Abstract
Background
Several studies have shown the association of solute carrier family 30 (zinc transporter) member 8 (SLC30A8) rs13266634 with type 2 diabetes (T2D). However, the association of alternative variants and haplotypes of SLC30A8 with T2D have not been studied in different populations. The aim of this study is to assess the association of the alternative SLC30A8 variants, rs7002176 and rs1995222 as well as the most common variant, rs13266634 and haplotypes with glutamic acid decarboxylase antibodies (GADA) negative diabetes in Malaysian subjects.
Methods
Single nucleotide polymorphisms (SNPs) of SLC30A8; rs7002176, rs1995222 and rs13266634 were genotyped in 1140 T2D and 973 non-diabetic control subjects. Of these, 33 GADA positive diabetic subjects and 353 metabolic syndrome (MetS) subjects were excluded from subsequent analysis.
Results
The recessive genetic model controlled for age, race, gender and BMI shows that the alternative SLC30A8 variant, rs1995222 is associated with GADA negative diabetes (OR = 1.29, P = 0.02) in Malaysian subjects. The most common variant, rs13266634 is also associated with GADA negative diabetes (OR = 1.45, P = 0.001). This association is more pronounced among Malaysian Indians (OR = 1.93, P = 0.001). Moreover, the CG haplotype and CG-CG diplotype have been equally associated with increased diabetic risk (OR = 1.67, P = 8.6 × 10-5).
Conclusions
SLC30A8 SNPs and haplotypes are associated with GADA negative diabetes in Malaysian subjects, and this association is markedly higher among Malaysian Indian subjects.
Keywords: T2D, GADA negative diabetes, SLC30A8, Haplotypes, Alternative variants
Background
Type 2 diabetes (T2D) is a serious public health problem with its prevalence rapidly increases globally. In the next two decades, Asian countries will be hit hardest, particularly China and India where the diabetic populations will more than double [1-3]. In Malaysia, more than 2.1 million of the adult population have diabetes (11.7%) [3]. T2D is a complicated metabolic disorder, characterized by insulin resistance and/or pancreatic β-cell dysfunction resulting from both genetic and environmental factors [2,4,5]. Among T2D patients, latent autoimmune diabetes of adults (LADA) occurs in 2-12% of individuals [6-9]. LADA can be distinguished by the presence glutamic acid decarboxylase antibodies (GADA) in adult diabetic patients who clinically are similar to T2D subjects at diagnosis [7,10-12] and are insulin independent for at least in the first six months [13].
The interaction of a stable genetic background with the rapidly changing environment has resulted in rapid changes in the prevalence of T2D observed over recent decades [14]. Insulin resistance has been proposed to be a major driver of progression to T2D. However, most of the validated genetic variants are involved in β-cell function. The genome-wide association studies (GWAS) approach has dramatically increased the number of T2D susceptibility loci, expanding the list from five loci in 2007 to more than 60 loci in 2012. The association to T2D of more than 20 newly reported loci in Asians [15,16] and Europeans [17] needs to be studied in other populations as well. A consistent association of T2D risk with variants of the pancreatic β-cell–specific zinc transporter gene SLC30A8 has been discovered in European subjects [18-23]. This has been also reported in multi-ethnic case-control studies, including Asian [2,24-32], Arabian [33], European [17] and American populations [34]. The link between impaired β-cell function and Zn transport activity by SLC30A8 has been reported in several studies [35-39]. The consensus is that SLC30A8 is crucial for insulin processing and secretion, and the major contribution of the SLC30A8 SNPs to T2D is mediated through defects in insulin secretion rather than action. The SLC30A8 gene encodes the ZnT-8 zinc transporter, which is exclusively expressed in pancreatic β-cells and co-localized with insulin-containing secretory granules [40,41]. SLC30A8 variants impair islet ZnT8 expression, insulin secretion, or glucose homeostasis [39,42,43]. In addition, these variants are associated with the production of a less active zinc transporter protein, suggesting less efficiency of zinc accumulation and insulin crystallization [44]. ZnT-8 is thought to be a key protein for insulin secretion by regulating the homeostasis of zinc, which is an essential metal ion for insulin storage and secretion into intracellular vesicles [45,46].
Studies on the association of variants of the SLC30A8 gene with T2D have been concerned with the most common SNP rs13266634. However, the association of alternative SLC30A8 SNPs and haplotypes with T2D has not been studied in different populations. The aim of this investigation was to study the association of alternative SLC30A8 SNPs (rs7002176, rs1995222) and haplotypes with GADA negative diabetes, and further to replicate the association of the most common variant, rs13266634 in Malaysian subjects.
Methods
1 Subjects and data collection
This hospital-based case-control study was been conducted at the University Malaya Medical Centre (UMMC), Kuala Lumpur. Patients previously diagnosed with T2D (FPG ≥ 7.0 mmol/l), who attended the UMMC for treatment were invited to participate in this study (the case group). For the control group, subjects who were enrolled for general health screening at UMMC (FPG ≤ 6.1 mmol/l) were approached to participate in this study. GADA positive diabetic patients and metabolic syndrome (MetS) subjects were excluded from this study. Data collection for this study took place between 2009 and 2011. Details regarding the study design, including subjects, data collection, demographic parameters, biochemical analysis and quality control have been reported previously [47]. This study has been approved by the Medical Ethics Committee of University Malaya Medical Centre. A written informed consent was obtained from each participant in the study.
2 SNPs selection and genotyping
SLC30A8 SNPs, rs7002176, rs1995222 and rs13266634 were been selected based on previous studies [18,48]. Genomic DNA was isolated from peripheral blood leukocytes using Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s protocol. The SNPs were genotyped by pre-designed Taqman genotype assays (C_29002970_10, C_1421536_10 and C_357888_10 respectively, Applied Biosystems Inc, Foster City, USA) according to the manufacturer’s protocol using StepOnePlus Real-Time PCR system (Applied Biosystems Inc, Foster City, USA). No-template controls (NTCs) were included together with samples in each batch. The genotype call rates for SNPs, rs7002176, rs1995222 and rs13266634 were 98.5% (1130 diabetes; 951 non-diabetes), 97.1% (1108 diabetes; 943 non-diabetes) and 99.1% (1136 diabetes; 960 non-diabetes) respectively. The concordance rate, based on blind duplicate comparisons (10% of the samples that were blindly re-genotyped) was 100%.
3 Statistical analysis
Deviation of genotypes from Hardy-Weinberg Equilibrium was assessed with the DeFinetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl from the Institute of Human Genetics). The linkage disequilibrium (LD) between SNPs and the construction of haplotypes and diplotypes of related SNPs were performed with SNP & Variation Suite v7.x program (Golden Helix, Bozeman, MT, USA). Social Package of Statistical Science (SPSS version 11.5, LEAD Technologies; Inc. USA) was used to study the associations of SNPs of SLC30A8 using recessive, dominant and additive genetic models with T2D. These associations were evaluated by logistic regression analysis controlled for age, gender and body mass index. Significance was inferred when P < 0.05.
Results and discussion
The study included 1140 T2D and 973 non-diabetic control subjects. GADA positive diabetic subjects are classified as LADA since the genetic causes of this class of diabetes are similar to type 1 diabetes. To minimize variation among the diabetic group, the 33 GADA positive diabetic subjects were excluded from the study. Application of the new metabolic syndrome (MetS) criteria [49] on non-diabetic control subjects revealed that 353 subjects had MetS. Metabolic syndrome is a strong risk factor for diabetes which may affect the association study and Hardy-Weinberg Equilibrium. Hence, subjects in the control group with MetS were excluded from subsequent analysis. The metabolic and diabetic parameters were been significantly different between the target and control groups (Table 1).
Table 1.
Demographic and biochemical characterizations of participants
| Demography and biochemical profiles |
Normal control |
GADA negative diabetes |
|
|---|---|---|---|
| n = 620 | n = 1107 | ||
| Gender % |
Male/female |
61.6/38.4 |
67.6/32.4 |
| Races % |
Malay |
41.0 |
38.4 |
| Chinese |
32.9 |
26.5 |
|
| Indian |
26.1 |
35.1 |
|
| Age (years) |
49.5(48.6–50.3) |
51.3(50.8–51.9) |
|
| P-value |
|
0.001
|
|
| Body mass index (kg/m2) |
24.6(24.3–25.0) |
27.4(27.1–27.7) |
|
| P-value |
|
<0.001
|
|
| Waist circumference (cm) |
85.2(84.2–86.2) |
96.1(95.4–96.8) |
|
| P-value |
|
<0.001
|
|
| Systolic blood pressure (mmHg) |
128(127–130) |
134(133–135) |
|
| P-value |
|
<0.001
|
|
| Diastolic blood pressure (mmHg) |
80(79–80) |
81(80–82) |
|
| P-value |
|
0.005
|
|
| Fasting plasma glucose (mmol/l) |
4.98(4.94–5.02) |
7.89(7.73–8.05) |
|
| P-value |
|
<0.001
|
|
| Fasting plasma insulin (pmol/l) |
59.65(56.35–63.15) |
98.15(94.08–102.39) |
|
| P-value |
|
<0.001
|
|
| HOMA-β |
111.35(107.19–115.67) |
70.84(67.41–74.44) |
|
| P-value |
|
<0.001
|
|
| HOMA-IR |
1.27(1.20–1.35) |
2.45(2.34–2.55) |
|
| P-value |
|
<0.001
|
|
| High density lipoprotein (mmol/l) |
1.34(1.32–1.37) |
1.09(1.07–1.10) |
|
| P-value |
|
<0.001
|
|
| Triglyceride (mmol/l) |
1.13(1.09-1.17) |
1.58(1.53–1.63) |
|
| P-value | <0.001 | ||
The results presented represent geometric means (95% confidence interval of the mean), which evaluated by ANOVA. Bolded values are significant. GADA: glutamic acid decarboxylase antibodies.
1 Association of SLC30A8 SNPs with GADA negative diabetes
SNPs of SLC30A8, rs7002176, rs1995222 and rs13266634 showed no deviation from Hardy-Weinberg Equilibrium in the control group (P-value = 0.36, 0.54, 0.40 respectively). The alternative SLC30A8 SNPs; rs7002176 and rs1995222 were evaluated in this study. The logistic regression model (adjusted for age, race, gender and BMI) for rs1995222 showed association (recessive genetic model, OR = 1.29; P = 0.02) with GADA negative diabetes in Malaysian subjects. This finding is in contrast with findings in Pima Indian [48]. However, the SNP rs7002176 showed no association with GADA negative diabetes, a finding which is in agreement with Rong et al.[48]. The finding of non-association might be explained by the differences in genetic background, genetic model used and ethnicities between the populations [17,26] (Table 2).
Table 2.
Association of SLC30A8 polymorphism with GADA negative diabetes among Malaysian subjects and ethnic groups
| SLC30A8 SNPs |
Control |
GADA negative diabetes |
Recessive |
Dominant |
Additive |
||
|---|---|---|---|---|---|---|---|
|
OR (95% CI) | |||||||
| Freq. | 11/12/22 | Freq. | 11/12/22 | P-value | |||
|
rs7002176 (A > T) | |||||||
| Combined* |
0.42 |
113/284/208 |
0.45 |
222/541/335 |
1.12(0.85–1.46) 0.43 |
1.17(0.93–1.47) 0.18 |
1.11(0.95–1.29) 0.19 |
| Malay# |
0.37 |
35/115/100 |
0.42 |
79/191/150 |
1.61(1.00–2.59) 0.05 |
1.16(0.80–1.67) 0.43 |
1.23(0.96–1.57) 0.11 |
| Chinese# |
0.48 |
46/98/55 |
0.47 |
56/160/75 |
0.70(0.43–1.14) 0.15 |
1.06(0.67–1.65) 0.81 |
0.89(0.67–1.19) 0.43 |
| Indian# |
0.43 |
32/71/53 |
0.47 |
87/190/110 |
1.12(0.70–1.78) 0.65 |
1.32(0.88–1.99) 0.18 |
1.22(0.93–1.60) 0.15 |
|
rs1995222 (A < G) | |||||||
| Combined* |
0.63 |
238/273/87 |
0.67 |
498/440/138 |
1.29(1.03–1.62)
0.02
|
1.17(0.85–1.60) 0.34 |
1.18(1.01–1.39)
0.039
|
| Malay# |
0.60 |
88/116/40 |
0.64 |
167/189/54 |
1.22(0.85–1.77) 0.28 |
1.55(0.93–2.58) 0.10 |
1.24(0.95–1.61) 0.11 |
| Chinese# |
0.54 |
57/97/43 |
0.54 |
87/133/66 |
1.16(0.75–1.79) 0.51 |
0.97(0.60–1.56) 0.89 |
1.05(0.80–1.38) 0.73 |
| Indian# |
0.78 |
93/60/4 |
0.80 |
244/118/18 |
1.26(0.85–1.87) 0.25 |
0.27(0.06–1.22) 0.09 |
1.11(0.79–1.56) 0.53 |
|
rs13266634 (C > T) | |||||||
| Combined* |
0.60 |
223/284/105 |
0.65 |
506/415/181 |
1.45(1.16–1.81)
0.001
|
0.99(0.75–1.32) 0.96 |
1.18(1.01–1.37)
0.03
|
| Malay# |
0.58 |
84/127/42 |
0.60 |
158/192/73 |
1.16(0.80–1.66) 0.44 |
0.85(0.53–1.35) 0.49 |
1.02(0.79–1.30) 0.90 |
| Chinese# |
0.50 |
56/90/56 |
0.49 |
85/115/91 |
1.14(0.74–1.76) 0.55 |
0.93(0.61–1.44) 0.76 |
1.03(0.79–1.33) 0.84 |
| Indian# | 0.74 | 83/67/7 | 0.82 | 263/108/17 | 1.93(1.31–2.85) 0.001 | 0.81(0.31–2.14) 0.68 | 1.58(1.14–2.17) 0.006 |
Risk allele frequency (Freq.) and genotype counts in individuals with GADA negative diabetes and control subjects. 11, homozygous of major allele; 12, heterozygous; 22, homozygous of minor allele. GADA, glutamic acid decarboxylase antibodies. Risk allele is denoted in boldface. Bolded values are significant. *Controlled for age, race, gender and BMI. #Controlled for age, gender and BMI. The outliers (studentized residual is greater than 2.0 or less than −2.0) were excluded.
The most common variant, rs13266634 showed significant association with GADA negative diabetes in Malaysian subjects (recessive genetic model, OR = 1.45, P = 0.001). This finding is in agreement with previous studies in various Asian populations [2,24-32]. However, the association of this SNP with GADA was different for subjects of the three main Malaysian races (Malay, Chinese and Indian). The rs13266634 SNP was strongly associated with GADA negative diabetes among Malaysian Indian subjects (recessive genetic model, OR = 1.93, P = 0.001), but not with Malaysian Chinese and Malay subjects (Table 2). Tan et al.[30] have shown similar association with diabetes among Singaporean Malay subjects, whereas, their finding among Singaporean Chinese and Indian subjects have shown no association. This discrepancy in association of this SNP with GADA among Asian populations might be due to the small sample size used in the analysis. In addition, the environmental risk profile, lifestyle, body composition and linkage disequilibrium patterns might be involved. Likewise, the association between rs13266634 in the SLC30A8 gene locus and susceptibility to T2D has been demonstrated in Caucasian populations [19,20,22,23]. The rs13266634 SNP is a nonsynonymous Arg325 → Trp325 variant in the zinc transporter SLC30A8 [19,50]. Since SLC30A8 encodes a zinc transporter expressed solely in the secretory vesicles of β-cells, and is implicated in the final stages of insulin biosynthesis involving co-crystallization with zinc [18], its association with T2D is to be expected.
Previous studies had suggested that the major contribution of the SLC30A8 SNPs to T2D was mediated through defects in insulin secretion rather than action [35]. However, neither of these SNPs showed an association with HOMA-β nor HOMA-IR (Additional file 1: Table S1). Similar outcomes have been reported in other studies [2,26]. These conflicting results on the role of SLC30A8 in insulin secretion might be explained by the different genetic background between different populations. Moreover, other gene interactions, that may also have contributed in the metabolism and regulation of insulin activity [46].
2 Association of SLC30A8 haplotypes and diplotypes with GADA negative diabetes
Two-SNP haplotype and diplotype blocks (rs1995222 and rs13266634) with significant LD were identified (Figure 1). There was a significant linkage between SNPs rs13266634 and rs1995222 (r2 = 0.20), although, the distance between the two SNPs is approximately 45 kb. This finding is similar to that reported in Europeans (r2 = 0.19). However, SNP rs13266634 is near to SNP rs7002176, the distance between the two SNPs is approximately 2.7 kb, but there is no significant linkage between them. The haplotypes and diplotypes with frequency < 2% of the combined races were been excluded from subsequent analysis. The logistic regression model (adjusted for age, race, gender and BMI) showed that the haplotype CG and the diplotype CG-CG (containing the risk alleles of the SNPs) are equal risk factors for T2D in the combined races (OR = 1.67, P = 8.6 × 10-5), and this risk is higher in diabetic Indian subjects (OR = 1.93, P = 0.001) (Table 3). The haplotype (CG) and diplotype (CG-CG) showed a stronger association with GADA negative diabetes than single individual SNPs. This strength could be attributed to the possible epistatic interaction between these SNPs in determining overall risk of T2D. The haplotypes and diplotypes are not associated with GADA negative diabetes in Malay and Chinese subjects except the diplotype AC-AT is associated with protection against diabetes in Malay subjects (OR = 0.26, P = 0.0002) (Additional file 1: Table S2). The frequencies of haplotype GT and diplotype CG-GT are higher in Indian subjects without diabetes compared to Indian subjects with diabetes (OR = 0.51; 0.48, P = 0.008; 0.005, respectively) (Table 3). This study is a hospital-based, and the sampling method is non-probability. Thus, the sampling for this study has limited its generalization to the whole Malaysian population. Sub-grouping of subjects according to races resulted in small sample size, which likely gives insufficient power to provide an evidence for association within the ethnic subgroups.
Figure 1.
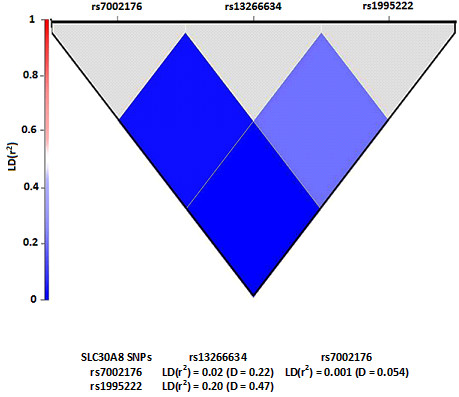
Pairwise linkage disequilibrium among SLC30A8 SNPs in Malaysian subjects.
Table 3.
Association of SLC30A8 common haplotypes and diplotypes with GADA negative diabetes among combined races and Indian subjects
| rs13266634, rs1995222 |
Combined races |
Indians |
||||||
|---|---|---|---|---|---|---|---|---|
|
Frequency |
P-value | OR (95% CI) |
Frequency |
P-value | OR (95% CI) | |||
| Control | GADA negative diabetes | Control | GADA negative diabetes | |||||
|
Haplotypes | ||||||||
| AT |
0.44 |
0.39 |
0.10 |
0.83(0.67–1.03) |
0.23 |
0.20 |
0.28 |
0.78(0.50–1.23) |
| CG |
0.21 |
0.32 |
8.6 × 10
-5
|
1.67(1.29–2.15) |
0.36 |
0.51 |
0.001
|
1.93(1.31–2.85) |
| GT |
0.17 |
0.14 |
0.08 |
0.77(0.58–1.03) |
0.22 |
0.12 |
0.008
|
0.51(0.31–0.84) |
| AC |
0.14 |
0.13 |
0.59 |
0.92(0.67–1.25) |
0.15 |
0.15 |
0.91 |
0.97(0.57–1.65) |
|
Diplotypes | ||||||||
| CG-CG |
0.21 |
0.32 |
8.6 × 10
-5
|
1.67(1.29–2.15) |
0.36 |
0.51 |
0.001
|
1.93(1.31–2.85) |
| CG-AT |
0.24 |
0.22 |
0.36 |
0.89(0.69–1.14) |
0.20 |
0.16 |
0.15 |
0.70(0.43–1.13) |
| CG-GT |
0.14 |
0.11 |
0.09 |
0.76(0.55–1.05) |
0.20 |
0.11 |
0.005
|
0.48(0.29–0.80) |
| CG-AC |
0.12 |
0.11 |
0.62 |
0.92(0.67–1.27) |
0.14 |
0.13 |
0.94 |
1.02(0.58–1.79) |
| GT-AT |
0.08 |
0.07 |
0.62 |
0.90(0.60–1.36) |
0.02 |
0.01 |
0.30 |
0.48(0.12–1.91) |
| AC-AT | 0.07 | 0.04 | 0.001 | 0.43(0.27–0.70) | 0.006 | 0.01 | NA | NA |
Controlled for age, gender, race and BMI. The outliers (studentized residual is greater than 2.0 or less than−2.0) were excluded. GADA, glutamic acid decarboxylase antibodies.
Conclusions
The most common SLC30A8 variant, rs13266634 is associated with GADA negative diabetes in Malaysian subjects, and this association is more pronounced among Malaysian Indian subjects. In addition, the alternative SLC30A8 variant, rs1995222 is significantly linked with rs13266634 (r2 = 0.2) and shows a mild association with GADA negative diabetes. The risk of these SNPs is strengthened by the haplotypes and diplotypes containing the SNPs risk alleles.
Abbreviations
SLC30A8: Solute carrier family 30 (zinc transporter), member 8; T2D: Type 2 diabetes; SNP: Single nucleotide polymorphisms; GADA: Glutamic acid decarboxylase antibodies; OR: Odds ratio; GWAS: Genome-wide association studies; UMMC: University Malaya Medical Centre; NTC: No template controls; LD: Linkage disequilibrium; MetS: Metabolic syndrome; BMI: Body mass index; HOMA-β: Homeostasis model assessment of β-cell function; HOMA-IR: Homeostasis model assessment of insulin resistance.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SDS conceived the design of the study, collected the data, performed the experiments, statistical analyses and drafted the manuscript. RSA participated in the design of the study, statistical analyses and drafted the manuscript. ISI and ZA participated in collection of data. SM participated in the design of the study and reviewed the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Impact of SLC30A8 SNPs, haplotypes and diplotypes on beta-cell function (HOMA-β) and insulin resistance (HOMA-IR) in normal Malaysian subjects. Table S2. Association of SLC30A8 common haplotypes and diplotypes with GADA negative diabetes among Malaysian Malay and Chines subjects.
Contributor Information
Sameer D Salem, Email: sameersalem@yahoo.com.
Riyadh Saif-Ali, Email: reyadh70@yahoo.com.
Ikram S Ismail, Email: ikram@ummc.edu.my.
Zaid Al-Hamodi, Email: zalhamodi@yahoo.com.
Sekaran Muniandy, Email: sekaran@um.edu.my.
Acknowledgements
We thank all the participants of this research and all nursing and medical staff at UMMC for their dedication in this study. The authors wish to thank University of Malaya for supporting this research grant; IPPP (PV029/2012A) and UMRG (RG350/11 HTM).
References
- Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, Lam VK, Ma RC, So WY, Cho YS. et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57:2226–2233. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Diabetes Federation. Diabetes Atlas Update. 5. 2012. http://www.idf.org/sites/default/files/WP_5E_Update_Country.pdf. [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura Y. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–710. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- Qian Y, Lu F, Dong M, Lin Y, Li H, Chen J, Shen C, Jin G, Hu Z, Shen H. Genetic variants of IDE-KIF11-HHEX at 10q23.33 associated with type 2 diabetes risk: a fine-mapping study in Chinese population. PLoS One. 2012;7:e35060. doi: 10.1371/journal.pone.0035060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik RG, Brooks-Worrell BM, Palmer JP. Latent autoimmune diabetes in adults. J Clin Endocrinol Metab. 2009;94:4635–4644. doi: 10.1210/jc.2009-1120. [DOI] [PubMed] [Google Scholar]
- Turner R, Stratton I, Horton V, Manley S, Zimmet P, Mackay IR, Shattock M, Bottazzo GF, Holman R. UKPDS 25: autoantibodies to islet-cell cytoplasm and glutamic acid decarboxylase for prediction of insulin requirement in type 2 diabetes. Lancet. 1997;350:1288–1293. doi: 10.1016/S0140-6736(97)03062-6. [DOI] [PubMed] [Google Scholar]
- Monge L, Bruno G, Pinach S, Grassi G, Maghenzani G, Dani F, Pagano G. A clinically orientated approach increases the efficiency of screening for latent autoimmune diabetes in adults (LADA) in a large clinic-based cohort of patients with diabetes onset over 50 years. Diabet Med. 2004;21:456–459. doi: 10.1111/j.1464-5491.2004.01177.x. [DOI] [PubMed] [Google Scholar]
- Roh M-O, Jung C-H, Kim B-Y, Mok J-O, Kim C-H. The prevalence and characteristics of latent autoimmune diabetes in adults (LADA) and its relation with chronic complications in a clinical department of a university hospital in Korea. Acta Diabetolo. 2010;47:1–6. doi: 10.1007/s00592-010-0228-y. [DOI] [PubMed] [Google Scholar]
- Groop LC, Bottazzo GF, Doniach D. Islet cell antibodies identify latent type I diabetes in patients aged 35-75 years at diagnosis. Diabetes. 1986;35:237–241. doi: 10.2337/diab.35.2.237. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenström G, Gottsäter A, Bakhtadze E, Berger B, Sundkvist G. Latent autoimmune diabetes in adults. Diabetes. 2005;54:S68–S72. doi: 10.2337/diabetes.54.suppl_2.S68. [DOI] [PubMed] [Google Scholar]
- Tuomi T, Carlsson A, Li H, Isomaa B, Miettinen A, Nilsson A, Nissén M, Ehrnström BO, Forsén B, Snickars B. et al. Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes. 1999;48:150–157. doi: 10.2337/diabetes.48.1.150. [DOI] [PubMed] [Google Scholar]
- Petrie JR, Pearson ER, Sutherland C. Implications of genome wide association studies for the understanding of type 2 diabetes pathophysiology. Biochem Pharmacol. 2011;81:471–477. doi: 10.1016/j.bcp.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia K-S, Dimas AS, Hassanali N. et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Chen C-H, Hu C, Long J, Hee Ong RT, Sim X, Takeuchi F, Wu Y, Go MJ, Yamauchi T. et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Elbers Clara C, Guo Y, Peter I, Gaunt Tom R, Mega Jessica L, Lanktree Matthew B, Tare A, Castillo Berta A, Li Yun R. et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Consortium TWTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, Froguel P, Kadowaki T. Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50:2461–2466. doi: 10.1007/s00125-007-0827-5. [DOI] [PubMed] [Google Scholar]
- Hu C, Zhang R, Wang C, Wang J, Ma X, Lu J, Qin W, Hou X, Wang C, Bao Y. et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS One. 2009;4:e7643. doi: 10.1371/journal.pone.0007643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Li X, Xu M, Hong J, Huang Y, Tan J, Lu X, Dai M, Yu B, Ning G. Zinc transporter-8 gene (SLC30A8) is associated with type 2 diabetes in Chinese. J Clin Endocrinol Metab. 2008;93:4107–4112. doi: 10.1210/jc.2008-0161. [DOI] [PubMed] [Google Scholar]
- Xu M, Bi Y, Xu Y, Yu B, Huang Y, Gu L, Wu Y, Zhu X, Li M, Wang T. et al. Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PLoS One. 2010;5:e14022. doi: 10.1371/journal.pone.0014022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li P, Cai L, Zhang B, Tang X, Zhang X, Li Y, Xian Y, Yang Y, Wang L. et al. Association study of genetic variants in eight genes/loci with type 2 diabetes in a Han Chinese population. BMC Med Genet. 2010;11:97. doi: 10.1186/1471-2350-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Kang ES, Kim SH, Han SJ, Kim CH, Kim HJ, Ahn CW, Cha BS, Nam M, Nam CM. et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Humn Genet. 2008;53:991–998. doi: 10.1007/s10038-008-0341-8. [DOI] [PubMed] [Google Scholar]
- Tan JT, Ng DPK, Nurbaya S, Ye S, Lim XL, Leong H, Seet LT, Siew WF, Kon W, Wong TY. et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab. 2010;95:390–397. doi: 10.1210/jc.2009-0688. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li H, Loos RJF, Yu Z, Ye X, Chen L, Pan A, Hu FB, Lin X. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57:2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kifagi C, Makni K, Boudawara M, Mnif F, Hamza N, Abid M, Granier C, Ayadi H. Association of genetic variations in TCF7L2, SLC30A8, HHEX, LOC387761, and EXT2 with Type 2 diabetes mellitus in Tunisia. Genet Test Mol Biomarkers. 2011;15:399–405. doi: 10.1089/gtmb.2010.0199. [DOI] [PubMed] [Google Scholar]
- Waters KM, Stram DO, Hassanein MT, Le Marchand L, Wilkens LR, Maskarinec G, Monroe KR, Kolonel LN, Altshuler D, Henderson BE. et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:E1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorus FK, Goubert P, Semakula C, Vandewalle CL, De Schepper J, Scheen A, Christie MR, Pipeleers DG. IA-2-autoantibodies complement GAD65-autoantibodies in new-onset IDDM patients and help predict impending diabetes in their siblings: the Belgian diabetes registry. Diabetologia. 1997;40:95–99. doi: 10.1007/s001250050648. [DOI] [PubMed] [Google Scholar]
- Prentki M, Corkey BE. Are the beta-cell signaling molecules malonyl-CoA and cystolic long-chain acyl-CoA implicated in multiple tissue defects of obesity and NIDDM? Diabetes. 1996;45:273–283. doi: 10.2337/diab.45.3.273. [DOI] [PubMed] [Google Scholar]
- Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P. et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger H, Machicao F, Stefan N, Tschritter O, Thamer C, Kantartzis K, Schafer SA, Kirchhoff K, Fritsche A, Haring HU. Polymorphisms within novel risk loci for type 2 diabetes determine beta-cell function. PLoS One. 2007;2:e832. doi: 10.1371/journal.pone.0000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoni S, Nettleton JA, Hivert M-F, Ye Z, van Rooij FJA, Shungin D, Sonestedt E, Ngwa JS, Wojczynski MK, Lemaitre RN. et al. Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant. Diabetes. 2011;60:2407–2416. doi: 10.2337/db11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimienti F, Devergnas S, Pattou F, Schuit F, Garcia-Cuenca R, Vandewalle B, Kerr-Conte J, Van Lommel L, Grunwald D, Favier A. et al. In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci. 2006;119:4199–4206. doi: 10.1242/jcs.03164. [DOI] [PubMed] [Google Scholar]
- Kirchhoff K, Machicao F, Haupt A, Schafer SA, Tschritter O, Staiger H, Stefan N, Haring HU, Fritsche A. Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia. 2008;51:597–601. doi: 10.1007/s00125-008-0926-y. [DOI] [PubMed] [Google Scholar]
- Palmer ND, Goodarzi MO, Langefeld CD, Ziegler J, Norris JM, Haffner SM, Bryer-Ash M, Bergman RN, Wagenknecht LE, Taylor KD. et al. Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the insulin resistance atherosclerosis family study. Diabetes. 2008;57:1093–1100. doi: 10.2337/db07-1169. [DOI] [PubMed] [Google Scholar]
- Nicolson TJ, Bellomo EA, Wijesekara N, Loder MK, Baldwin JM, Gyulkhandanyan AV, Koshkin V, Tarasov AI, Carzaniga R, Kronenberger K. et al. Insulin storage and glucose homeostasis in mice null for the granule zinc transporter ZnT8 and studies of the type 2 diabetes-associated variants. Diabetes. 2009;58:2070–2083. doi: 10.2337/db09-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chausmer AB. Zinc, Insulin and Diabetes. J Am Coll Nutr. 1998;17:109–115. doi: 10.1080/07315724.1998.10718735. [DOI] [PubMed] [Google Scholar]
- Chimienti F. Zinc, pancreatic islet cell function and diabetes: new insights into an old story. Nutr Res Rev. 2013;26:1–11. doi: 10.1017/S0954422412000212. [DOI] [PubMed] [Google Scholar]
- Salem SD, Saif-Ali R, Ismail IS, Al-Hamodi Z, Poh R, Muniandy S. IGF2BP2 alternative variants associated with glutamic acid decarboxylase antibodies negative diabetes in Malaysian subjects. PLoS ONE. 2012;7:e45573. doi: 10.1371/journal.pone.0045573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Hanson RL, Ortiz D, Wiedrich C, Kobes S, Knowler WC, Bogardus C, Baier LJ. Association analysis of variation in/Near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes. 2009;58:478–488. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; national heart, lung, and blood institute; american heart association; world heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53:2330–2337. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Impact of SLC30A8 SNPs, haplotypes and diplotypes on beta-cell function (HOMA-β) and insulin resistance (HOMA-IR) in normal Malaysian subjects. Table S2. Association of SLC30A8 common haplotypes and diplotypes with GADA negative diabetes among Malaysian Malay and Chines subjects.


