Abstract
Plasma cell leukemia (PCL) is an aggressive and rare hematological malignancy that originates either as primary disease (pPCL) or as a secondary leukemic transformation (sPCL) of multiple myeloma (MM). We report here the genetic aberrations and survival of 80 patients with pPCL or sPCL and make comparisons with 439 cases of MM. pPCL presents a decade earlier than sPCL (54.7 vs 65.3 years) and is associated with longer median overall survival (11.1 vs 1.3 months; P<0.001). 14q32 (IgH) translocations are highly prevalent in both sPCL and pPCL (82–87%); in pPCL IgH translocations almost exclusively involve 11q13 (CCND1), supporting a central etiological role, while in sPCL multiple partner oncogenes are involved, including 11q13, 4p16 (FGFR3/MMSET) and 16q23 (MAF), recapitulating MM. Both show ubiquitous inactivation of TP53 (pPCL 56%; sPCL 83%) by coding mutation or 17p13 deletion; complemented by p14ARF epigenetic silencing in sPCL (29%). Both show frequent N-RAS or K-RAS mutation. Poor survival in pPCL was predicted by MYC translocation (P=0.006). Survival in sPCL was consistently short. Overall pPCL and sPCL are different disorders with distinct natural histories, genetics and survival.
Keywords: translocation, deletion, p53, cyclin D, RAS, MYC
Introduction
Plasma cell leukemia (PCL) is the most aggressive presentation of the plasma cell neoplasms and is characterized by circulating plasma cells >2 × 109 l−1 in peripheral blood1 or by a relative plasmacytosis >20% of blood leukocytes.2 While primary disease (pPCL) presents as de novo leukemia, secondary leukemic transformation (sPCL) arises from pre-existing multiple myeloma (MM)2,3 probably as a consequence of clonal transformation. The relationship between pPCL and MM is unclear; however, pPCL is not currently distinguished from MM or sPCL as a distinct entity in the WHO classification of hematopoietic tumors.4
PCL is rare. sPCL occurs in only 1.8–4% of MM patients and pPCL occurs with a comparable incidence. Consequently, few large studies of PCL patients have been reported2,3,5–7 and the molecular basis of PCL remains poorly understood. Nevertheless, previous studies indicate that pPCL tumors are often hypodiploid and suggest a greater incidence of t(11;14) (33%) or t(14;16) (13%) than that found in MM tumors.5,7
The prognosis of PCL is poor.2,3,8,9 To further understand the genetic basis of PCL and to correlate genetic variants with survival, we examined and report a cohort of 80 patients with pPCL or sPCL seen at Mayo Clinic over the past four decades, representing the largest PCL cohort reported to date.
Materials and methods
Patients
We searched for patients diagnosed with PCL at our institution and included cases that met common diagnostic criteria.1,2 As a minimum, patients required a peripheral plasmacytosis >2×109 l−1 or >20% of the total leukocyte count. Patient data and samples were accessed following Mayo Clinic Institutional Review Board sanction and in accordance with the principles laid down in the Declaration of Helsinki. Patients were characterized for approximately 60 demographic, clinical and laboratory variables. In addition, karyotype studies and or material for fluorescence in situ hybridization (FISH) or PCR were obtained for 41 patients, including 18 with pPCL and 23 with sPCL. We compared clinical and genetic data from these patients with data from a large published cohort of 439 MM patients.10–12
Tumor specimens
Samples were processed for cytogenetic studies and plasma cell labeling index13–15 at the time of clinical procurement. Additional tumor material (cells or DNA) from 33 patients was stored.
Interphase FISH
Twenty-six archived mononuclear cell pellets were analyzed by FISH. We used validated probes to detect IgH (CHVH) and IgL (CLVL) segregation (‘break apart’),16 Δ13,11,17 t(4;14) (p16.3q32),17,18 t(11;14) (q13;q32),19,20 t(14;16) (q32;q23)21 and 17p13.1.12 A c-myc probe from Vysis (Downers Grove, IL, USA) and probes 5′ and 3′ of c-myc (RPCI 11.C 645E10; RPCI 11.C 259L23) from Invitrogen (Carlsbad, CA, USA) and IgH (CH) and IgL (CL) probes16 were used to detect c-myc amplification, 3′- or 5′-myc break apart and CH-myc or CL-myc fusion. Bacterial artificial chromosome probes from Invitrogen were used to enumerate PTEN (RPCI 11.C 380G5) and MDM2 (RPCI 11.C 775J10) loci; these were labeled with SpectrumGreen or SpectrumOrange by nick translation and were validated on normal metaphases before use.
Methylation-sensitive PCR of p16/p14 promoters
Bone marrow DNA from 27 patients (16 sPCL, 11 pPCL) was modified with sodium bisulfite using CpGenome DNA Modification Kit (Intergen, NY, USA) and assayed by methylation-modification sensitive PCR using a HotStartTaq Kit from Qiagen, (Valencia, CA, USA). Primer sequences and reaction conditions for p16 and p14 have previously been reported.22,23 Controls included universal methylated human genomic DNA (Intergen) and healthy donor peripheral blood DNA. Products were analyzed by electrophoresis.
Mutational analysis of TP53 and N- or K-RAS
Bone marrow-derived DNA was screened for RAS or TP53 mutations using conformation sensitive gel electrophoresis.24,25 Abnormal samples were assessed by DNA sequencing. For TP53, exons 1–11 were sequenced. Both N-RAS and K-RAS were screened; exons 1 and 2 were sequenced, targeting the codons most commonly mutated in hematological malignancy (12, 13 and 61).26–29 Primer sequences are listed in Supplementary Table S1 (online).
Statistical analysis
Groups of continuous data with similar variance were compared using a Student’s t-test. Categorical data across groups were tested by Fisher’s exact test. Overall survival (OS) was assessed using the Kaplan–Meier method. Survival associations with categorical variables were assessed by log-rank test; continuous variables were assessed by parametric regression modeling or by reduction to ordinal data and log-rank test. Test statistics were calculated using a two-sided assessment and P<0.05 was considered significant. JMP v6.0 Statistical Discovery (Cary, NC, USA) software was used.
Results
Patient demographics
We identified 86 patients with a diagnosis of PCL, of whom 80 fulfilled our study criteria. These included 41 patients with pPCL and 39 with sPCL (Table 1). The six excluded patients had a plasmacytosis below the common diagnostic threshold. The number of PCL cases identified was equivalent to 1.3% of MM patients seen during the same period. Median age was 59.2 years, although patients with pPCL were a decade younger (median 54.5 years) than patients with sPCL (65.7 years) or MM (66 years) (P<0.001) suggesting differences in pPCL and sPCL biology.
Table 1.
PCL patient characteristics
| Characteristics | Indice | Normal values | MM | sPCL | pPCL | P-value† |
|---|---|---|---|---|---|---|
| Patient numbers | n | 439 | 39 | 41 | ||
| Age (years) | Median | 66 | 65.7 | 54.5 | < 0.001* | |
| Range | 20–92 | 48–88 | 31–82 | — | ||
| Sex | Female (%) | 41 | 40 | 41 | 1 | |
| Symptoms (days)—leukemic | Median | — | 15.5 | 30 | 0.025* | |
| Time to progression, MM to sPCL (months) | Median | — | 20.8 | — | — | |
| Extraosseous plasmacytoma | Cases (%) | 6 | 22 | 0.21 | ||
| Osteolytic lesions | Cases (%) | 81 | 53 | 35 | 0.19 | |
| Fractures | Cases (%) | 35 | 35 | 18 | 0.15 | |
| Splenomegaly | Cases (%) | — | 8 | 18 | 0.29 | |
| Hepatomegaly | Cases (%) | — | 11 | 32 | 0.04* | |
| Adenopathy | Cases (%) | — | 3 | 6 | 0.6 | |
| WBC ( × 109l−1 ) | Median | 3.8–10.4 | 5.8 | 15.7 | 21.5 | 0.04* |
| PB plasmacytosis (%) | Median | 0 | 0 | 52 | 46 | 0.017* |
| PB plasmacytosis (× 109l−1) | Median | 0 | 0 | 6.3 | 7.2 | 0.19 |
| Range | 0 | — | 2.3–76 | 1.5–164 | — | |
| Hgb (g per 100ml) | Median | 11.5–17.5 | 10.9 | 9.1 | 9.4 | 0.05* |
| Platelets (× 109l−1) | Median | 149–375 | 242 | 53 | 98 | 0.05* |
| Serum creatinine (mg per 100 ml) | Median | 0.6–1.3 | 1.2 | 1.4 | 1.9 | 0.03* |
| Serum calcium (mg per 100 ml) | Median | 8.9–10.1 | 9.5 | 9.8 | 10 | 0.25 |
| Serum phosphate (mg per 100 ml) | Median | 2.5–4.5 | — | 4.35 | 4.8 | 0.05* |
| ALP(Ul1) | Median | 45–142 | — | 141 | 163 | 0.09 |
| AST (Ul−1) | Median | 18–49 | — | 43.5 | 26 | 0.008* |
| LDH (U1−1) | Median | 122–222 | — | 434 | 459 | 0.47 |
| Total bilirubin (mg per 100 ml) | Median | 0.1–1.1 | 0.4 | 0.5 | 0.5 | 0.177 |
| CRP (mg per 100 ml) | Median | 0.020–0.80 | 0.32 | 0.46 | 0.4 | 0.11 |
| β2M (µgl−1) | Median | 0.70–1.80 | 3.6 | 3.5 | 6.8 | 0.02* |
| IgG | Cases (%) | 52 | 38 | 28 | 0.47 | |
| IgA | Cases (%) | 21 | 28 | 13 | 0.16 | |
| IgD | Cases (%) | 1 | 5 | 2 | 1 | |
| IgE | Cases (%) | 0 | 2 | 0 | 1 | |
| Light-chain only | Cases (%) | 16 | 31 | 41 | 0.36 | |
| Non-secretor | Cases (%) | 7 | 0 | 8 | 0.11 | |
| κ (excluding non-secretors) | Cases (%) | 66 | 58 | 42 | 0.24 | |
| λ(excluding non-secretors) | Cases (%) | 34 | 42 | 58 | 0.24 | |
| PCLI | Median | 0.4 | 3.0 | 1.4 | 0.47 | |
| BM plasmacytosis | BM (%) | < 5 | 46 | 63 | 78 | 0.11 |
| Survival (months) | Median | 34 (48)a | 1.3 | 11.1 | — |
Abbreviations: BM, bone marrow; MM, multiple myeloma; PCL, plasma cell leukemia; pPCL, primary PCL; PB, peripheral blood; sPCL, secondary PCL.
Significant difference between sPCL and pPCL (P< 0.05).
P-values compare means of sPCL and pPCL.
Survival of nonhyperdiploid and hyperdiploid MM, respectively.
Clinical features
In sPCL, the median time to leukemic progression from MM was approximately midway (20.8 months) between diagnosis and median survival of MM patients, indicating that transformation from MM to sPCL is not a late event in MM but occurs both early and late. Extramedullary disease was more evident at diagnosis among pPCL than sPCL cases (Table 1). pPCL presented with more hepatomegaly (32 vs 11%, P=0.04), splenomegaly (18 vs 8%, P=0.29), extra-osseous plasmacytoma (22 vs 6%, P=0.21) and adenopathy (6 vs 3%). By contrast, bone disease was more common in sPCL with a higher prevalence of osteolytic lesions (sPCL 53% vs pPCL 35%, P=0.19) and more fractures (sPCL 35% vs pPCL 18%), consistent with its origin from MM.
Surprisingly, despite their younger age and shorter clinical prodrome, patients with pPCL presented with more renal dysfunction (median creatinine 1.90mg per 100 ml) than patients with sPCL (median creatinine 1.40mg per 100 ml) (P=0.03). Both leukemias presented with more renal impairment than newly diagnosed MM (median creatinine 1.20 mg per 100 ml). Perhaps because of renal impairment, pPCL patients had higher β2-microglobulin (6.80 µg ml−1) than patients with sPCL (3.50 µg ml−1) (P=0.022) or MM (3.6 µg ml−1). A monoclonal paraprotein was detectable in 94% of PCL. However, light-chain only secreting tumors were substantially increased in PCL (pPCL 41%, sPCL 31%) compared with MM (16%). Tumors expressing λ light chain were over-represented in pPCL (56%) and in sPCL (41%) compared with MM (34%).
Survival and treatment in PCL
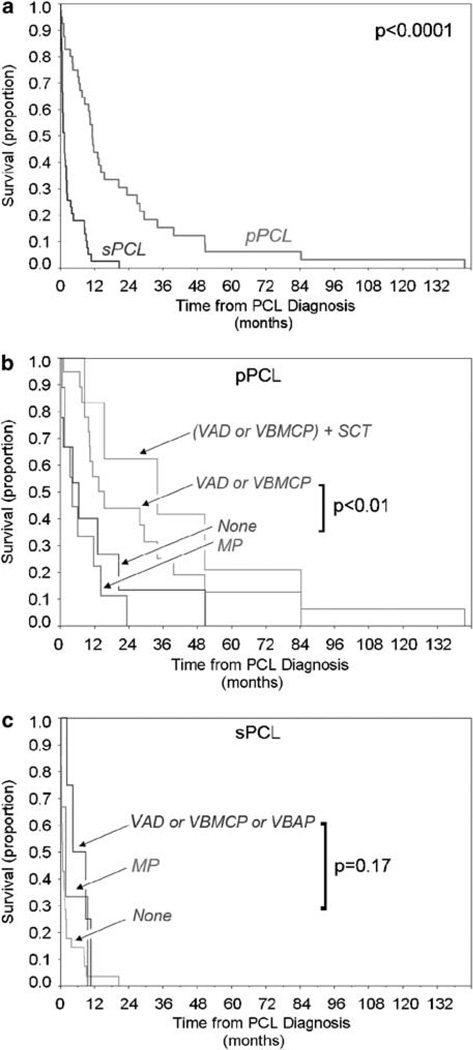
Overall survival was extremely short among patients diagnosed with sPCL (median 1.3 months), even though many cases (>50%) of sPCL occurred within the first 24 months of MM diagnosis. OS in pPCL was also short (median 11.2 months); however, it was significantly longer than survival in sPCL (P<0.0001) (Figure 1).
Figure 1.
Overall survival (OS) in pPCL and sPCL. (a) OS in PCL showing superior survival of pPCL vs sPCL from the time of leukemia diagnosis. (b) OS in pPCL stratified by initial treatment with multiagent chemotherapy (VAD or VBMCP), MP, or no treatment. The survival of patients treated with multiagent chemotherapy and SCT is also shown. (c) OS in sPCL stratified by treatment with combination chemotherapy (VAD, VBMCP or VBAP), MP or no treatment MP, melpahalan and prednisone; PCL, plasma cell leukaemia; pPCL, primary PCL; SCT, stem cell transplant; sPCL, secondary PCL.
The treatments administered were heterogeneous and are reported here in a descriptive fashion. Of 41 pPCL patients, 21 were treated with multiagent intravenous chemotherapy (VAD or VBMCP), and 20 received oral melphalan and prednisone (MP) or no standard therapy. OS of pPCL patients treated with VAD or VBMCP (median 15.4 months) was substantially longer (two to threefold) than patients who received MP (4.1 months) or no therapy (6.4 months) (P=0.007) (Figure 1b). Retrospective treatment analyses can incur bias; however, comparison of the pretreatment characteristics of patients in these treatment arms revealed few significant pretreatment differences between patients treated with oral or intensive therapy apart from less hepatosplenomegaly in the later group (Supplementary Table S2). Organomegaly was not accompanied by higher rates of liver test abnormalities and did not independently correlate with survival (median OS 11.7 months with organomegaly, vs 11.2 months without organomegaly, P=0.49). Of pPCL patients treated with VAD or VBMCP, 30% were treated later with hematopoietic stem cell transplant (SCT). In this setting, SCT was associated with a survival advantage of 22 months (OS 34 vs 11.3 months); however, the true attributable benefit from SCT on OS in pPCL is probably less than 22 months due to the selection bias for SCT favoring younger patients (47.3 vs 54.3 years, P=0.09) and possibly chemotherapy responsive patients (enabling stem-cell collection).
Patients with sPCL received less therapy in our cohort than patients with pPCL. Eight (21%) patients received VAD, VBMCP or VBAP. Three patients received MP alone. The 11 sPCL patient who received VAD, VBMCP, VBAP or MP were distinguished from nontreated patients by younger age (60.9 years vs 66.4 years) and by lack of bone disease (0 vs 65%, P<0.01); however, despite this bias, median survival was short with these treatments and was not significantly better than no treatment (Figure 1c).
Cytogenetics in PCL
Standard cytogenetic results were available for 38 PCL patients. An abnormal karyotype was obtained in 27 cases (Table 2). Most PCL karyotypes were complex and hypodiploid or pseudodiploid. Only two tumors, both sPCL, were hyperdiploid. One pPCL tumor had 84–88 chromosomes, consistent with 4N hypodiploidy. As summarized in Table 3, when compared with MM there is a clear trend among PCL tumors to hypodiploidy that is most pronounced in pPCL.
Table 2.
Conventional cytogenetics in pPCL and sPCL
| Patient | Age (years) |
Sex | Prior MM |
Cytogenetics | Survival (months) |
|---|---|---|---|---|---|
| 1 | 57.5 | F | N | Complex, includes t(2;6); full report not available | 27.8 |
| 2 | 55.9 | F | N | 39–43,X,−X,−1,−4,add(8)(p11.2),−13,−14,add(16)(q22),der(?)t(1;?)(q23;?),der(?)t(1;?) (q23;?)+mar[cp4]/46,XX[16] | 11.2 |
| 3 | 62.2 | M | N | 42–45,XY,−1,−13,−14,−15,−16,−18,+der(14) t(11;14)(q13;q32),+3–10mar[AN][24]/46,XY[1] | 3.4 |
| 4 | 61.8 | F | N | 41–42,XX,−1,add(5)(q13),−8–9–10,add(11)(p13),−13,add(14)(q32),add(15)(p13),−16,+add(18)(q23),add(19)(q13.3),del(20)(q11.2),−22,+2mar[cp16]/46,XX[4] | 8.6 |
| 5 | 59.5 | M | N | 44–47,XY,add(1)(p13),−5,−11,−13,der(14)t(11;14)(q13;q32),−17,−18,−20,−22,+4–8mar[cp3] /46XY[9] | |
| 6 | 50.8 | M | N | 44,XY,add(1)(p13),add(2)(p21),−6,add(8)(p23),t(11;14)(q13;q32),add(12)(q24.1),−13,−14,+mar[4]/46,XY[16] | 10.2 |
| 7 | 49.3 | F | N | 45,X,−X,del(1)(p22p32),t(1;19)(q11;q13.1),del(4)(q27),add(5)(p15.1),add(6)(p23),−7,del(7)(q22q34),−13,add(14)(q32),+der(19)t(1;19)(q11;q13.1),del(20)(q11.2), +mar[cp20] | 6.8 |
| 8 | 42.6 | F | N | 84–88,XX, −X,−X,add(1 )(p13)x2, −3, −6,l(6)(p10),add(7)(q32)x2,−12, −12,del(13)(q12q14)x2, add(14)(q32)x2,add(16)(p13.3)x2,−20,−20,add(20)(p13)x2,?add(22)(q11.2)x2,+3–6mar[cp9]/46,XX[11] | 34.0 |
| 11 | 31.7 | M | N | 46,XY,t(11 ;14)(q13;q32)[4]/46,XY[36] | 84.3 |
| 14 | 73.2 | M | N | 42–44,XY,add(1 )(q21 ),dup(1 )(q12q25),add(6)(q25),der(8;12)(q10;q10),+add(11) (p11.2), add(14)(q32),−15,del(16)(q22)+mar[cp6]/46,XY[14] | 4.3 |
| 16 | 45.1 | F | N | 46,XX[20] | 11.1 |
| 18 | 46.8 | M | N | 46,XY[20] | 141.7 |
| 20 | 59.2 | M | N | 46,X,−Y,del(1)(p13),+dic(1;21)(q10;p10),t(1;4;?)(q42;q12;?),t(3;17)(q21;q25),add(5) (q13), del (6)(q25),+9,der(14)t(11 ;14)(q13;q32),add(15)(p11.2),del(16)(q22),i(21 )(q10)[6]/ 46,X,−Y,del(1)(p13),+dic(1;21)(q10;p10),t(1;4;?)(q42;q12;?),t(3;17)(q21;q25),add(5)(q13), del(6)(q25),+9,der(14)t(11 ;14)(q13;q32),add(15)(p11.2),del(16)(q22),i(21) (q10),+mar[2] | 1.1 |
| 23 | 43.5 | M | N | 45,XY; full report not available | 6.5 |
| 25 | 52.7 | M | N | 42–44,XY; full report not available | 14.3 |
| 27 | 52.9 | F | N | 45,X,−X,+del(1)(p13p31),−6,t(11;14)(q13;q32)−20,+mar[3]/46,XX[27] | 22.5 |
| 28 | 52.1 | M | N | 46,XY,t(11:14)(q13;q32),−20,+der(?)t?;1 )(?;q12)[2]/46,XY[12] | 15.4 |
| 29 | 42.5 | M | N | 46,XY[20] | 50.7 |
| 30 | 64.1 | M | N | 46,XY[35] | 11.4 |
| 33 | 56.3 | F | N | 36–45,XX[7]/46,XX[4]; full report not available | 1.6 |
| 37 | 82.7 | M | N | 46,XY[20] | 13.5 |
| 38 | 51.0 | M | N | 46,XY | 26.7 |
| 42 | 78.4 | F | Y | 46,XX[14] | 8.5 |
| 43 | 62.5 | M | Y | 44,XY,−3,−21 [1]/45,XY,−22[1]/46,XY,t(1 ;2)(p11 ;p11)[1]/46,XY[4] | 9.5 |
| 44 | 54.3 | M | Y | 46–50,XY,+3,−5,+7,+9,−13,−14,−17,−18,+21,tdic(1;5)(p11;q35),tdic(1;18) (p11 ;q23),+1–2mar[AN][19]/46,XY[1] | 2.5 |
| 45 | 77.8 | M | Y | 42,XY,−13,−17,−21,der(3)t(3;?)(p2?5;?)der(14)t(14;?)(q32;?)[2]/43,XY, −5,−12,−21, −22,der(3)t(3;?)(q2?5;?)der(4)t(4;?)(q2?5;?),del(20)(q11.2q13.3),+mar[2]/16, XY[AN] | 0.2 |
| 46 | 62.6 | M | Y | 45,XY,der(1)del(1)(p13;p22)t(1;?)(q21;?),−2,−5,der(7)t(7;?)(q?22;?),−8×2,+12,del(12) (p12)×2,−14,der(14)t(14;?)(q32;?),−16,−17,−19,−20,+22,+1−6mar,+r[cp19]/46XY[1] | 0.8 |
| 47 | 68.2 | M | Y | 44–48,XY,add(1)(p11),−12,−22,+5–7mar[cp3]/46,XY[17] | 1.3 |
| 48 | 80.7 | M | Y | 46,XY | 0.9 |
| 49 | 73.2 | M | Y | 46,XY[13] | 3.7 |
| 50 | 67.2 | F | Y | 46,XX | 0.5 |
| 51 | 68.9 | M | Y | 51–56,X,−Y,+3,+5,−6,+7,−8,+15,+21,+21,del(1)(p22p34),+[der(6)t(1;6)(q12;q15)=2],+der(9)t(8;9) (q13;p22),mar18[AA] | 0.4 |
| 52 | 64.9 | F | Y | 47–48,XX,+1,+9,+12,−13,−14,+19,tdic(1 ;?)(p13;?),der(1)t(1;?)(q23;?),der(1)t(1;19)(p11; p11),der(2),t(2;?)(p?15;?),der(8)t(8;?)(q24.1;?),del(13)(q12q14),der(15) t(1; 15)(q23; p11), der(19)t(19;?)(q11 ;?),[del(21 )(q22)=2],del(22)(q1 ?1 q1 ?3)[AN][9]/46,XX[11 ] | 0.6 |
| 53 | 59.4 | M | Y | 46,X,inv(Y)(p11.2q11.2),t(6;?)(q23;?),del(13)(q22)[2]/46, X, inv(Y)(p11.2q11.2)[14] | 20.5 |
| 54 | 77.5 | F | Y | 40–45,XX,add(1)(p13),add(1)(p22),add(4)(q27),del(5)(q15q33),add(6)(q13),−8,−10, add(11)(p13),del(12)(p11.2),−13,−14,add(16)(q24),i(18)(q10),del(20)(p11.2),−21,+1,−3mar [cp20] | 1.7 |
| 55 | 71.1 | F | Y | 40,X-X,i(1)(q10),del(1)(q32),add(2)(p15),−4,der(7;13)(q10;q10),−10,−13,−14,−22,der(22) t(13;22)(q14;p12)+mar[?] | 0.5 |
| 56 | 50.2 | F | Y | 45–46,XX,t(11;14)(q13;q32),−13,der(14)t(11;14)(q13;q32),der(16)t(1;16)(q23;q32)+0−1mar[cp13]/46,XX[7] | 0.5 |
| 57 | 56.4 | M | Y | 31,X,−Y,−1,add(1)(q32),−2,add(3)(q21),−4,add(5)(q11.2), −6,−8,−10,−12,−13,−14, −16,−17–, 19,−20,del(22)(q13)[19]/46,XY[1] | 0.5 |
Abbreviations: pPCL, primary PCL; sPCL, secondary PCL.
Table 3.
Ploidy in PCL and MM tumors (%)a
| Ploidy | MM | sPCL | pPCL |
|---|---|---|---|
| Hypodiploid (< 45, > 75) | }40 | 42 | 60 |
| Pseudo/diploid (45–47) | 42 | 40 | |
| Hyperdiploid (48–75) | 60 | 17 | 0 |
Abbreviations: MM, multiple myeloma; PCL, plasma cell leukemia.
Percentages derived from abnormal karyotypes only.
Seven pPCL tumors had 14q32 translocations detected by standard cytogenetics. Strikingly, of these, all seven (100%) were to 11q13 (CCND1). By contrast, only three IgH translocations were detected in sPCL tumors by standard cytogenetics and of these, only one was to 11q13; in the two remaining cases the translocation partner was not identified.
14q32 translocation and partner oncogenes
Conventional cytogenetic results were next supplemented with targeted genetic studies on 33 PCL tumor samples for which material was available. Samples were examined for a broad spectrum of genetic abnormalities relevant to MM or hematological malignancy (Table 4). By FISH, 14q32 translocations were found in a substantial 87% of pPCL and 82% sPCL tumors. Notably, a significant correlation between IgH translocation and nonhyperdiploidy in plasma cell neoplasms has previously been described by us and others20,30,31 and our findings in PCL are consistent with this relationship. Samples with identified IgH translocations were re-examined by FISH to define the translocation partner, probing oncogenes targeted in MM. By this approach, 75% of pPCL and 100% of sPCL 14q32 translocation partners were identified. Notably, all pPCL samples with IgH translocation and an identified FISH partner (n=12) again showed translocation to the cyclin D1 (CCND1) locus on 11q13, highlighting the importance of this rearrangement in the etiology of pPCL. Of sPCL tumors with 14q32 rearrangement, 3/5 (60%) were to 11q13 (CCND1) and 1/5 (20%) each were to 4p16.3 (FGFR3/MMSET) or 16q23 (MAF).
Table 4.
Genetic aberrations in pPCL and sPCL prevalence and association with OS
| Abnormality | Tested (n)a | Prevalence (%) | OS: pPCL (median) | OS: sPCL (median) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pPCL | sPCL | P-valueb | Negative | Positive | P-valuec | Negative | Positive | P-valuec | ||
| IgH translocationsd | ||||||||||
| 14q32 break apart | 26 | 87 | 82 | 0.62 | 27.8 | 13.5 | 0.91 | 0.50 | 0.53 | 0.26 |
| t(11;14)(q13;q32) | 21 | 65 | 49 | 0.60 | 11.2 | 13.4 | 0.79 | 0.53 | 0.53 | 0.56 |
| t(4;14)(p16.3;q32) | 20 | 0 | 16 | 0.35 | 13.5 | — | — | 0.53 | 1.67 | 0.11 |
| t(14;16)(q32;q23) | 20 | 0 | 16 | 0.35 | 13.5 | — | — | 0.53 | 0.53 | 0.99 |
| CCND3/14q32 | 7 | 0 | 0 | — | 11.2 | — | — | 0.50 | — | — |
| t(11;14) by FISH or by informative karyotypee | 30 | 71 | 23 | 0.03a | 11.2 | 11.4 | 0.76 | 1.33 | 0.53 | 0.18 |
| p53 pathway | ||||||||||
| Allelic TP53 mutation | 25 | 25 | 23 | — | 11.4 | 13.5 | 0.52 | 0.53 | 0.93 | 0.96 |
| Allelic TP53 deletion (17p13.1) | 20 | 50 | 75 | 0.37 | 27.8 | 11.4 | 0.76 | 0.83 | 0.53 | 0.31 |
| Combined all cause TP53 loss (mutation or deletion) | 15 | 56 | 83 | 0.58 | 22.5 | 13.5 | 0.81 | 0.53 | 0.83 | 0.48 |
| Biallelic TP53 loss | 15 | 11 | 33 | 0.52 | 27.8 | 11.4 | 0.28 | 0.53 | 2.5 | 0.20 |
| MDM2 amplification | 18 | 0 | 0 | — | 13.5 | — | — | 0.50 | — | — |
| —13q | 19 | 85 | 67 | 0.56 | 15.4 | 11.4 | 0.89 | 0.83 | 0.53 | 0.62 |
| c-Myc locus | ||||||||||
| Myc amplification | 18 | 8 | 17 | 1.0 | 15.4 | 8.6 | 0.18 | 0.53 | 0.83 | 0.68 |
| CH/myc fusion | 18 | 0 | 0 | — | 13.5 | — | — | 0.53 | — | — |
| CL/myc fusion | 18 | 0 | 0 | — | 13.5 | — | — | 0.53 | — | — |
| 3′/myc breakapart | 18 | 33 | 33 | — | 27.8 | 8.6 | 0.006a | 0.53 | 1.67 | 0.05 |
| 5′/myc breakapart | 18 | 8 | 17 | 1.0 | 15.4 | 11.2 | 0.35 | 0.53 | 0.83 | 0.68 |
| PTEN deletion | 18 | 8 | 33 | 0.24 | 15.4 | 8.6 | 0.18 | 0.83 | 0.5 | 0.018a |
| RAS mutation | 24 | 27 | 15 | 0.63 | 27.8 | 10.2 | 0.069 | 0.83 | 10.5 | 0.21 |
| p16 methylation | 24 | 27 | 38 | 0.68 | 13.5 | 27.8 | 0.37 | 0.53 | 1.67 | 0.67 |
| p14 methylation | 27 | 0 | 29 | 0.10 | 11.4 | — | — | 1.3 | 0.93 | 0.34 |
Abbreviations: FISH, fluorescence in situ hybridization; OS, overall survival; pPCL, primary disease; sPCL, secondary leukemic transformation.
Limitations on banked material prevented application of all tests to all samples.
Fisher’s exact P-value.
Log-rank P-value.
Only patients who tested positive by FISH for 14q32 break apart were tested for specific 14q32 rearrangements. Prevalence of specific IgH translocations was calculated as (fraction of patients with 14q32 break apart) × (fraction of 14q32 break aparts with the specific translocation).
Includes all patients positive or negative for t(11;14) by informative cytogenetics or by FISH.
p53 inactivation
Deletion of 17p13.1, causing allelic loss of TP53, was detected in 50% of pPCL tumors and a remarkable 75% of sPCL tumors. Moreover, TP53 deletion was complemented by functionally relevant TP53 coding mutations in 24% of PCL patients tested (Table 5), contributing to a substantial prevalence of allelic TP53 inactivation of 56% in pPCL and 83% in sPCL (Table 4). The high prevalence of TP53 inactivation in de novo pPCL is surprising; in MM 17p13.1, deletion is a late event found only in 10% of tumors12,35,36 and TP53 coding mutations are rare (3%).37 Eleven percent of pPCL and 33% of sPCL tumors showed biallelic TP53 inactivation with simultaneous allelic deletion and mutation. Interestingly, monoallelic or biallelic inactivation of TP53 did not correlate significantly with survival in sPCL (P= 0.2–0.96), unlike MM, where −17p13.1 predicts adverse survival.12,36 Lack of correlation between TP53 status and survival may reflect ubiquitous targeting of the p53 pathway in sPCL.
Table 5.
Coding mutations in TP53 and N- or K-RAS in PCL that influence protein function and prevalence in other tumors
| TP53 mutations | |||||||||||||
| Leukemia | TP53 exon | Codon change | Predicted amino-acid change | p53 structural motifa | Structure function predictionb,c | Mutant p53 transactivation activityb,d | Number of mutations at codonb,e | Percentage of known TP53b,e substitutions | Cancers in which codon is commonly mutatedb,f(% of TP53 mutations at codon) | ||||
| pPCL | 5 | TGC>TGG | C141W | NDBL/β-sheets | Nonfunctional | Nonfunctional | 160 | 0.8 | colon, ovarian, oesophagus (>1%); breast, hematopoietic (>0.5%) | ||||
| pPCL | 5 | CTG>CCG | L145P | NDBL/β-sheets | Nonfunctional | Nonfunctional | 59 | 0.3 | prostate (>1%); lymphoid, pancreas, stomach (>0.5%) | ||||
| pPCL | 6 | TAT>TGT | Y220C | NDBL/β-sheets | Nonfunctional | Nonfunctional | 334 | 1.7 | oropharyngeal, biliary, ovarian, pancreas, breast, hematopoietic, lymphoid, others (all >1%) | ||||
| pPCL | 7 | TAC>TGC | Y234C | NDBL/β-sheets | Nonfunctional | Nonfunctional | 178 | 0.9 | bone, oropharyngeal, kidney, cervix, brain, ovary, lymphoid, hematopoietic, others (all > 1 %) | ||||
| sPCL | 8 | CGT>TGT | R273C | L1/S/H2 | Nonfunctional | Nonfunctional | 1471 | 7.4 | penis (83%), nasophyngeal (27%), thyroid (15%), brain (15%), pancreas (12%); haematopoitic (6.1%), lymphoid (5.3%) | ||||
| sPCL | 8 | TTT>TAT | F270Y | NDBL/β-sheets | Nonfunctional | Functional | 101 | 0.5 | lymph node (1.1%), oesophagus (0.8%), lung (0.75%), hematopoitic (0.4%) | ||||
| sPCL | 11 | AAG > -AG (373delA) | FS | NA | Nonfunctional | NA | NA | NA | NA | ||||
| N- and K- RAS mutations | |||||||||||||
| Leukemia | RAS | Exon | Codon change | Predicted aa change | No.identical amino-acid substitutions in Sanger databaseg | Cancers with identical mutation (no. of identical mutations per cancer in COSMIC databaseg) | |||||||
| pPCL | K | 1 | GGT>GAT | G12V | 2422 | Colon (966), pancreas (787), lung (384), ovary (90) | |||||||
| pPCL | K | 2 | CCA>CAC | Q61H | 74 | Colon (22), lung (21), pancreas (7), hematopoietic and lymphoid (6) | |||||||
| pPCL | N | 2 | CCA>CGA | Q61R | 386 | Skin (208), thyroid (97), hematopoietic and lymphoid (41) | |||||||
| sPCL | K | 1 | GGT>GCT | G12R | 471 | Pancreas (325), colon (53), lung (44), hematopoietic and lymphoid (6) | |||||||
| sPCL | K | 2 | CCA>CAC | Q61H | 74 | Colon (22), lung (21), pancreas (7), hematopoietic and lymphoid (6) | |||||||
| sPCL | N | 2 | CCA>CGA | Q61R | 386 | Skin (208), thyroid (97), hematopoietic and lymphoid (41) | |||||||
Abbreviations: COSMIC, Catalog of Somatic Mutations in Cancer; FS, frameshift; IARC, International Agency for Research on Cancer; PCL, plasma cell leukemia; pPCL, primary PCL; sPCL, secondary PCL; WT, wild type.
Data were obtained from the IARC p53 website (http://www-p53.iarc.fr/index.html).
Functional predictions were derived using a model that takes into account the three-dimensional structure of WTand mutant proteins and is trained on the transactivaton dataset from Kato et al.34
Functional classification of TP53 mutations based on the overall transcriptional activity on eight different promoters as measured by Kato et al.34
Number and percent of TP53 mutations that occur at the specified codon in the IARC database of 19 796 TP53 mutations.
Cancers that commonly target the specified codon for mutation and, per cancer, the percent of TP53 missense or nonsense mutations that involve the codon.
The data were obtained from the Sanger COSMIC, http://www.sanger.ac.uk/genetics/CGP/cosmic.
As inactivation of p53 can also occur through overexpression of the regulatory protein, Mdm2, or by suppression of the CDKN2A locus transcript, p14ARF,38,39 as well as by inactivation of TP53, we screened PCL samples for MDM2 amplification and for epigenetic silencing of p14ARF. No focal amplicons of MDM2 were detected indicating that p53 pathway inactivation in PCL is rare, if ever caused by MDM2 gene copy number change. However, the upstream tumor suppressor p14ARF, whose product directly binds Mdm2 to regulate p53, and whose expression is regulated by CpG island methylation,40,41 was targeted by promoter methylation in 29% of sPCL cases tested, demonstrating a second mechanism by which p53 activity can be inhibited in PCL.
13q deletion
Deletion of chromosome 13q is a prognostic marker in MM, with a higher prevalence among nonhyperdiploid MM. Therefore, we assessed its frequency in PCL. Loss of 13q by FISH was ubiquitous in pPCL (85%) and more common than in MM (54%) (P=0.02); however, it was not significantly increased between MM and sPCL (67%) (P=0.53). Interestingly, like p53 inactivation, deletion 13q did not influence survival in PCL (P>0.45), perhaps because of its ubiquity or over-riding molecular prognostic factors.
MYC translocation
Rearrangement of MYC was identified by 3′ FISH break apart in 33% of pPCL and sPCL tumors and was complemented by MYC amplification or 5′ MYC translocations in 8 and 17% of patients, respectively. MYC rearrangements were associated with a trend toward inferior OS in pPCL (for example, for 3′ MYC break apart median OS was 8.6 months vs 27.8 months without, P=0.006). The translocation partner was not identified by FISH.
RAS mutation
Mutations of K-RAS or N-RAS at codons 12, 13 or 61, previously characterized as functionally activating,26–29 were found in 27% pPCL and 15% sPCL (Table 5). Activating mutations of RAS were associated with a trend toward poorer outcome in pPCL (P=0.069). However, the prevalence of K- or N-RAS mutation in sPCL was comparable to that reported in MM (21%),28 suggesting little, if any, selective pressure for RAS activation in secondary leukemic transformation from MM.
PTEN deletion
Deletion of PTEN, which causes Akt activation,42–45 was found in 33% of sPCL tumors and in 8% of pPCL (P=0.24). These data compare with a previous report of interstitial PTEN deletion in 6% of primary myeloma and in 2 of 10 cases (20%) of PCL46 suggesting that PTEN loss may occur in the transition from myeloma to sPCL.
Discussion
pPCL and sPCL are related yet distinct biological entities with differing modes of presentation and survival. pPCL patients are younger than sPCL patients and present without an MM prodrome with more extramedullary disease and higher creatinine plus β2M than sPCL patients. In contrast, sPCL patients have more bone disease and worse prognosis. pPCL patients treated with VAD or VBMCP with and without SCT in our cohort had significantly longer OS than that of pPCL patients treated with oral MP or no therapy. In contrast, in sPCL survival was almost uniformly poor with no apparent benefit from conventional chemotherapy, even though more than half of sPCL cases occurred within 2 years of MM diagnosis.
Although pPCL and sPCL have overlapping genetic characteristics, these are not identical. pPCL in our series is exclusively nonhyperdiploid, while sPCL tumors, like MM, are sometimes hyperdiploid. 14q32 (IgH) translocations are ubiquitous in PCL and in primary leukemias are almost exclusively targeted at 11q13(CCND1), belying a fundamental importance in establishing the pPCL phenotype, while in sPCL, 14q32 rearrangements target not only 11q13 but also 4p16.3(FGFR3/MMSET) and 16q23(MAF), recapitulating a skewed spectrum of translocations observed in MM.
An increased prevalence of t(11;14) translocations in pPCL has previously been described.5,47 However, the prevalence of t(11;14) (q13;q32) in our cohort of pPCL patients (71%), is substantially greater than has previously been reported (33%). This could reflect regional variations in leukemogenesis or in delineation between pPCL and sPCL. Although small, a third study47 found 4/5 (80%) of pPCL tumors had t(11;14) compared with only 1/9 (11%) of sPCL tumors, supporting the high prevalence of t(11;14) in pPCL described here.
sPCL tumors resemble MM tumors in their spectrum of IgH translocations and ploidy but like pPCL tumors, are prejudiced in favor of nonhyperdiploid t(11;14) (q13;q32) tumors. Conspicuously, t(11;14) is a favorable prognostic factor in MM.12 However, its prevalence in PCL suggests that this translocation, when combined with other mutations (such as TP53 inactivation), is more amenable than other 14q32 rearrangements observed in MM to promoting leukemia, perhaps because it directly uncouples the cyclin D early cell-cycle checkpoint from cellular pathways that interpret the surrounding growth factor environment.
Monoallelic and biallelic TP53 abnormalities are strikingly common in both pPCL and sPCL suggesting that the impairment of the p53 tumor suppressor pathway is an important contributor to extramedullary tumor expansion. However, in established sPCL, unlike in MM, TP53 inactivation does not influence survival, raising the possibility that sPCL tumors lacking TP53 abnormalities; instead, silence other p53 pathway factors (such as p14ARF) to similar effect and that loss of p53 activity may be virtually universal in sPCL. Although loss of p53 may predispose MM patients to sPCL, it may not be the final leukemia-inducing event. Of two assessable sPCL patients with −17p13.1 both were found to have similar deletions in their MM cells 7 or 35 months before leukemia diagnosis (not shown). Instead, reduction of p53 surveillance may be a prerequisite for plasma cell survival allowing dysregulation of oncogenes such as RAS and MYC or escape from the bone marrow microenvironment.
Supplementary Material
Acknowledgements
This work was supported in part by the Donaldson Charitable fund Trust, Public Health Service grant no. R01 CA83724-01 (RF) and P01 CA62242 (RF, AD, RAK, PRG) from the National Cancer Institute. RF and PRG are supported by the CI-5 Cancer Research Fund-Lilly Clinical Investigator Award of the Damon Runyon Cancer Research Fund. PRG and RF are also supported by the ECOG Grant CA21115-25C from the National Cancer Institute. RET is supported by a fellowship from AMGEN and the Haematology Society of Australia and New Zealand (HSANZ).
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Kyle RA, Maldonado JE, Bayrd ED. Plasma cell leukemia. Report on 17 cases. Arch Intern Med. 1974;133:813–818. doi: 10.1001/archinte.133.5.813. [DOI] [PubMed] [Google Scholar]
- 2.Noel P, Kyle RA. Plasma cell leukemia: an evaluation of response to therapy. Am J Med. 1987;83:1062–1068. doi: 10.1016/0002-9343(87)90942-9. [DOI] [PubMed] [Google Scholar]
- 3.Dimopoulos MA, Palumbo A, Delasalle KB, Alexanian R. Primary plasma cell leukaemia. Br J Haematol. 1994;88:754–759. doi: 10.1111/j.1365-2141.1994.tb05114.x. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe ES, Harris NL, Stein H, Vardiman JW. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press; 2001. [Google Scholar]
- 5.Avet-Loiseau H, Daviet A, Brigaudeau C, Callet-Bauchu E, Terre C, Lafage-Pochitaloff M, et al. Cytogenetic, interphase, and multicolor fluorescence in situ hybridization analyses in primary plasma cell leukemia. Blood. 2001;97:822–825. doi: 10.1182/blood.v97.3.822. [DOI] [PubMed] [Google Scholar]
- 6.Cai ZJ. Primary plasma cell leukemia—a comprehensive analysis of 44 cases. Zhonghua Zhong Liu Za Zhi. 1990;12:314–317. [PubMed] [Google Scholar]
- 7.Garcia-Sanz R, Orfao A, Gonzalez M, Tabernero MD, Blade J, Moro MJ, et al. Primary plasma cell leukemia: clinical, immunophenotypic, DNA ploidy, and cytogenetic characteristics. Blood. 1999;93:1032–1037. [PubMed] [Google Scholar]
- 8.Costello R, Sainty D, Bouabdallah R, Fermand JP, Delmer A, Divine M, et al. Primary plasma cell leukaemia: a report of 18 cases. Leuk Res. 2001;25:103–107. doi: 10.1016/s0145-2126(00)00102-8. [DOI] [PubMed] [Google Scholar]
- 9.Christou L, Hatzimichael E, Chaidos A, Tsiara S, Bourantas KL. Treatment of plasma cell leukemia with vincristine, liposomal doxorubicin and dexamethasone. Eur J Haematol. 2001;67:51–53. doi: 10.1034/j.1600-0609.2001.067001051.x. [DOI] [PubMed] [Google Scholar]
- 10.Oken MM, Leong T, Lenhard RE, Jr, Greipp PR, Kay NE, Van Ness B, et al. The addition of interferon or high dose cyclophosphamide to standard chemotherapy in the treatment of patients with multiple myeloma: phase III eastern cooperative oncology group clinical trial EST 9486. Cancer. 1999;86:957–968. [PubMed] [Google Scholar]
- 11.Fonseca R, Harrington D, Oken MM, Dewald GW, Bailey RJ, Van Wier SA, et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: an eastern cooperative oncology group study. Cancer Res. 2002;62:715–720. [PubMed] [Google Scholar]
- 12.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 13.Drewinko B, Alexanian R, Boyer H, Barlogie B, Rubinow SI. The growth fraction of human myeloma cells. Blood. 1981;57:333–338. [PubMed] [Google Scholar]
- 14.Drewinko B, Alexanian R. Growth kinetics of plasma cell myeloma. J Natl Cancer Inst. 1977;58:1247–1253. doi: 10.1093/jnci/58.5.1247. [DOI] [PubMed] [Google Scholar]
- 15.Kyle RA. Multiple myeloma. An update on diagnosis and management. Acta Oncol. 1990;29:1–8. doi: 10.3109/02841869009089984. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA, et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood. 2002;100:1417–1424. [PubMed] [Google Scholar]
- 17.Fonseca R, Oken MM, Greipp PR. The t(4;14)(p16.3;q32) is strongly associated with chromosome 13 abnormalities in both multiple myeloma and monoclonal gammopathy of undetermined significance. Blood. 2001;98:1271–1272. doi: 10.1182/blood.v98.4.1271. [DOI] [PubMed] [Google Scholar]
- 18.Hayman SR, Bailey RJ, Jalal SM, Ahmann GJ, Dispenzieri A, Gertz MA, et al. Translocations involving the immunoglobulin heavy-chain locus are possible early genetic events in patients with primary systemic amyloidosis. Blood. 2001;98:2266–2268. doi: 10.1182/blood.v98.7.2266. [DOI] [PubMed] [Google Scholar]
- 19.Fonseca R, Hoyer JD, Aguayo P, Jalal SM, Ahmann GJ, Rajkumar SV, et al. Clinical significance of the translocation (11;14)(q13;q32) in multiple myeloma. Leuk Lymphoma. 1999;35:599–605. doi: 10.1080/10428199909169625. [DOI] [PubMed] [Google Scholar]
- 20.Fonseca R, Debes-Marun CS, Picken EB, Dewald GW, Bryant SC, Winkler JM, et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood. 2003;102:2562–2567. doi: 10.1182/blood-2003-02-0493. [DOI] [PubMed] [Google Scholar]
- 21.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidl S, Ackermann J, Kaufmann H, Keck A, Nosslinger T, Zielinski CC, et al. DNA-methylation analysis identifies the E-cadherin gene as a potential marker of disease progression in patients with monoclonal gammopathies. Cancer. 2004;100:2598–2606. doi: 10.1002/cncr.20295. [DOI] [PubMed] [Google Scholar]
- 24.Ganguly A. An update on conformation sensitive gel electrophoresis. Hum Mutat. 2002;19:334–342. doi: 10.1002/humu.10059. [DOI] [PubMed] [Google Scholar]
- 25.Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA. 1993;90:10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahuja HG, Foti A, Bar-Eli M, Cline MJ. The pattern of mutational involvement of RAS genes in human hematologic malignancies determined by DNA amplification and direct sequencing. Blood. 1990;75:1684–1690. [PubMed] [Google Scholar]
- 27.Bezieau S, Devilder MC, Avet-Loiseau H, Mellerin MP, Puthier D, Pennarun E, et al. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum Mutat. 2001;18:212–224. doi: 10.1002/humu.1177. [DOI] [PubMed] [Google Scholar]
- 28.Ortega MM, Faria RM, Shitara ES, Assis AM, Albuquerque DM, Oliveira JS, et al. N-RAS and K-RAS gene mutations in Brazilian patients with multiple myeloma. Leuk Lymphoma. 2006;47:285–289. doi: 10.1080/10428190500300969. [DOI] [PubMed] [Google Scholar]
- 29.Liang DC, Shih LY, Fu JF, Li HY, Wang HI, Hung IJ, et al. K-Ras mutations and N-Ras mutations in childhood acute leukemias with or without mixed-lineage leukemia gene rearrangements. Cancer. 2006;106:950–956. doi: 10.1002/cncr.21687. [DOI] [PubMed] [Google Scholar]
- 30.Smadja NV, Leroux D, Soulier J, Dumont S, Arnould C, Taviaux S, et al. Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid cases. Genes Chromosomes Cancer. 2003;38:234–239. doi: 10.1002/gcc.10275. [DOI] [PubMed] [Google Scholar]
- 31.Saez B, Martin-Subero JI, Guillen-Grima F, Odero MD, Prosper F, Cigudosa JC, et al. Chromosomal abnormalities clustering in multiple myeloma reveals cytogenetic subgroups with nonrandom acquisition of chromosomal changes. Leukemia. 2004;18:654–657. doi: 10.1038/sj.leu.2403256. [DOI] [PubMed] [Google Scholar]
- 32.Cho Y, Gorina S, Jeffrey P, Pavletich N. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 33.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 34.Kato S, Han S-Y, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. PNAS. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultheis B, Kramer A, Willer A, Hegenbart U, Goldschmidt H, Hehlmann R. Analysis of p73 and p53 gene deletions in multiple myeloma. Leukemia. 1999;13:2099–2103. doi: 10.1038/sj.leu.2401609. [DOI] [PubMed] [Google Scholar]
- 36.Gertz MA, Lacy MQ, Dispenzieri A, Greipp PR, Litzow MR, Henderson KJ, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and −17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837–2840. doi: 10.1182/blood-2005-04-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, et al. Clinical significance of TP53 mutation in myeloma. Leukemia. 2007;21:582–584. doi: 10.1038/sj.leu.2404524. [DOI] [PubMed] [Google Scholar]
- 38.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eymin B, Gazzeri S, Brambilla C, Brambilla E. Mdm2 over-expression and p14(ARF) inactivation are two mutually exclusive events in primary human lung tumors. Oncogene. 2002;21:2750–2761. doi: 10.1038/sj.onc.1205359. [DOI] [PubMed] [Google Scholar]
- 40.Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, Watkins DN, et al. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res. 2001;61:2816–2821. [PubMed] [Google Scholar]
- 41.Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci USA. 2001;98:4990–4995. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahia PL, Aguiar RC, Alberta J, Kum JB, Caron S, Sill H, et al. PTEN is inversely correlated with the cell survival factor Akt/PKB and is inactivated via multiple mechanismsin haematological malignancies. Hum Mol Genet. 1999;8:185–193. doi: 10.1093/hmg/8.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Hyun T, Yam A, Pece S, Xie X, Zhang J, Miki T, et al. Loss of PTEN expression leading to high Akt activation in human multiple myelomas. Blood. 2000;96:3560–3568. [PubMed] [Google Scholar]
- 44.Leslie NR, Bennett D, Gray A, Pass I, Hoang-Xuan K, Downes CP. Targeting mutants of PTEN reveal distinct subsets of tumour suppressor functions. Biochem J. 2001;357:427–435. doi: 10.1042/0264-6021:3570427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stiles B, Gilman V, Khanzenzon N, Lesche R, Li A, Qiao R, et al. Essential role of AKT-1/protein kinase B alpha in PTEN-controlled tumorigenesis. Mol Cell Biol. 2002;22:3842–3851. doi: 10.1128/MCB.22.11.3842-3851.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H, Qi XY, Claudio J, Zhuang L, Patterson B, Stewart AK. Analysis of PTEN deletions and mutations in multiple myeloma. Leuk Res. 2006;30:262–265. doi: 10.1016/j.leukres.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Chang H, Sloan S, Li D, Patterson B. Genomic aberrations in plasma cell leukemia shown by interphase fluorescence in situ hybridization. Cancer Genet Cytogenet. 2005;156:150–153. doi: 10.1016/j.cancergencyto.2004.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.