Abstract
Background
Simultaneous contribution of hundreds of electrocardiographic biomarkers to prediction of long-term mortality in post-menopausal women with clinically normal resting electrocardiograms (ECGs) is unknown.
Methods and Results
We analyzed ECGs and all-cause mortality in 33,144 women enrolled in Women’s Health Initiative trials, who were without baseline cardiovascular disease or cancer, and had normal ECGs by Minnesota and Novacode criteria. Four hundred and seventy seven ECG biomarkers, encompassing global and individual ECG findings, were measured using computer algorithms. During a median follow-up of 8.1 years (range for survivors 0.5–11.2 years), 1,229 women died. For analyses cohort was randomly split into derivation (n=22,096, deaths=819) and validation (n=11,048, deaths=410) subsets. ECG biomarkers, demographic, and clinical characteristics were simultaneously analyzed using both traditional Cox regression and Random Survival Forest (RSF), a novel algorithmic machine-learning approach. Regression modeling failed to converge. RSF variable selection yielded 20 variables that were independently predictive of long-term mortality, 14 of which were ECG biomarkers related to autonomic tone, atrial conduction, and ventricular depolarization and repolarization.
Conclusions
We identified 14 ECG biomarkers from amongst hundreds that were associated with long-term prognosis using a novel random forest variable selection methodology. These were related to autonomic tone, atrial conduction, ventricular depolarization, and ventricular repolarization. Quantitative ECG biomarkers have prognostic importance, and may be markers of subclinical disease in apparently healthy post-menopausal women.
Keywords: Electrocardiography, epidemiology, women, prognosis
Introduction
Amongst post-menopausal women quantitative electrocardiographic (ECG) biomarkers have a prognostic value.1–4 Prior studies focused on single ECG measures such as QRS width,5 small groups of measures such as ventricular repolarization abnormalities,1, 2 or categories of findings such as minor and major ECG abnormalities3. Modern digital ECG software has the ability to abstract hundreds of quantitative measures from a standard 12-lead ECG. To date there have been no studies exploring the prognostic value of such a large number of ECG measures in a non-parsimonious manner.
Risk stratification based on utilization of hundreds of quantitative ECG biomarkers presents several unique challenges, which make use of traditional regression methods difficult. First, ECG measures are highly correlated, making their simultaneous use in a regression model problematic. Second, ECG measures may have nonlinear effects that require complex transformations. Third, manual identification of two-way and three-way interactions among hundreds of variables is challenging. Fourth, regression models with hundreds of variables may be overfit, consequently performing poorly in testing scenarios. Random Forest (RF) methodology, a non-parametric decision tree based approach, has been proposed as a cutting-edge analytical method to address these issues.6–8 Recently, RF methodology has been extended to deal with time-to-event data, an approach termed Random Survival Forests (RSF).8
The objective of this study was to evaluate the prognostic importance of quantitative electrocardiographic biomarkers in post-menopausal women without known cardiovascular disease or cancer, who had normal baseline resting ECGs, utilizing a data-rich model. We studied women with normal ECGs as they have been shown to have a lesser risk of mortality compared to those with major or minor ECG abnormalities3. We used RSF methodology to classify women into subgroups of risk, and to identify clinical and ECG predictors of mortality. With this approach numerous decision trees were developed, and then used to: (1) identify the most important predictors (i.e., variable selection), and (2) construct risk stratification models.
Methods
Study population
The Women’s Health Initiative (WHI) Clinical Trial (http://www.whiscience.org/about/) enrolled 68,132 post-menopausal women (Supplementary Material Figure 1) between the ages of 50 and 79, into randomized trials testing three prevention strategies (hormone therapy, dietary modification, or calcium/vitamin D). Eligible women had a choice of enrolling into one, two, or all three components. At baseline demographic and clinical characteristics, physical measures, and a standard 12-lead ECG were collected. Exclusion criteria were component specific, and were related to competing risks, safety reasons, and adherence or retention reasons.9
We focused only on those women who had a baseline ECG available, of good quality, and without arm lead reversal. We excluded women who had any minor or major ECG abnormalities3 according to Minnesota10, 11 or Novacode12 criteria. The remaining 35,774 women had ECGs with sinus rhythm, normal AV conduction, no evidence of old myocardial infarction as suggested by Q-waves, normal QRS duration, normal ventricular repolarization, no left atrial enlargement, no right ventricular hypertrophy, no right atrial enlargement, and no fascicular block.
We further excluded 2,510 women who had suspected or known cardiovascular disease (history of angina, prior percutaneous coronary intervention, prior coronary artery bypass grafting, peripheral arterial disease, prior carotid endarterectomy, aortic aneurysm, or stroke), or a history of cancer (breast, ovarian, colon, cervical, liver, lung, brain, bone, or stomach cancer; or leukemia, lymphoma, or Hodgkin’s disease). Finally, 120 women had missing outcome values and were excluded. This resulted in 33,144 women without known cardiovascular disease or cancer, with normal baseline 12-lead ECGs who were analyzed.
Electrocardiographic Analysis
Standard 12-lead ECGs were recorded at baseline using standardized procedures.1, 3, 13 These ECGs were processed at a central laboratory (EPICORE Center, University of Alberta, Alberta, Edmonton, and later at EPICARE in Wake Forest University, Winston-Salem, NC) and classified by Minnesota code and Novacode criteria, with use of Marquette 12-SL program, 2001 version (General Electric, Menomonee Falls, Wisconsin).1, 2 Software also abstracted continuous duration and voltage measures by lead for the median beats in each lead, all of which were recorded simultaneously for 10 seconds.
Four hundred and seventy seven ECG measures abstracted by the Marquette program were studied, encompassing both global and individual ECG measures. Global measures included: ventricular rate, median PR duration, median QT duration, median QTc interval, median P-wave axis, median QRS axis, and median T-wave axis. Two measures of ultra-short heart rate variability were studied: standard deviation of the mean value of RR intervals over 10 second recording (SDNN), and the square root of the mean value of the squares of the differences between all adjacent RR intervals (RMS-SD).
The Marquette program assigned biphasic (i.e., first inflection above or below baseline, and second inflection in opposite polarity) P waves and T waves two sets of variables, where the second set of variables was termed “prime”. This is different and should not be confused with the term “prime” used in clinical ECG interpretation which refers to wave notching.
Individual ECG measures are as follows:
P wave measures included: P wave and P prime wave amplitudes, intrinsicoid times (i.e., time from onset to peak), durations, and areas in all 12-leads.
Q wave measures included: Q wave amplitudes, intrinsicoid times, durations, and areas in all 12-leads.
R wave measures included: R wave and R prime wave amplitudes, intrinsicoid times, durations, and areas in all 12-leads.
S wave measures included: S wave and S prime wave amplitudes, intrinsicoid times, durations, and areas in all 12-leads.
QRS complex measures included: QRS intrinsicoid times (time from onset of QRS complex to middle of QRS complex) in all 12-leads.
ST segment measures included: beginning of ST segment amplitudes (at J point), middle of ST segment amplitudes (at J + 1/16 average RR interval), end of ST segment amplitudes (at J point + 1/8 average RR interval), and ST segment amplitudes at J point + 60 msec in all 12-leads.
T wave measures included: T wave and T prime wave amplitudes, intrinsicoid times, and areas in all 12-leads. Amplitudes were recorded to the nearest 100th of a millivolt and times recorded to the nearest millisecond.
Outcome
All-cause mortality, a clinically relevant and unbiased end-point,14 was recorded centrally by the WHI Clinical Coordinating Center.15
Statistical Analysis
Random Survival Forests
Random survival forest (RSF) analysis8 was employed using all-cause mortality for the outcome. Candidate predictor variables included all 477 ECG measures described above in addition to 22 baseline demographic and clinical predictors (Table 1).
Table 1.
Baseline Characteristics
| Derivation (n=22,096) |
Validation (n=11,048) |
|
|---|---|---|
| Age, y | 61 (50–79) | 61 (50–79) |
| Ethnicity | ||
| White | 18,395 (83%) | 9,172 (83%) |
| Black | 1,792 (8%) | 925 (8%) |
| Hispanic | 975 (4%) | 511 (5%) |
| American Indian | 94 (0%) | 31 (0%) |
| Asian/Pacific Islander | 541 (2%) | 270 (2%) |
| Unknown | 299 (1%) | 139 (1%) |
| Smoking | ||
| Never smoked | 11,436 (52%) | 5,738 (52%) |
| Past smoker | 9,018 (41%) | 4,463 (40%) |
| Current smoker | 1,642 (7%) | 847 (8%) |
| Hypertension | 5,715 (26%) | 2,839 (26%) |
| Treated Diabetes | 628 (3%) | 344 (3%) |
| Systolic Blood Pressure, mmHg | 124 (113 to 135) | 124 (113 to 135) |
| Diastolic Blood Pressure, mmHg | 75 (70 to 81) | 75 (70 to 81) |
| Body Mass Index, kg/m2 | 27.5 (24.3 to 31.3) | 27.4 (24.4 to 31.5) |
| Statin Use | 1,116 (5%) | 538 (5%) |
| Other Antihyperlipidemic Medication Use | 1,304 (6%) | 634 (6%) |
| Aspirin Use | 4,013 (18%) | 1,987 (18%) |
| Bilateral Oophorectomy | 3,370 (17%) | 1936 (18%) |
| Hysterectomy | 8,430 (38%) | 4,308 (39%) |
| Waist-to-Hip Ratio | 0.80 (0.76 to 0.85) | 0.80 (0.76 to 0.85) |
| Pregnancy | ||
| Never Pregnant | 1864 (8%) | 929 (8%) |
| 1 | 1,534 (7%) | 751 (7%) |
| 2–4 | 13,129 (59%) | 6,550 (59%) |
| 5+ | 5,569 (25%) | 2,818 (26%) |
| HRT Usage Status | ||
| Never Used | 10,210 (46%) | 5,089 (46%) |
| Past User | 3,763 (17%) | 1,810 (16%) |
| Current User | 8,123 (37%) | 4,149 (38%) |
| Income | ||
| Less than $10,000 | 807 (4%) | 373 (3%) |
| $10,000 to $19,999 | 2,285 (10%) | 1,206 (11%) |
| $20,000 to $34,999 | 4,997 (23%) | 2,446 (22%) |
| $35,000 to $49,999 | 5,198 (24%) | 2,575 (23%) |
| $50,000 to $74,999 | 4,410 (20%) | 2,233 (20%) |
| $75,000 to $99,999 | 2,023 (9%) | 1,012 (9%) |
| $100,000 to $149,999 | 1,288 (6%) | 639 (6%) |
| $150,000 or more | 623 (3%) | 285 (3%) |
| Unknown | 465 (2%) | 279 (3%) |
| Alcoholic Drinks Per Week | 0.4 (0-2.7) | 0.4 (0–2.7) |
| Marital Status | ||
| Never Married | 908 (4%) | 463 (4%) |
| Divorced / Separated | 3490 (16%) | 1,813 (16%) |
| Widowed | 3270 (15%) | 1,685 (15%) |
| Presently married / Living as married | 14428 (65%) | 7,087 (64%) |
| Medical Insurance | 20,716 (94%) | 10,372 (94%) |
| Education | ||
| 0–8 Years | 293 (1%) | 158 (1%) |
| Some high school | 715 (3%) | 342 (3%) |
| High School Diploma / GED | 3,958 (18%) | 1,871 (17%) |
| School After High School | 8,714 (39%) | 4,283 (39%) |
| College Degree Or Higher | 8,416 (38%) | 4,394 (40%) |
Continuous variables are medians (25th to 75th percentile), except for age which is median (range)
Derivation and validation subsets
Two-thirds of the women were randomly selected for primary analysis (derivation cohort, n = 22,096, deaths=819) and the remainder were selected for external validation (validation cohort, n = 11,048, deaths=410). When randomly selecting the derivation and validation cohorts we stratified according to event type (death or censoring) to ensure a similar event rate in both cohorts. The mortality rates in these cohorts were similar (Supplementary Material Figure 2).
Forest analysis
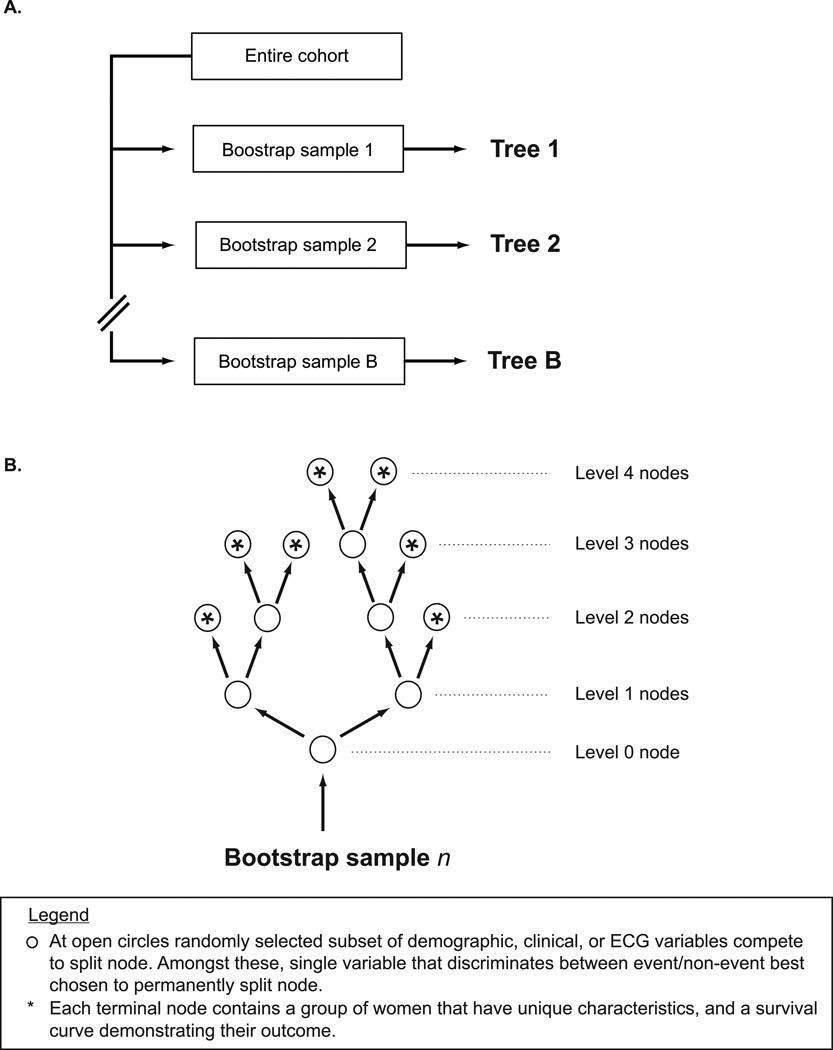
Using the derivation cohort, an RSF of 1000 trees was constructed, each tree from an independent and unique bootstrap sample of the data (Figure 1A). At each node of the tree, we randomly selected a subset of candidate variables (Figure 1B). For example, the variable occupying the level 0 branch/node was chosen through a “competition” of 22 randomly selected variables; the number of variables randomly selected is the square root of the number of total candidate variables (in this case the square root of 499 ≈ 22). For each of the 22 variables, we split the bootstrap sample into two groups, constructed Kaplan-meier survival curves, and calculated a log-rank statistic. The variable whose split yielded the highest log-rank value “won the competition”, and was thus chosen to occupy the node. We split categorical variables according to their natural categories, and continuous variables at 10 randomly selected cut points.
Figure 1.
Approach to constructing a Random Survival Forest. (A.) One thousand bootstrap samples of women were derived from full cohort, and (B.) each sample was then used to construct a unique and independent decision tree.
For each subsequent node of the tree we repeated the same process: random selection of candidate variables, splitting of each variable with construction of survival plots and calculation of log-rank statistic, and selection of the best splitting variable. The process continued down each branch of the tree until we reached a unique subset which contained no fewer than 3 deaths8, i.e., a terminal node. This approach yielded extensively grown trees on average having 143 terminal nodes, where each terminal node included a group of women having similar characteristics and survival outcomes.
Maximal subtrees for identification of predictive variables
As we have described elsewhere16, the most important variables for prediction were identified as those that most frequently split nodes nearest to the trunks of the trees (i.e., the root node). Figure 2 demonstrates a random tree with color coding of “maximal subtrees”. A maximal subtree for a variable v is the largest subtree whose lowest branch is split using v (i.e., no other parent branches of the subtree are split using v). There may be no maximal subtree, or there may be several. The shortest distance from the tree trunk to the root of a maximal subtree of v is the minimal depth of v. For example in Figure 2, income splits the tree trunk and has a minimal depth of zero, while age occupies the root of two yellow subtrees with minimal depths of 3 and 6 respectively. The most predictive variables are those whose minimal depth (averaged over the forest) is smaller than a threshold value determined under the null hypothesis that a variable is unrelated to the survival distribution.16 For variables like age, in which there are more than one maximal subtrees, we used only the lowest value of minimal depth for calculating average minimal depth across the forest. We have previously shown that this variable approach successfully identifies the strongest predictors with no loss of overall model accuracy due to excessive parsimony.16
Figure 2.
Example of one decision tree from forest. Depth of a branch (node) is indicated by numbers 0–10. Highlighted are maximal subtrees (i.e., largest subtree whose lowest branch is split using variable of interest) for the variables income (blue), and age (yellow). Income has one maximal subtree at minimal depth 0. Age has two maximal subtrees at minimal depths 3 and 6.
Construction of prediction models
We constructed 8 different prediction models using the derivation cohort: (Model 1) RSF using all 499 demographic, clinical, and ECG variables, (Model 2) Cox regression using all 499 variables, (Model 3) I1-penalized Cox regression using all 499 variables, (Model 4) AIC-penalized Cox regression. (Model 5) RSF using the 20 variables identified by the maximal subtree algorithm, (Model 6) Cox regression using the 20 variables identified by the maximal subtree algorithm, (Model 7) I1-penalized Cox regression using the top 100 RSF variables with lasso parameter selected by 10-fold cross-validation, and (Model 8) AIC-penalized Cox regression using the top 50 RSF variables.
The choices of 100 variables for Model 7, and 50 variables for Model 8, were arbitrary but necessary in order for these penalized Cox regression methods to converge.
Validation of prediction models
Predictive accuracy for all models was assessed using Harrell’s concordance index, both internally (using OOB cross-validation in the derivation cohort) and externally (using the validation cohort).
We assessed the individual predictiveness of the top variables identified by the maximal subtrees algorithm by constructing a sequence of nested models and then calculating measures of discrimination (Harrell’s concordance index) and calibration (Continuous Ranked Probability Score (CRPS)17), defined as the area under the prediction error curve using the Brier score) for each. Values were calculated using OOB cross-validation.
We investigated interactions amongst our top 20 variables using linkage hierarchical clustering anaylsis. Specifics regarding methods and results can be found in Supplementary Material.
Missing data imputation
Data was missing on 32 of the 499 variables, although very few of these data were missing (maximum amount missing for a variable: 14.3%; average missed per variable: 1.5%). Missing data was imputed using the forest method8 such that imputed data was not guided by outcomes (i.e., survival behavior of patients did not bias imputation).
Computational methods
Data assembly was performed with SAS version 9.1.3 (SAS Institute Inc., Cary, NC). Analyses were performed using R version 2.7.2 (www.r-project.org), using the publically available randomSurvivalForest library18, 19 written by two of the authors (H.I., U.B.K.). I1-penalization was performed using the coxnet function in the glmnet library (http://cran.r-project.org/web/packages/glmnet/), and AIC penalization and fitting was performed using stepAIC from the MASS library (http://cran.r-project.org/web/packages/MASS/).
Results
Characteristics and outcomes
Table 1 shows the baseline characteristics of the derivation and validation cohorts. Global ECG measures are shown in Table 2, and all other individual ECG measures are shown in Table 3.
Table 2.
Global ECG measures
| Derivation (n=22,096) |
Validation (n=11,048) |
|
|---|---|---|
| Ventricular rate (beats per minute) | 65 (59, 71) | 65 (59, 71) |
| Median PR duration (ms) | 158 (144, 172) | 156 (144, 172) |
| Median QT duration (ms) | 400 (382, 418) | 400 (382, 418) |
| Median QTc interval (ms) | 413 (406, 423) | 413 (406, 423) |
| Median P wave axis (degrees) | 54 (42, 65) | 55 (42, 65) |
| Median QRS axis (degrees) | 27 (8, 48) | 27 (8, 48) |
| Median T wave axis (degrees) | 40 (28, 51) | 40 (28, 51) |
| SDNN (ms) | 16 (11, 25) | 17 (11, 25) |
| RMS-SD (ms) | 17 (11, 26) | 17 (11, 26) |
Variables are medians (25th to 75th percentile)
Table 3.
Lead-specific ECG quantitative measures
| I | II | III | aVL | aVR | aVF | V1 | V2 | V3 | V4 | V5 | V6 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-wave amplitude (µV) | Q1 | 63 | 92 | 39 | −24 | − | 63 | 24 | 39 | 48 | 53 | 53 | 48 |
| Q2 | 78 | 117 | 58 | −14 | −97 | 87 | 34 | 53 | 63 | 63 | 63 | 58 | |
| Q3 | 92 | 141 | 83 | 53 | −83 | 112 | 48 | 73 | 78 | 78 | 73 | 73 | |
| P-wave duration (ms) | Q1 | 98 | 98 | 67 | 52 | 98 | 96 | 39 | 80 | 98 | 98 | 98 | 98 |
| Q2 | 106 | 106 | 98 | 90 | 106 | 104 | 46 | 102 | 106 | 106 | 106 | 106 | |
| Q3 | 114 | 114 | 11 | 10 | 114 | 112 | 55 | 110 | 114 | 114 | 114 | 114 | |
| P-wave area (µV * ms) | Q1 | 156 | 254 | 60 | −34 | - | 151 | 27 | 80 | 135 | 148 | 148 | 140 |
| Q2 | 200 | 330 | 13 | −10 | - | 229 | 50 | 122 | 172 | 183 | 181 | 170 | |
| Q3 | 247 | 404 | 221 | 10 | - | 305 | 76 | 163 | 210 | 221 | 216 | 203 | |
| P-wave intrinsicoid duration (ms) | Q1 | 50 | 44 | 28 | 26 | 46 | 36 | 20 | 26 | 34 | 38 | 44 | 46 |
| Q2 | 60 | 50 | 40 | 44 | 54 | 46 | 26 | 34 | 42 | 46 | 52 | 54 | |
| Q3 | 66 | 58 | 50 | 64 | 62 | 54 | 32 | 40 | 52 | 58 | 66 | 66 | |
| P’-wave amplitude (µV) | Q1 | 0 | 0 | −24 | 0 | 0 | 0 | −48 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | −34 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P’-wave duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 59 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 27 | 48 | 0 | 0 | 68 | 0 | 0 | 0 | 0 | 0 | |
| P’-wave area (µV * ms) | Q1 | 0 | 0 | −16 | 0 | 0 | 0 | −81 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | −51 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| P’-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 64 | 68 | 0 | 0 | 64 | 0 | 0 | 0 | 0 | 0 | |
| Q-wave amplitude (µV) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 24 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34 | |
| Q3 | 53 | 43 | 68 | 63 | 688 | 39 | 0 | 0 | 0 | 0 | 48 | 63 | |
| Q-wave duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 13 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | |
| Q3 | 18 | 16 | 21 | 19 | 51 | 16 | 0 | 0 | 0 | 0 | 16 | 18 | |
| Q-wave area (µV * ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 10 | 0 | 0 | 10 | 483 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | |
| Q3 | 27 | 20 | 44 | 32 | 871 | 18 | 0 | 0 | 0 | 0 | 23 | 33 | |
| Q-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 6 | 0 | 0 | 8 | 32 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | |
| Q3 | 10 | 10 | 14 | 12 | 36 | 10 | 0 | 0 | 0 | 0 | 10 | 12 | |
| R-wave amplitude (µV) | Q1 | 600 | 590 | 73 | 24 | 14 | 219 | 73 | 273 | 551 | 937 | 100 | 800 |
| Q2 | 781 | 771 | 15 | 43 | 34 | 410 | 126 | 424 | 815 | 120 | 124 | 996 | |
| Q3 | 991 | 976 | 37 | 66 | 63 | 629 | 195 | 629 | 112 | 150 | 150 | 121 | |
| R-wave duration (ms) | Q1 | 48 | 47 | 20 | 40 | 6 | 39 | 20 | 28 | 40 | 42 | 42 | 48 |
| Q2 | 63 | 60 | 29 | 55 | 15 | 52 | 24 | 34 | 45 | 47 | 49 | 63 | |
| Q3 | 74 | 75 | 51 | 68 | 20 | 70 | 28 | 40 | 50 | 52 | 59 | 72 | |
| R-wave area (µV * ms) | Q1 | 673 | 637 | 38 | 26 | 0 | 215 | 44 | 215 | 583 | 974 | 104 | 907 |
| Q2 | 913 | 895 | 11 | 51 | 16 | 458 | 88 | 374 | 882 | 127 | 132 | 116 | |
| Q3 | 120 | 120 | 41 | 82 | 38 | 766 | 152 | 603 | 121 | 162 | 167 | 145 | |
| R-wave intrinsicoid duration (ms) | Q1 | 26 | 28 | 12 | 24 | 8 | 24 | 12 | 18 | 26 | 28 | 28 | 28 |
| Q2 | 34 | 34 | 23 | 32 | 12 | 32 | 14 | 22 | 30 | 32 | 34 | 36 | |
| Q3 | 38 | 40 | 40 | 40 | 42 | 40 | 18 | 28 | 34 | 36 | 38 | 40 | |
| R’-wave amplitude (µV) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| R’-wave duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| R’-wave area (µV * ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| R’-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S-wave amplitude (µV) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 527 | 644 | 405 | 190 | 24 | 0 |
| Q2 | 0 | 19 | 14 | 0 | 590 | 53 | 712 | 874 | 605 | 346 | 131 | 0 | |
| Q3 | 73 | 131 | 41 | 12 | 844 | 175 | 917 | 113 | 825 | 527 | 263 | 63 | |
| S-wave duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 52 | 40 | 30 | 23 | 7 | 0 |
| Q2 | 0 | 7 | 26 | 0 | 40 | 15 | 59 | 48 | 38 | 33 | 27 | 0 | |
| Q3 | 27 | 28 | 51 | 34 | 66 | 34 | 65 | 56 | 45 | 40 | 36 | 25 | |
| S-wave area (µV * ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 693 | 676 | 300 | 115 | 9 | 0 |
| Q2 | 0 | 8 | 92 | 0 | 0 | 25 | 977 | 103 | 537 | 267 | 87 | 0 | |
| Q3 | 47 | 96 | 46 | 97 | 943 | 150 | 128 | 145 | 834 | 472 | 218 | 42 | |
| S-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 46 | 52 | 52 | 46 | 0 |
| Q2 | 0 | 30 | 38 | 0 | 0 | 44 | 42 | 50 | 54 | 56 | 56 | 0 | |
| Q3 | 58 | 60 | 50 | 54 | 40 | 58 | 46 | 54 | 58 | 60 | 60 | 58 | |
| S’-wave amplitude (µV) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S’-wave duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S’-wave area (µV * ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| S’-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| QRS intrinsicoid duration (ms) | Q1 | 34 | 36 | 38 | 34 | 36 | 38 | 40 | 42 | 34 | 34 | 34 | 36 |
| Q2 | 38 | 38 | 44 | 40 | 38 | 42 | 42 | 48 | 38 | 36 | 38 | 38 | |
| Q3 | 40 | 42 | 48 | 44 | 40 | 46 | 46 | 52 | 50 | 40 | 40 | 42 | |
| ST segment at J point amplitude | Q1 | 4 | 4 | −15 | −5 | −35 | −5 | −20 | −10 | −15 | −10 | −5 | −4 |
| Q2 | 14 | 19 | 4 | 4 | −20 | 14 | −5 | 14 | 9 | 9 | 9 | 19 | |
| Q3 | 29 | 39 | 24 | 24 | −10 | 29 | 9 | 39 | 29 | 29 | 29 | 34 | |
| Middle ST segment amplitude (µV) | Q1 | 4 | 9 | −5 | −5 | −35 | 4 | 14 | 43 | 29 | 14 | 9 | 4 |
| Q2 | 14 | 24 | 9 | 4 | −20 | 14 | 24 | 63 | 48 | 34 | 19 | 14 | |
| Q3 | 29 | 39 | 19 | 14 | −10 | 29 | 39 | 92 | 78 | 53 | 39 | 24 | |
| End ST segment amplitude (µV) | Q1 | 19 | 24 | −10 | 4 | −59 | 9 | 9 | 73 | 63 | 39 | 29 | 14 |
| Q2 | 34 | 43 | 9 | 14 | −44 | 24 | 29 | 112 | 97 | 68 | 48 | 29 | |
| Q3 | 53 | 63 | 24 | 29 | −25 | 43 | 48 | 161 | 141 | 102 | 78 | 48 | |
| ST 60 msec after J point amplitude | Q1 | 7 | 12 | −4 | −3 | −32 | 4 | 12 | 43 | 31 | 17 | 9 | 4 |
| Q2 | 17 | 24 | 7 | 4 | −21 | 16 | 25 | 67 | 53 | 35 | 23 | 14 | |
| Q3 | 28 | 39 | 19 | 14 | −11 | 27 | 40 | 96 | 79 | 56 | 39 | 26 | |
| T-wave amplitude (µV) | Q1 | 166 | 209 | −29 | 48 | - | 112 | −92 | 219 | 273 | 263 | 234 | 180 |
| Q2 | 219 | 263 | 53 | 92 | - | 156 | −34 | 332 | 380 | 366 | 317 | 239 | |
| Q3 | 278 | 327 | 11 | 14 | - | 209 | 63 | 458 | 507 | 483 | 415 | 312 | |
| T-wave area (µV * ms) | Q1 | 930 | 119 | −68 | 22 | - | 609 | - | 139 | 166 | 153 | 130 | 985 |
| Q2 | 120 | 150 | 23 | 46 | - | 872 | - | 203 | 226 | 206 | 173 | 129 | |
| Q3 | 151 | 183 | 60 | 73 | - | 118 | 351 | 275 | 296 | 269 | 225 | 166 | |
| T-wave intrinsicoid duration (ms) | Q1 | 102 | 106 | 72 | 88 | 104 | 104 | 62 | 82 | 94 | 98 | 102 | 104 |
| Q2 | 114 | 116 | 10 | 10 | 116 | 118 | 100 | 96 | 106 | 112 | 114 | 116 | |
| Q3 | 126 | 128 | 12 | 12 | 128 | 130 | 120 | 110 | 118 | 124 | 126 | 128 | |
| T’-wave amplitude (µV) | Q1 | 0 | 0 | 4 0 | 4 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| T’-wave area (µV * ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| T’-wave intrinsicoid duration (ms) | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Definitions
- Q1 is 25th percentile, Q2 is 50th percentile or median, and Q3 is 75th percentile
- P-wave intrinsicoid duration = time from P onset to peak of P
- P’-wave intrinsicoid duration = time from P’ onset to peak of P’, where P’ is a second deflection of the P-wave that is opposite in polarity to the original P-wave
- Q-wave intrinsicoid duration = time from Q onset to peak of Q
- R-wave intrinsicoid duration = time from Q onset to peak of R
- S-wave intrinsicoid duration = time from Q onset to peak of S
- S’-wave intrinsicoid duration = time from Q onset to peak of S’
- R’-wave intrinsicoid duration = time from Q onset to peak of R’
- T-wave intrinsicoid duration = time from end of ST segment to peak of T
- T’-wave intrinsicoid duration = time from end of ST segment to peak of T’, where T’ is a second deflection of the T-wave that is opposite in polarity to the original T-wave
- QRS intrinsicoid duration = time from onset of QRS complex to middle of QRS complex
During a median follow-up time of 8.1 years (range for survivors 0.5–11.2 years), 1,229 women (3.7%) died. Causes of death included cardiovascular diseases (n=251, 20%), cancer (n=664, 54%), homicide/suicide (n=13, 1%), accident/injury (n=42, 3%), other/unknown (n=259, 21%).
Identification of predictors
In the derivation cohort using all demographic, clinical, and ECG predictors, the 20 variables identified by RSF that were most predictive of long-term all-cause mortality (Figure 3) were the following:
- ECG variables representing autonomic tone
-
○Ventricular variability (SDNN, RMS-SD)
-
○Ventricular rate
-
○
- ECG variables representing atrial conduction
-
○P wave durations (P wave intrinsicoid duration in leads V3 and V4, P wave duration in lead V2)
-
○P wave areas (P wave area in lead V2)
-
○P wave amplitude (P wave amplitude in lead I)
-
○P wave axis (median of all leads)
-
○
- ECG variables representing ventricular depolarization and repolarization
-
○QT duration (median of all leads)
-
○
- ECG variables representing ventricular repolarization
-
○T wave areas (T wave area in lead I, T wave area in lead aVL)
-
○T wave amplitude (T wave amplitude in lead I)
-
○T wave axis (median in all leads)
-
○
- Traditional variables
-
○Age
-
○Waist-to-hip ratio
-
○Smoking
-
○Income
-
○Systolic blood pressure
-
○Body mass index
-
○
Figure 3.
Minimal depth (variable importance) for (A.) all variables averaged out from all trees in forest (1,000 trees), and (B.) zoomed in on top 20 variables. Dashed blue line is threshold for filtering variables: variables to left of line are predictive. On y-axis is ranking of variables where age is most predictive, then waist-to-hip ratio, and so forth.
External validation
We used the validation subset (n=11,048) to externally validate eight RSF and Cox prediction models (Table 4). The Cox regression models (Models 2–4) utilizing all 499 variables did not converge. The RSF and Cox regression models constructed with covariates selected by various variable selection methods demonstrated similar discriminative accuracy in the derivation and validation datasets. Hazard ratios and 95% confidence intervals derived from Cox model (Model 6) are shown in Supplementary Material Table 1.
Table 4.
C-index values
| # variables in model |
Derivation Cohort | Validation Cohort | ||
|---|---|---|---|---|
|
n Deaths |
22,097 819 (3.7%) |
11,048 410 (3.7%) |
||
| Prediction models utilizing all covariates | ||||
| Model 1 | RSF | 499 | 0.6815 | 0.6710 |
| Model 2 | Cox | 499 | Did not converge | |
| Model 3 | I1-penalized Cox | 499 | Did not converge | |
| Model 4 | AIC-penalized Cox | 499 | Did not converge | |
| Prediction models utilizing covariates selected by variable selection methods | ||||
| Model 5 | RSF | 20 | 0.6992 | 0.6934 |
| Model 6 | Cox | 20 | 0.6954 | 0.6975 |
| Model 7 | I1-penalized Cox | 59 | 0.7003 | 0.6978 |
| Model 8 | AIC-penalized Cox | 22 | 0.7005 | 0.6980 |
Models 1–4 utilize all 499 demographic, clinical, and ECG variables available.
Models 5–6 utilize 20 variables selected by RSF variable selection method (Demographic/clinical: age, waist-to-hip ratio, smoking, income, systolic blood pressure, body mass index. ECG: SDNN, ventricular rate, T-wave area (lead I), P-wave intrinsicoid duration (leads V3, V4), P-wave duration (lead V2), T-wave amplitude (lead I), RMS-SD, T-wave axis, P-wave axis, P-wave amplitude (lead I), T-wave area (lead aVL), QT duration, P-wave area (lead V2))
Model 7 utilizes 59 variables selected by lasso approach from top 100 RSF variables (Demographic/clinical: age, waist-to-hip ratio, smoking, systolic blood pressure, income, body mass index, hypertension, education, diastolic blood pressure, marital status, alcoholic drinks per week, treated diabetes. ECG: SDNN, P-wave intrinsicoid duration (leads I, aVL, V2, V4, V5, V6), ventricular rate, P-wave duration (leads I, aVL, V2, V3, V6), RMS-SD, T-wave axis, P-wave axis, R-wave duration (leads aVF, V1, V4), P-wave area (leads I, V1), QRS intrinsicoid duration (lead I), T-wave intrinsicoid duration (leads I, III, aVL), P-wave amplitude (leads I, aVL, V5), T-wave area (leads aVR, V3), R-wave amplitude (leads II, V1, V5, V6), R-wave intrinsicoid duration (leads II, aVF, V1, V3, V4), P’-wave area (lead V1), R-wave area (leads III, aVL, V3, V6), T-wave amplitude (lead V1), QTc duration, P’-wave amplitude (lead V2))
Model 8 utilizes 22 variables selected by AIC stepwise approach from top 50 RSF variables (Demographic/clinical: age, waist-to-hip ratio, smoking, systolic blood pressure, income, body mass index, hypertension, education, marital status. ECG: ventricular rate, P-wave duration (lead V2), T-wave axis, P-wave axis, R-wave duration (lead V4), P-wave area (lead I), P’-wave intrinsicoid duration (lead aVL), QRS intrinsicoid duration (lead I), T-wave area (leads aVR, aVL), P-wave amplitude (lead I), R-wave intrinsicoid duration (lead aVF), R-wave area (lead V5))
We assessed the individual contribution of 20 variables (6 demographic / clinical variables, and 14 ECG variables) selected by RSF variable selection method to discrimination (c-index) and calibration (CRPS) in sequential nested RSF models, where the first model utilized only age, the second age and waist-to-hip ratio, the third age, waist-to-hip ratio, and smoking, and so forth. Figure 4 shows that these performance measures stabilized in the range of 15–20 variables, near the size of the model identified by the primary analysis (Figure 3, Table 4).
Figure 4.
Measures of (A.) discrimination and (B.) calibration using validation cohort for nested models with variables ordered by increasing minimal depth for top 20 variables. First model included top variable (age), second model included top two variables (age and waist-to-hip ratio), third model included top three variables (age, waist-to-hip ratio, and smoking), and so forth.
Discussion
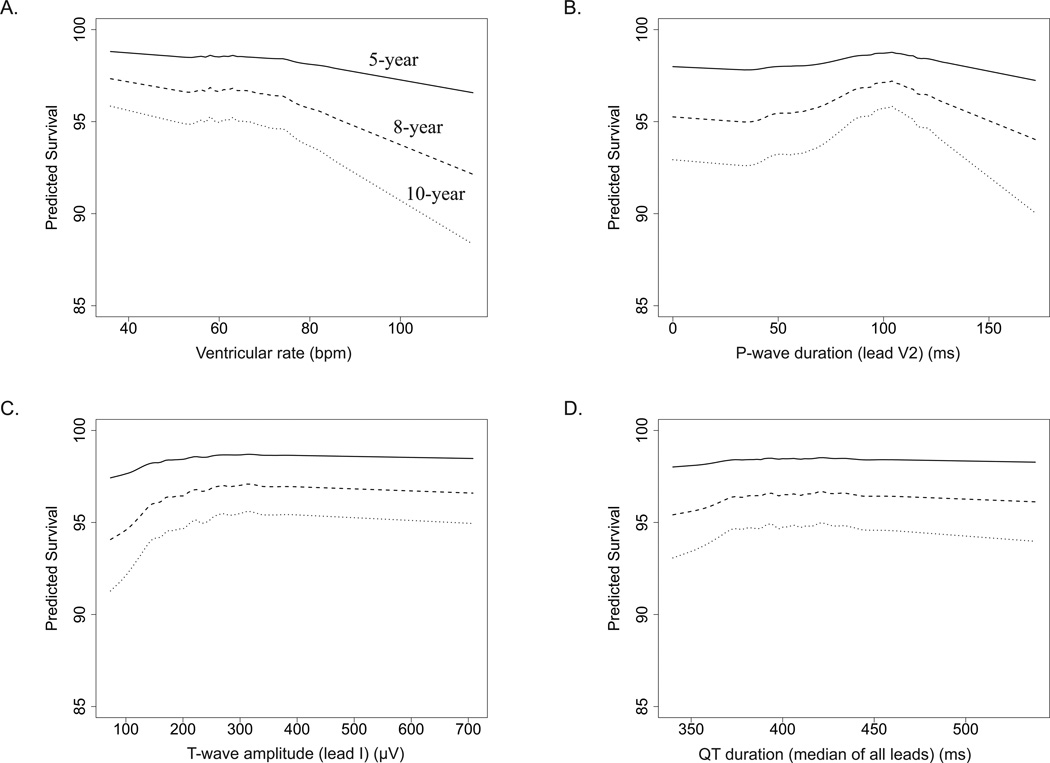
Among 33,144 post-menopausal women without known cardiovascular disease or cancer, with normal resting electrocardiograms by Minnesota and Novacode criteria, we found that 20 variables were independently predictive of long-term mortality, 14 of which were electrocardiographic biomarkers representing autonomic tone (ventricular rate and variability), atrial conduction (P wave durations and areas), ventricular depolarization (QT duration), and ventricular repolarization (T wave axis, amplitude, and areas). Selected plots demonstrating adjusted predicted survival for an ECG biomarker from each one of these four categories are shown in Figure 5 (all others shown in as Supplementary Material Figure 3). Further, we found that parsimonious prediction models incorporating these ECG measures, along with demographic and clinical characteristics selected by an RSF variable selection procedure, yielded better predictive accuracy than non-parsimonious RSF model using all variables (Table 4). Lastly, parsimonious RSF model populated by RSF variable selection procedure was sparser (i.e., containing less covariates) than parsimonious regression models populated by various other variable selection approaches, but performed similarly well in terms of prediction (Table 4). While other investigators have reported on the predictive utility of ECG findings in women1–3, we are the first to use an algorithmic approach to simultaneously assess hundreds of digitally measured ECG variables without the bias of pre-selection.
Figure 5.
Adjusted-predicted survival (%) at 5, 8, and 10 years for (A.) ventricular rate, (B.) P-wave duration (lead V2), (C.) T-wave amplitude (lead I), and (D.) QT duration (median of all leads).
Utilizing hundreds of electrocardiographic measures for prediction modeling presents a unique challenge. Many of these variables are highly correlated, may have complex interactions that are difficult to detect, and may have non-linear associations with outcome. Traditional regression and variable selection methods perform poorly under these types of conditions, and tend to produce biased results.6 Our findings confirm these challenges. When we attempted to employ standard Cox modeling we were unable to generate models that converged (Table 4). Additionally, for the penalized Cox regression modeling (Model 7 and Model 8), it was necessary to restrict the selection of the model variables in an arbitrary manner in order for these methods to converge. To address these challenges we used RSF methodology both for risk modeling and variable selection.
Machine learning, the scientific discipline from which RSF methodology is derived, is a field concerned with the design and development of algorithms that allow computers to change behavior based on data.20 This approach assumes that “nature produces data in a black box whose insides are complex, mysterious, and at least, partly unknowable.” 6 As such, instead of attempting to model data from the black box (i.e., traditional regression), machine learning is concerned with iterative algorithms such as RSF that are intensely focused on prediction.
Unlike classification and regression trees (CART) where only a single tree is constructed, RSF uses a large number of survival trees for prediction and variable selection.8 Growing extensive trees with hundreds of decision branches is a general principle of RF methodology.7 Doing so yields trees with low bias (i.e., prediction models that better estimate the predictor being estimated). To ensure low variance (i.e., amount of variation within predicted results), trees must be decorrelated. This is accomplished by introducing two forms of randomization when growing a tree (Figure 1). First, trees are grown using independent bootstrap samples of data. Second, each tree is grown by randomly selecting a subset of candidate variables for splitting at each node. Employing this two-stage randomization yields stable and accurate inference and resolves the instability of CART.21 RF has been shown to be accurate, and comparable to state-of-the-art predictors such as bagging22, boosting23, and support vector machines24. Further, RF has been shown to be highly effective in problems involving large numbers of correlated variables.7, 16, 25–27 Examples in the literature include genetics28, 29, environmental science30, and rheumatology31.
We believe that RSF analysis may have potential future applications in clinical practice. RSF prediction model can be stored as an object in the statistical software, and then be used at a later time on external datasets. This is possible because the random seed chain used to generate the original model is stored. Thus once a model is generated, it can be used repeatedly on test data sets and will yield identical results if repeated on the same data set. Further, if the original data is used on the restored model, the results will be identical to that of the original analysis. Moreover, this applies even when the training and/or test data have missing values because we also store the seed chain used to impute missing data values. Thus when the model is restored the seed chain used to impute data is re-initialized and the original forest and its imputation mechanism are reproduced exactly as before. These properties may allow RSF to be used as a prediction tool in clinical settings. It is technologically feasible to create web-based or even hand-held RSF "calculators" that could be used in practice.
Our study has several important limitations. First, the WHI clinical trials enrolled mostly white, highly-educated women, and may therefore have limited generalizability. Second, many of the clinical variables were by self-report, and data regarding standard blood biomarkers were lacking. Lastly, we did not have an external dataset (replication cohort) with which to validate our prediction models, although we attempted to do so by setting a portion of our data aside for validation. We are not aware of a similar cohort of post-menopausal women with detailed ECG data to allow such replicaton/validation. It is possible that several other NHLBI cohorts, including the Framingham Heart Study, may soon digitize ECG data and make it available to investigators.
In summary, we found that electrocardiographic biomarkers representing autonomic tone, atrial conduction, and ventricular depolarization and repolarization were independently predictive of long-term mortality in post-menopausal women who had no known cardiovascular disease or cancer, and had normal ECGs by standard clinical criteria. These findings suggest that further research will be necessary to identify underlying pathophysiological mechanisms and potential therapeutic implications. Additionally, we introduced RSF, a machine learning approach to data analysis, which may be of utility in other complex data problems in cardiovascular medicine.
Supplementary Material
What is Known
Prior studies demonstrated that amongst post-menopausal women single ECG measures, or small groups of ECG measures, are prognostic of long-term mortality.
Simultaneous contribution of hundreds of ECG measures to prediction of mortality in this population has not been studied.
What the Study Adds
We use random survival forests, a novel “machine learning” statistical approach, to demonstrate that amongst apparently healthy post-menopausal women with clinically normal ECGs, ECG biomarkers related to autonomic tone, atrial conduction, and ventricular depolarization and repolarization have long-term prognostic significance.
Acknowledgements
For a list of the WHI investigators, see http://www.whi.org/about/investigators.php.
Funding Sources
Supported by National Heart, Lung, and Blood Institute CAN #8324207 (EZG, MSL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures
None.
References
- 1.Rautaharju PM, Kooperberg C, Larson JC, LaCroix A. Electrocardiographic abnormalities that predict coronary heart disease events and mortality in postmenopausal women: the Women's Health Initiative. Circulation. 2006;113:473–480. doi: 10.1161/CIRCULATIONAHA.104.496091. [DOI] [PubMed] [Google Scholar]
- 2.Rautaharju PM, Kooperberg C, Larson JC, LaCroix A. Electrocardiographic predictors of incident congestive heart failure and all-cause mortality in postmenopausal women: the Women's Health Initiative. Circulation. 2006;113:481–489. doi: 10.1161/CIRCULATIONAHA.105.537415. [DOI] [PubMed] [Google Scholar]
- 3.Denes P, Larson JC, Lloyd-Jones DM, Prineas RJ, Greenland P. Major and minor ECG abnormalities in asymptomatic women and risk of cardiovascular events and mortality. Jama. 2007;297:978–985. doi: 10.1001/jama.297.9.978. [DOI] [PubMed] [Google Scholar]
- 4.Zhang ZM, Prineas RJ, Eaton CB. Evaluation and comparison of the Minnesota Code and Novacode for electrocardiographic Q-ST wave abnormalities for the independent prediction of incident coronary heart disease and total mortality (from the Women's Health Initiative) The American journal of cardiology. 2010;106:18–25. doi: 10.1016/j.amjcard.2010.02.007. e12. [DOI] [PubMed] [Google Scholar]
- 5.Iuliano S, Fisher SG, Karasik PE, Fletcher RD, Singh SN. QRS duration and mortality in patients with congestive heart failure. American heart journal. 2002;143:1085–1091. doi: 10.1067/mhj.2002.122516. [DOI] [PubMed] [Google Scholar]
- 6.Breiman L. Statistical Moderling: The Two Cultures. Statistical Science. 2001;16:199–231. [Google Scholar]
- 7.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 8.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random Survival Forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 9.Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C, Rossouw JE. The Women's Health Initiative recruitment methods and results. Annals of epidemiology. 2003;13:S18–S77. doi: 10.1016/s1047-2797(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 10.Prineas RJ, Crow RS, Blackburn H. The Minnesota Code Manual of Electrocardiographic Findings. John Wright PSB; Boston, MA: 1982. p. 203. [Google Scholar]
- 11.Prineas RJ, Crow RS, Zhang ZM. The Minnesota Code Manual of Electrocardiographic Findings (Second edition) Published by Springer-London; 2009. pp. 277–324. [Google Scholar]
- 12.Rautaharju PM, Park LP, Chaitman BR, Rautaharju F, Zhang ZM. The Novacode criteria for classification of ECG abnormalities and their clinically significant progression and regression. Journal of electrocardiology. 1998;31:157–187. [PubMed] [Google Scholar]
- 13.Design of the Women's Health Initiative Clinical Trial and Observational Study. Controlled Clinical Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 14.Lauer MS, Blackstone EH, Young JB, Topol EJ. Cause of death in clinical research: time for a reassessment? Journal of the American College of Cardiology. 1999;34:618–620. doi: 10.1016/s0735-1097(99)00250-8. [DOI] [PubMed] [Google Scholar]
- 15.Curb JD, McTiernan A, Heckbert SR, Kooperberg C, Stanford J, Nevitt M, Johnson KC, Proulx-Burns L, Pastore L, Criqui M, Daugherty S. Outcomes ascertainment and adjudication methods in the Women's Health Initiative. Annals of epidemiology. 2003;13:S122–S128. doi: 10.1016/s1047-2797(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 16.Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-Dimensional Variable Selection for Survival Data. J Am Stat Assoc. 2010;105:205–217. [Google Scholar]
- 17.Gerds TA, Cai T, Schumacher M. The performance of risk prediction models. Biom J. 2008;50:457–479. doi: 10.1002/bimj.200810443. [DOI] [PubMed] [Google Scholar]
- 18.Ishwaran H, Kogalur UB. [Accessed on May 5, 2009];RandomSurvivalForest 3.5.1 R Package. Available at: http://cran.r-project.org. [Google Scholar]
- 19.Ishwaran H, Kogalur UB. Random Survival Forests for R. Rnews. 2007;7:25–31. [Google Scholar]
- 20.Wikipedia contributors. Machine learning. [Accessed February 3, 2010];Wikipedia, The Free Encyclopedia. 2010 Jan;24 at 20:58. Available at: http://en.wikipedia.org/wiki/Machine_learning. [Google Scholar]
- 21.Breiman L. Heuristics of instability and stabilization in model selection. Ann Statist. 1996;24:2350–2383. [Google Scholar]
- 22.Breiman L. Bagging predictors. Machine Learning. 1996;1996:123–140. [Google Scholar]
- 23.Freund Y, Shapire RE. Experiments with a new boosting algorithm. Proc of the 13th Int Conf on Machine Learning. 1996:148–156. [Google Scholar]
- 24.Cortes C, Vapnik VN. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 25.Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, Van Eerdewegh P. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 26.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunetta KL, Hayward LB, Segal J, Van Eerdewegh P. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang H, Lin A, Holford M, Enerson BE, Lu B, Lawton MP, Floyd E, Zhao H. Pathway analysis using random forests classification and regression. Bioinformatics. 2006;22:2028–2036. doi: 10.1093/bioinformatics/btl344. [DOI] [PubMed] [Google Scholar]
- 29.Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, Foekens JA, van de Vijver M, Massague J. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkhurst DF, Brenner KP, Dufour AP, Wymer LJ. Indicator bacteria at five swimming beaches-analysis using random forests. Water Res. 2005;39:1354–1360. doi: 10.1016/j.watres.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Ward MM, Pajevic S, Dreyfuss J, Malley JD. Short-term prediction of mortality in patients with systemic lupus erythematosus: classification of outcomes using random forests. Arthritis and rheumatism. 2006;55:74–80. doi: 10.1002/art.21695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.