zhao J, Yonezawa T, Hasegawa M, Crabbe MJ, Li J, Zhong Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (chenopodiaceae) PLoS One. 2013;8(3):e58747. doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Young GJ, Palmer JD. Plant genetics: gene transfer from parasitic to host plants. Nature. 2004;432(7014):165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- Davis CC, Anderson WR, Wurdack KJ. Gene transfer from a parasitic flowering plant to a fern. Proc Biol Sci/R Soc. 2005;272(1578):2237–2242. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Hao W, Gummow JS, Jain K, Ahmed D, Palmer JD. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010;8:150. doi: 10.1186/1741-7007-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Maruyama S, Nozaki H, Shirasu K. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science. 2010;328(5982):1128. doi: 10.1126/science.1187145. [DOI] [PubMed] [Google Scholar]
- Xi Z, Bradley RK, Wurdack KJ, Wong K, Sugumaran M, Bomblies K, Rest JS, Davis CC. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fernandez-Aparicio M, Wafula EK, Das M, Jiao Y, Wickett NJ, Honaas LA, Ralph PE, Wojciechowski MF, Timko MP. et al. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol Biol. 2013;13:48. doi: 10.1186/1471-2148-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss GM. Correlated evolution of life history and host range in the nonphotosynthetic parasitic flowering plants Orobanche and Phelipanche (Orobanchaceae) J Evol Biol. 2007;20(2):471–478. doi: 10.1111/j.1420-9101.2006.01273.x. [DOI] [PubMed] [Google Scholar]
- Parker C. Parasitic weeds: a world challenge. Weed Sci. 2012;60(2):269–276. doi: 10.1614/WS-D-11-00068.1. [DOI] [Google Scholar]
- Dawson JH, Musselman LJ, Wolswinkel P, Dorr I. Biology and control of Cuscuta. Rev Weed Sci. 1994;6:265–317. [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, dePamphilis CW, Das M, Fernandez-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI. The parasitic plant genome project: New tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012;60(2):295–306. doi: 10.1614/WS-D-11-00113.1. [DOI] [Google Scholar]
- Tian J, Sun G, Ding Q, Huang J, Oruganti S, Xie B. The third International Conference on Bioinformatics and Computational Biology (BICoB): 23–25 March 2011. New Orleans, Louisiana: The third International Conference on Bioinformatics and Computational Biology (BICoB); 2011. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644. doi: 10.1038/nbt.1883. U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Muller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25(10):3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80(1):75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18(3):500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol. 2013;30(7):1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Westwood JH, Roney JK, Khatibi PA, Stromberg VK. RNA translocation between parasitic plants and their hosts. Pest Manag Sci. 2009;65(5):533–539. doi: 10.1002/ps.1727. [DOI] [PubMed] [Google Scholar]
- Leblanc M, Kim G, Patel B, Stromberg V, Westwood J. Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant Cuscuta pentagona. New Phytol. 2013;200(4):1225–1233. doi: 10.1111/nph.12439. [DOI] [PubMed] [Google Scholar]
- Vallenback P, Jaarola M, Ghatnekar L, Bengtsson BO. Origin and timing of the horizontal transfer of a PgiC gene from Poa to Festuca ovina. Mol Phylogenet Evol. 2008;46(3):890–896. doi: 10.1016/j.ympev.2007.11.031. [DOI] [PubMed] [Google Scholar]
- Igarashi A, Yamagata K, Sugai T, Takahashi Y, Sugawara E, Tamura A, Yaegashi H, Yamagishi N, Takahashi T, Isogai M. et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology. 2009;386(2):407–416. doi: 10.1016/j.virol.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Leblanc M, Kim G, Westwood JH. RNA trafficking in parasitic plant systems. Front Plant Sci. 2012;3:203. doi: 10.3389/fpls.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly R, Hamamouch N, Abu-Nassar J, Wolf S, Joel DM, Eizenberg H, Kaisler E, Cramer C, Gal-On A, Westwood JH. Movement of protein and macromolecules between host plants and the parasitic weed Phelipanche aegyptiaca Pers. Plant Cell Rep. 2011;30(12):2233–2241. doi: 10.1007/s00299-011-1128-5. [DOI] [PubMed] [Google Scholar]
- Haupt S, Oparka KJ, Sauer N, Neumann S. Macromolecular trafficking between Nicotiana tabacum and the holoparasite Cuscuta reflexa. J Exp Bot. 2001;52(354):173–177. doi: 10.1093/jexbot/52.354.173. [DOI] [PubMed] [Google Scholar]
- Jiang L, Qu F, Li Z, Doohan D. Inter-species protein trafficking endows dodder (Cuscuta pentagona) with a host-specific herbicide-tolerant trait. New Phytol. 2013;198(4):1017–1022. doi: 10.1111/nph.12269. [DOI] [PubMed] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S. Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot. 2006;57(4):911–921. doi: 10.1093/jxb/erj076. [DOI] [PubMed] [Google Scholar]
- Pribylova J, Spak J. Dodder transmission of phytoplasmas. Methods Mol Biol. 2013;938:41–46. doi: 10.1007/978-1-62703-089-2_4. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Bock R. Exchange of genetic material between cells in plant tissue grafts. Science. 2009;324(5927):649–651. doi: 10.1126/science.1170397. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA. 2012;109(7):2434–2438. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp N, Zenk MH. Homogeneous strictosidine synthase from cell-suspension cultures of rauvolfia-serpentina. Phytochemistry. 1988;27(12):3811–3815. doi: 10.1016/0031-9422(88)83023-1. [DOI] [Google Scholar]
- Kutchan TM, Hampp N, Lottspeich F, Beyreuther K, Zenk MH. The cDNA clone for strictosidine synthase from Rauvolfia serpentina. DNA sequence determination and expression in Escherichia coli. FEBS Lett. 1988;237(1–2):40–44. doi: 10.1016/0014-5793(88)80167-4. [DOI] [PubMed] [Google Scholar]
- McKnight TD, Roessner CA, Devagupta R, Scott AI, Nessler CL. Nucleotide sequence of a cDNA encoding the vacuolar protein strictosidine synthase from Catharanthus roseus. Nucleic Acids Res. 1990;18(16):4939. doi: 10.1093/nar/18.16.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Liao ZH, Chen M, Wang Q, Yang CX, Yang YJ. Molecular cloning and characterization of the Strictosidine synthase Gene from Rauwolfia verticillata. Russ J Plant Phys. 2008;55(5):670–675. doi: 10.1134/S1021443708050117. [DOI] [Google Scholar]
- Lu Y, Wang H, Wang W, Qian Z, Li L, Wang J, Zhou G, Kai G. Molecular characterization and expression analysis of a new cDNA encoding strictosidine synthase from Ophiorrhiza japonica. Mol Biol Rep. 2009;36(7):1845–1852. doi: 10.1007/s11033-008-9389-y. [DOI] [PubMed] [Google Scholar]
- Ma X, Panjikar S, Koepke J, Loris E, Stockigt J. The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed beta-propeller fold in plant proteins. Plant Cell. 2006;18(4):907–920. doi: 10.1105/tpc.105.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockigt J, Barleben L, Panjikar S, Loris EA. 3D-Structure and function of strictosidine synthase–the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol Biochem. 2008;46(3):340–355. doi: 10.1016/j.plaphy.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Hicks MA, Barber AE 2nd, Giddings LA, Caldwell J, O’Connor SE, Babbitt PC. The evolution of function in strictosidine synthase-like proteins. Proteins. 2011;79(11):3082–3098. doi: 10.1002/prot.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Jones JT, Danchin EG. Horizontal gene transfer in nematodes: a catalyst for plant parasitism? MolPlant Microbe Interact. 2011;24(8):879–887. doi: 10.1094/MPMI-03-11-0055. [DOI] [PubMed] [Google Scholar]
- Richards TA, Soanes DM, Jones MD, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA. 2011;108(37):15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithofer A, Boland W. Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol. 2012;63:431–450. doi: 10.1146/annurev-arplant-042110-103854. [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- Ramakrishna A, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav. 2011;6(11):1720–1731. doi: 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33(Web Server issue):W677–W680. doi: 10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98(2):152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet–next generation sequence assembly visualization. Bioinformatics. 2010;26(3):401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78(5):364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic outline of the identification of horizontally transferred genes in O. aegyptiaca transcriptomes.
Gene loci and the structures of the Brassicaceae-specific SSL genes.
The gene Structure and its two mRNA isoforms in CaSSL .
The AtSSL1 expression levels of A. thaliana in different developmental stages calculated by GENEVESTIGATOR.
The AtSSL1 expression changes of A. thaliana upon different treatments calculated by GENEVESTIGATOR.
Primer sequences of four candidate reference genes and their ranking orders of table expression evaluated by RefFinder.
Results of positive selection using site models.
Results of positive selection using branch-site models.
Sites undergone positive selection in the OaSSL gene of O. aegyptiaca.